Abstract
Cell migration is orchestrated by molecular networks supporting motility. The scaffolds ERC1/ELKS and Liprin-α1 sustain cell migration and invasion by assembling dynamic plasma membrane-associated platforms. ERC1/ELKS forms cytoplasmic condensates with liquid–like behavior. In this study we tested whether the ability of ERC1 to form condensates is relevant to its function in cell motility. We identified the shortest N-terminal region of ERC1 sufficient to drive phase separation in vitro and in cells. Fluorescence recovery after photobleaching confirmed the dynamic behavior of ERC1(1-244) condensates. Surprisingly, deletion of ERC1(1-244) including an intrinsically disordered region did not abolish the ability of ERC1DΔN to form condensates. Although the interactions of ERC1ΔN with partners were unaffected, the biophysical properties of ERC1ΔN condensates were altered, with consequences on cell motility. These findings highlight the importance of ERC1/ELKS to assemble functional networks, and show that altering the properties of ERC1–driven condensates interferes with tumor cell motility.
Similar content being viewed by others
Introduction
Cell motility is crucial for physiological and disease-related processes1, including the formation of metastases that relies on the ability of tumor cells to disseminate from primary tumors to establish secondary tumors2. Cell motility requires coordination and continuous rearrangement of adhesion and cytoskeleton to promote protrusion of the cell front3,4. Adhesion is necessary to exert forces that allow cell protrusion, and continuous rearrangement of adhesions at the cell front is required to avoid inhibition of cell motility5. It is essential to analyse how intracellular protein networks affect the organization of the cell periphery to promote movement. Integrin-mediated cell migration on extracellular matrix has become a major paradigm to address the regulatory mechanisms of cell motility.
The scaffolds ERC1/ELKS6, and its interacting partners Liprin-α17 and LL5α/β8,9 are part of a stable plasma membrane–associated platform (PMAP) linking microtubules to the cell cortex and to integrin-mediated focal adhesions, and directing constitutive secretion10,11,12. We found that in migrating cells, the same network shows a highly dynamic behavior linked to fast turnover of adhesions and protrusive activity of the cell front13,14,15. Endogenous ERC1/ELKS, Liprin-α1, and LL5 proteins colocalize near the cell front, defining dynamic, highly polarized cytoplasmic PMAPs specific to migrating cells13. ERC1/ELKS and its interacting partners stabilize the front of migrating cells, promoting extension and the internalization of active integrins13,14.
Liquid-liquid phase separation (LLPS) has been proposed as a fundamental mechanism to organize cytoplasm and nucleoplasm16. Membraneless organelles enriched in specific proteins and RNAs form spontaneously as a consequence of liquid-liquid demixing from a phase transition occurring above a critical concentration of the macromolecules involved. Growing experimental evidence supports the hypothesis that LLPS underlies the formation of many biomolecular condensates that are able to concentrate biological macromolecules at specific sites and times17. The dynamic nature of PMAPs near the front of migrating cells suggests that PMAPs are driven by phase separation18,19. This hypothesis has been supported by the finding that the PMAP component ERC1/ELKS forms condensates with liquid-like properties20. Work in neurons and in vitro has confirmed that mammalian ERC1/ELKS and the C. elegans orthologue ELKS-1 undergo phase separation21,22. ERC1/ELKS condensates host by direct interaction the clients LL5α/β and Liprin-α1 that influence the dynamic behavior of condensates, and focal adhesion regulators like GIT1, an ArfGAP directly interacting with Liprin-α120,23.
In this study, we show that the N-terminal portion of ERC1/ELKS, including an intrinsically disordered region (IDR, residues 1-142) is sufficient to induce the formation of condensates in vitro and in cells. Strikingly, deletion of this region does not abrogate the formation of condensates, but modulates their biophysical properties. Although the homotypic and heterotypic interactions of ERC1ΔN with PMAP partners Liprin-α1 and LL5 are not affected, as well as the formation of dynamic PMAPs at the front of migrating tumor cells, expression of the ERC1ΔN affects cell spreading, motility, and invasion by tumor cells. Endogenous levels of ERC1ΔN can still allow the formation of dynamic PMAPs at the front of migrating tumor cells. Our studies reveal that altering the fine modulation of the dynamic properties of ERC1/ELKS–driven supramolecular scaffolds perturbs cell motility and that this mechanism constitutes an attractive target for the interference with tumor metastatic processes.
Results
The N-terminal region ERC1(1-244) is sufficient to induce the formation of condensates with liquid-like properties in cellulo and in vitro
Condensates formed by either ERC1 or ERC1-N (residues 1-676) exhibit liquid–like properties typical of condensates formed by LLPS20. Here we confirmed that full length ERC1 and ERC1-N formed cytoplasmic condensates (Fig. 1a), while the complementary C-terminal ERC1-C (residues 677-1116) didn’t (Fig. 1b). To address the requirements for the formation of intracellular condensates by ERC1, a series of GFP-tagged fragments of ERC1 were designed to gradually eliminate specific regions, based on predictions of IDRs by DisEMBL24 and coiled coils by Paircoil225 (Fig. 1a). The ability of the fragments to form cytoplasmic condensates was compared to full-length ERC1 and ERC1-N (positive controls) and to ERC1-C (negative control) (Fig. 1d). Progressive deletions from the C-terminal side of the polypeptide led to the identification of ERC1(1-244) as the shortest ERC1 fragment efficiently forming intracellular condensates. ERC1(1-244), including the predicted IDR (residues 1-142) and two short predicted coiled coils (Fig. 1a), formed round-shaped condensates (Fig. 1c, d) visible also in the nucleus, probably due to their small size.
a Scheme of the ERC1 N-terminal constructs used in this study. IDRs and coiled coil regions of ERC1 were predicted by the DisEMBL24 and Paircoil2 program25, respectively. Shorter N-terminal constructs were designed to progressively remove predicted disordered (in yellow) or coiled-coil (in light blue) regions. b, c Immunofluorescence of COS7 cells transfected with the indicated constructs. Scale bars, 20 μm. Line profiles for the GFP signal in the selected cells are shown in the lower part of each panel. d Percentage of transfected cells with condensates (n = 72–428 cells). e Left: Fluorescence images of condensates formed by either ERC1 or ERC1(1-244). Scale bar, 20 µm. Center: selected condensates (top) were approximated by an ellipse (bottom). Right: A.R. of condensates (means ± SEM; n = 485-500 condensates). Unpaired Student’s t-test, Mann–Whitney correction. 12 data points are outside the axis limits.
Dimeric GFP may influence the formation of condensates by the protein under study26. To exclude the contribution of dimeric GFP in the formation of condensates, we expressed mGFP-ERC1(1-244) tagged with monomeric GFP27. The mGFP-ERC1(1-244) fragment could still form intracellular condensates in a large fraction of transfected cells (Supplementary Fig. S1), supporting the conclusion that ERC1(1-244) is sufficient to promote LLPS.
ERC1(1-244) includes two short predicted coiled coils, CC1 (residues 142-182) and CC2 (residues 206-241). Deletion of CC2 abolished the formation of condensates by ERC1(1-188) (Fig. 1c, d). On the other side, deletion of the first 60 residues of the IDR abolished the ability of ERC1(60-420) to form condensates compared to ERC1(1-420), including the full IDR (Fig. 1c, d). These results show that ERC1(1-244) is the shortest N-terminal portion of ERC1 sufficient to induce the formation of intracellular condensates: the full IDR and the coiled coils included in ERC1(1-244) are both necessary for this process to occur in cells.
ERC1(1-244) condensates exhibited distinct morphology compared to full-length ERC1 condensates. We measured the aspect ratio (A.R.) of condensates to quantify the morphological difference28. ERC1 condensates had an A.R. of 1.62 ± 0.03 (SEM), while those formed by ERC1(1-244) displayed a more spherical A.R. of 1.27 ± 0.01 (SEM) (Fig. 1e). These significant differences in A.R. between ERC1 and ERC1(1-244) condensates may arise from variations in the molecular properties of the two types of condensates and include the quality of the intermolecular interactions that lead to their formation.
Fluorescence recovery after photobleaching (FRAP) was applied to characterize the liquid behavior and biophysical properties of different types of condensates. Full-length ERC1 condensates display a large mobile fraction with rapid fluorescence recovery20. The recovery of fluorescence after full bleach (ROI of 2.5 µm diameter) of GFP-ERC1 condensates was confirmed to be fast and efficient (t1/2 = 5.40 ± 1.04 s; 70% mobile fraction). ERC1(1-244) condensates showed a similarly fast and efficient recovery (t1/2 = 4.68 ± 0.82 sec; 74% mobile fraction) (Fig. 2a–c and Supplemantary Movie 1), confirming their liquid nature.
a Confocal imaging for FRAP analysis on a GFP-ERC1(1-244)–positive condensate (yellow arrow) in a COS7 cell (see also Supplemantary Movie 1). Scale bar, 10 μm. b Quantification of fluorescence recovery after full bleaching of GFP-ERC1 condensates and ERC1(1-244) condensates. c Half-life recovery (t½) extrapolated from FRAP data. Means ± SEM; n = 11-18 cells from three independent experiments. Unpaired Student’s t-test, Mann-Whitney correction. d His6X-Strep(2X)-GFP-ERC1(1-244) was passed through a Superdex 200 size exclusion column equilibrated in 20 mM Tris pH 8.5, 150 mM KCl in the presence of no additives, 1 M urea, or 0.5 M L-arginine in order to prevent non-specific adhesiveness of the polypeptide. e In vitro LLPS assay with purified GFP-ERC1(1-244). Spherical GFP-positive puncta form at 10 µM GFP-ERC1(1-244) in the presence of 10% PEG 3350. Scale bar 20 µm. f Left: FRAP analysis on ERC1(1-244) condensates reconstituted in vitro under the conditions described in (e) (10 µM purified protein in presence of 10% PEG 3350). Scale bar, 5 µm. Right: fluorescence recovery (means ± SEM; n = 3 droplets).
We tested the ability of purified a recombinant GFP-labeled(1-244) polypeptide to undergo phase separation in vitro, using L-arginine as a buffer additive to reduce non-specific stickiness/adhesivity of the concentrated recombinant protein29 (Fig. 2d). For in vitro LLPS, purified GFP-ERC1(1-244) was diluted in LLPS buffer (25 mM HEPES 7.5, 100 mM KCl) with different concentrations of polyethylene glycol (PEG) 3350 used as a crowding agent30. Formation of spherical condensates of ERC1(1-244) was observed at a protein concentration of 10 µM in the presence of PEG 3350 (Fig. 2e). FRAP analysis in vitro showed that the reconstituted condensates were in a liquid state, although somewhat less dynamic (t1/2 = 35 s) (Fig. 2f and Supplemantary Movie 2) than those observed in the cells. This is possibly due to changes in the viscoelastic properties of condensates induced by PEG31. Thus, ERC1(1-244) is sufficient to form liquid-like condensates both in cellulo and in vitro.
Time-lapse imaging on live COS7 cells expressing GFP-ERC1(1-244) showed that fusion events between ERC1(1-244) condensates were very rarely observed (Supplementary Movie 3). To further investigate this point we have assessed the morphology of ERC1(1-244) at 24 h, 48 h, and 72 h post-transfection to determine whether ERC1(1-244) condensates are smaller because of slower fusion times, or because they maintain the same distribution potentially indicating a distinct surface tension (Supplementary Fig. S2). We observed that ERC1(1-244) did not form extended condensates even at the longest times after transfection (72 h), indicating that the lack of extended condensates by this construct is not due to a slower fusion process, but rather to a distinct surface tension of the smaller ERC1(1-244) condensates. Interestingly, what is quite evident is the tendency over time of the small condensates to aggregate within cells.
Deletion of the N-terminal region affects the dynamic properties of ERC1 condensates
ERC1(1-244) represents the shortest N-terminal fragment of ERC1 that can form condensates. To test if this region was essential for the development of ERC1 condensates, we checked if its deletion would disrupt or alter the formation of condensates. To this end we generated the GFP-ERC1ΔN construct lacking the N-terminal fragment (Fig. 3a). Indeed, GFP-ERC1ΔN was still able generate condensates in cells (Fig. 3b, c). The C-terminal ERC1Δ375 fragment with a longer N-terminal deletion could still form large condensates in 25% of the transfected cells, while deletion of a larger N-terminal portion of the protein (residues 1-676) virtually abolished the ability of ERC1-C to form condensates.
a Scheme of the ERC1 C-terminal constructs used in this study. b Immunofluorescence of COS7 cells transfected with the indicated constructs. Scale bar, 20 μm. Line profiles for the GFP signal in the selected cells are shown on the right. c Percentage of transfected cells with condensates (n = 131–207 cells). d COS7 cells on fibronectin (2.5 μg/ml) were transfected for 4 h, then imaged for 18 h (one frame every 5 min) with a Live-Cell Imaging System equipped with 20x lens to follow the formation of ERC1ΔN condensates (see also Supplementary Movie 4). Scale bar, 20 μm. e GFP-ERC1 and GFP-ERC1ΔN expression induces the formation of condensates in a concentration-dependent manner in the cytoplasm of COS7 cells. In each transfected cell forming condensates, the cytoplasmic mean mrey Intensity (expressed as mean gray value, arbitrary units, a.u.) was measured every hour for 11 h, starting from the beginning of detectable GFP-ERC1 or GFP-ERC1ΔN expression, respectively (n = 18 cells/sample). The left graph shows the mean gray values ± SEM at each time point. In the two upper right graphs, for each cell the GFP-ERC1 or the GFP-ERC1ΔN expression levels (mean gray values) are plotted against the presence (purple dots) or absence (yellow dots) of detectable cytoplasmic condensates. The left bottom graph shows the average maximal mean gray values at which cells do not show condensates yet. f FRAP (spot bleaching, yellow arrows) on condensates in the cytoplasm of COS7 cells expressing either GFP-ERC1 or GFP-ERC1ΔN (see also Supplementary Movies 5–7). Bottom: 2.5× magnification. Scale bars, 10 μm. g Quantification of fluorescence recovery after spot bleaching. h Half-life recovery (t½) extrapolated from FRAP data. Means ± SEM; n = 17–24 cells from three independent experiments. One-Way ANOVA, Kruskal–Wallis test, Dunn’s correction.
Condensates induced by ERC1ΔN (Fig. 3b) were morphologically different from small spherical condensates produced by ERC1(1-244) (Fig. 1c). ERC1ΔN condensates occupied large portions of the cytoplasm and had irregular shapes in a high percentage of transfected cells (Fig. 3b, c). In support of the formation of extended cytoplasmic condensates by GFP-ERC1ΔN, we analyzed their formation by time-lapse imaging (Fig. 3d and Supplementary Movie 4). The extended cytoplasmic condensates formed by ERC1ΔN originated from growth and fusion of smaller condensates: as previously observed for GFP-ERC120, also, GFP-ERC1ΔN displayed a homogeneous distribution throughout the cytoplasm at low levels of expression. GFP-ERC1ΔN condensates appeared soon after transfection, and increased in number and size with time (Fig. 3d and Supplementary Movie 4). Analysis of the GFP-tagged proteins by time-lapse imaging after transfection shows that the levels of expression of GFP-ERC1 and GFP-ERC1ΔN in cells forming condensates are similar, and that the maximal expression levels without evident condensates reached by the cells expressing either GFP-ERC1 or GFP-ERC1ΔN protein are not significantly different (Fig. 3e). Moreover, in support of their origin by LLPS, we observed that both GFP-ERC1 and GFP-ERC1ΔN condensates in COS7 cells were sensitive to 1,6-hexanediol (Supplementary Fig. S3).
Therefore, deletion of the N-terminal portion of ERC1 including the predicted IDR did not prevent the formation of liquid-like condensates by the ERC1ΔN polypeptide, but affected their surface properties, ultimately leading to formation of larger, more dynamic structures.
FRAP analysis showed that the distinct morphological features of ERC1ΔN condensates corresponded to different biophysical properties compared to condensates formed by either full-length ERC1 or ERC1(1-244) (Fig. 3f and Supplementary Movies 5–7). Spot bleaching (2 µm diameter ROI) on large ERC1ΔN condensates showed a significantly faster and higher fluorescence recovery in the bleached area (t1/2 = 0.91 ± 0.10 s; 87% fluorescence recovery) compared to full length ERC1 (t1/2 = 3.11 ± 0.40 s; 82% fluorescence recovery), but lower than the diffuse cytosolic control ERC1-C (t1/2 = 0.65 ± 0.19 s; 100% fluorescence recovery) that is unable to form condensates (Fig. 3g, h). The results show that removal of the N-terminal region of ERC1 did not prevent the formation of condensates, but increased the liquid–like properties of ERC1ΔN condensates.
Deletion of the N-terminal region preserves ERC1 interactions within the PMAP network
PMAPs are necessary for efficient cell migration and invasion13,14,32. An important question is whether ERC1 phase separation is relevant for the formation of PMAPs and for their role in cell migration. The N-terminal portion of ERC1 is needed to form homodimers20,33 and to interact with PMAP components Liprin-α122,34 and LL5 proteins10 (Fig. 4a). Our data have shown that an extended N-terminal deletion (ERC1-C) is necessary to abrogate the capacity of ERC1 to form condensates (Fig. 3a). This large deletion disrupts homo- and heterotypic interactions between ERC1 and other components of the PMAPs (Fig. 4a). On the other hand, ERC1ΔN is expected to maintain its scaffolding function through the establishment of homo- and heterotypic interactions. We tested whether ERC1ΔN maintained the ability to oligomerize and to interact with its PMAPs partners. ERC1 forms parallel homodimers through extensive N-terminal coiled coil regions20, as predicted by Alphafold235 (Fig. 4b). Co-immunoprecipitation experiments showed that ERC1ΔN associated to full-length ERC1, showing that deletion of the N-terminal IDR and short coiled coils did not prevent the formation of oligomers. As expected, both ERC1(1-244) and ERC1-C did not interact virtually with full-length ERC1 (Fig. 4c, d).
a Scheme of the regions of interaction of ERC1 with the PMAP partners Liprin-α1 and LL5β. b Alphafold2 model of the N-terminal part (residues 1-676) of ERC1. c Aliquots of lysates (300 μg protein) from COS7 cells transfected with the indicated GFP– and FLAG–tagged ERC1 constructs were immunoprecipitated with GFP-Trap®–Agarose. Immunoprecipitates (IP) and lysates (30 μg) were immunoblotted to detect the FLAG–ERC1 construct (1) and the GFP-tagged constructs (2). d Quantification (n = 3 experiments) of the amount of FLAG-ERC1 co-precipitated with either GFP-ERC1 or GFP-ERC1ΔN. In each experiment, the amount of FLAG-ERC1 coprecipitating with GFP-ERC1 is considered as 100%. One way ANOVA, Kruskal Wallis; Dunn’s multiple comparisons. e Aliquots of lysates (300 μg protein) from COS7 cells co-transfected with GFP–tagged ERC1 constructs and mCherry-LL5β were immunoprecipitated with anti-GFP Abs. Immunoprecipitates (IP) and lysates (30 μg) were immunoblotted with anti-LL5 mAb 1H12 to detect mCherry-LL5β (1), and with anti-GFP to detect the GFP–tagged constructs (2). f Quantification (n = 3 experiments) of the amount of mCherry-LL5β co-precipitating with GFP–tagged ERC1 constructs. In each experiment, the amount of mCherry-LL5β coprecipitating with GFP-ERC1 is considered as 100%. One way ANOVA, Kruskal Wallis; Dunn’s multiple comparisons. g Aliquots (200 μg protein) of lysates from COS7 cells co-transfected with either GFP or GFP–tagged ERC1 constructs together with mCherry-Liprin-α1 were immunoprecipitated with GFP-Trap® Agarose. Immunoprecipitates (IP) and lysates (20 μg) were immunoblotted with anti-Liprin-α1 pAb to detect mCherry-Liprin-α1 (1) and with anti-GFP pAb to detect GFP and GFP-ERC1 constructs (2). h Quantification (n = 3 experiments) of the amount mCherry-Liprin-α1 co-precipitating with GFP–tagged constructs. In each experiment, the amount of mCherry-Liprin-α1 coprecipitating with GFP-ERC1 is considered as 100%. One way ANOVA, Kruskal Wallis; Dunn’s multiple comparisons.
The direct interaction of ERC1 with LL5β is relevant for cell motility9. Immunoblotting on immunoprecipitates of GFP–tagged ERC1 constructs revealed the specific co-immunoprecipitation of mCherry-LL5β with GFP-ERC1 and GFP-ERC1ΔN that includes the LL5β–interacting region (Fig. 4a), while no interaction was observed with ERC1-C that lacks the LL5β–interacting region (Fig. 4e, f).
Liprin-α1 has been described as a critical regulator of focal adhesion dynamics, invadosome function, and the formation of metastases by human breast cancer cells7. The interaction of ERC1 with Liprin-α1 is relevant to a number of important cellular processes14,34,36,37,38. Immunoprecipitation of either GFP-ERC1 or GFP-ERC1ΔN resulted in the specific co-immunoprecipitation of Liprin-α1 (Fig. 4g, h), although some non-specific binding of Liprin-α1 to GFP (negative control) could not be avoided by any of several experimental protocols tested. We therefore confirmed the interaction between GFP-ERC1ΔN and Liprin-α1 by their colocalization inside ERC1ΔN condensates. ERC1 interacts with the central ERC-binding region (EBR) of Liprin-α134, which is sufficient and necessary for the recruitment of Liprin-α1 inside ERC1 condensates20. Here, we confirmed that the ERC1-binding region Liprin-EBR was efficiently recruited inside ERC1ΔN condensates. As expected, the deletion of the EBR prevented the recruitment of the complementary Liprin-ΔEBR construct at ERC1ΔN condensates (Fig. 5). Liprin-ΔEBR was diffuse in the cytoplasm, and in contrast to Liprin-EBR showed no evident accumulation inside ERC1ΔN-positive condensates (Fig. 5b, c). These data show that ERC1ΔN can oligomerize and form heterotypic interactions with the PMAP partners, and suggest that deletion of the N-terminal region of ERC1 does not prevent the assembly of PMAPs.
a Scheme of Liprin-α1 constructs. b Confocal images of COS7 cells coexpressing GFP-tagged ERC1 constructs (green) with FLAG-tagged Liprin-α1 constructs (blue). Scale bar, 20 μm. Yellow asterisks indicate regions of ERC1-positive condensates. c Pearson’s correlation coefficient for the colocalization of Liprin-α1 and ERC1 constructs at condensates. Means ± SEM (n = 94–102 regions from 3 experiments). One-way Anova. d Frames from confocal time-lapses of MDA-MB-231 cells expressing the indicated proteins, migrating on fibronectin-coated substrates (see also related Supplementary Movies 8–10). Scale bar, 20 μm.
An increase in the local concentration of specific proteins may trigger phase separation39. We previously observed that ERC1 condensates appear when ERC1 has reached a certain level of protein expression in the cytoplasm20. Here, we observed that ERC1ΔN induced the formation of very large condensates at higher levels of protein expression in the cytoplasm (Fig. 3d and Supplementary Movie 4). On the other hand, at lower levels of expression, GFP-ERC1 localizes in PMAPs at the protruding edge of migrating MDA-MB-231 tumor cells, where ERC1 colocalizes with its interactors Liprin-α1 and LL5 proteins13. We tested whether the formation of ERC1-positive PMAPs was altered in MDA-MB-231 cells expressing low levels of the deletion mutant ERC1ΔN. The analysis of MDA-MB-231 cells migrating on fibronectin showed that both GFP-ERC1 and GFP-ERC1ΔN formed similar PMAPs at the front of the migrating tumor cells (Fig. 5d and Supplementary Movies 8–10).
Deletion of the N-terminal region of ERC1 enhances cell spreading
Measuring cell spreading on extracellular matrix represents an easy way to test effects on cell motility. PMAP proteins are required for effective integrin-mediated spreading of COS7 and MDA-MB-231 cells on fibronectin: depleting of either endogenous ERC1, Liprin-α1, or LL5 proteins inhibits the projected cell area, while overexpression of Liprin-α1 promotes cell spreading by increasing the projected area of different cell types, including breast cancer MDA-MB-231 cells13,40. To test whether altering ERC1 LLPS properties resulted in altered motility, we tested the effects of ERC1ΔN on MDA-MB-231 and COS7 cell spreading. Transfected cells replated on coverslips coated with a 10 µg/ml fibronectin were cultured for different times before fixation. While overexpression of full-length ERC1 did not affect COS7 cell spreading, ERC1ΔN enhanced spreading to an extent similar to that induced by Liprin-α1 expression. In contrast, expression of the ERC1(1-244) did not affect cell spreading (Fig. 6a, b). ERC1ΔN enhanced also spreading of MDA-MB-231 replated for 3–18 h on fibronectin to allow full spreading. Interestingly, neither full-length ERC1 nor shorter deletions limited to the N-terminal IDR of ERC1 (ERC1Δ51, ERC1Δ147) affected MDA-MB-231 cell spreading (Fig. 6c–e).
Cells were trypsinized and replated on fibronectin-coated substrates (10 µg/ml) for the indicated times. Projected cell areas were measured after fixation and immunostaining with anti-GFP and fluorescently–labeled phalloidin staining for F-actin. a COS7 cells expressing the indicated GFP-tagged proteins were allowed to spread for 1 h on fibronectin. b Quantification of the projected cell area of COS7 cells treated as in (a). Means ± SEM (n = 159-168 cells from 3 experiments). One-way ANOVA, Kruskal Wallis and Dunn’s post hoc. c Morphology of transfected MDA-MB-231 cells fixed after 18 h on fibronectin. Quantification of the projected area of MDA-MB-231 cells expressing the indicated GFP-tagged proteins that were allowed to spread on fibronectin for either 18 h (d) or 3 h (e). Means ± SEM (n = 148–183 cells from 3 experiments; 3 data points outside the axis limits). One-way ANOVA, Kruskal Wallis and Dunn’s post hoc. f GFP-ERC1ΔN rescues the inhibition of MDA-MB-231 cell spreading induced by silencing either endogenous Liprin-α1 or ERC1. Cells overexpressing either GFP or GFP-ERC1ΔN and silenced for the indicated endogenous proteins were allowed to spread for 18 h on fibronectin–coated substrate. Left: efficient silencing of endogenous Liprin-α1 or ERC1 detected by immunoblotting on 20 μg of protein lysate/lane. Right: Quantification of the projected cell area. Means ± SEM (n = 121–179 cells from 3 experiments; 5 data points outside the axis limits). One-way ANOVA, Kruskal Wallis and Dunn’s post hoc. g Silencing of endogenous LL5α/β proteins prevents the increase in MDA-MB-231 cell spreading induced by GFP-ERC1ΔN. Left: efficient silencing of endogenous LL5α and LL5β detected by immunoblotting on 30 μg of protein lysate/lane. Right: quantification of the projected cell area. Means ± SEM (n = 106–146 cells from 3 experiments). One-way ANOVA, Kruskal Wallis and Dunn’s post hoc. h Confocal immunofluorescence showing the subcellular localization of endogenous paxillin (red) and Liprin−α1 (blue) in COS7 cells transfected with the indicated GFP-tagged proteins (green) and after 1 h of spreading on fibronectin. Overexpression of GFP-ERC1, but not of GFP-ERC1ΔN, affects the subcellular distribution of endogenous Liprin-α1. Yellow arrows indicate the cytoplasmic regions shown in the 3.5x enlargements (right panels, showing paxillin in red and Liprin-α1 in blue). Scale bars in (a, c, h), 20 µm. i Quantification of the effects of the expression of the indicated constructs on the localization of endogenous Liprin-α1 near paxillin-positive focal adhesions. COS7 cells transfected with the indicated GFP-tagged constructs were allowed to spread for 1 h on 10 µg/ml fibronectin. Cells were fixed and immunostained with antibodies for endogenous Liprin-α1 and paxillin. Quantification was performed on selected areas (example at the top of the panel) including focal adhesions in the central area of the transfected cells (details in the Methods). Data are displayed as normalized mean density (ratio of Liprin-α1 volume to FA area) ±SEM; n = 102–117 selected regions per condition (4 data points fall outside the axis limits). Statistical analysis was performed using the Kruskal–Wallis test followed by Dunn’s post hoc correction.
Downregulation of endogenous PMAP proteins ERC1 and Liprin-α1 negatively affects cell spreading13,14. To test if the effect of ERC1ΔN on cell spreading was dependent on the endogenous PMAP proteins, we expressed ERC1ΔN in cells silenced for either endogenous ERC1 or Liprin-α1. ERC1ΔN is resistant to ERC1 silencing because it lacks the N-terminal siRNA targeting sequence. As expected, silencing of either endogenous protein inhibited cell spreading, and expression of ERC1ΔN was able to rescue the defect in spreading induced by silencing either Liprin-α1 or ERC1 (Fig. 6f). Therefore, endogenous Liprin-α1 and ERC1 are not required for the enhancement of cell spreading induced by ERC1ΔN. Interestingly, depletion of endogenous LL5α/β proteins significantly inhibited cell spreading both in control and ERC1ΔN cells (Fig. 6g). Therefore in contrast to endogenous Liprin-α1 and ERC1, endogenous LL5 proteins are required to support ERC1ΔN-dependent spreading.
In COS7 cells endogenous Liprin-α1 localizes around central paxillin-positive focal adhesions41. Overexpression of full-length ERC1 caused the redistribution of endogenous Liprin-α1 away from focal adhesions to the center of the cell (Fig. 6h). ERC1ΔN did not evidently perturb the localization of endogenous Liprin-α1 that was still found around focal adhesions (Fig. 6h). Quantification confirmed a significantly different effect of ERC1ΔN on the localization of endogenous Liprin-α1 compared to the effect observed with full length ERC1 (Fig. 6i). One hypothesis is that PMAPs organized by ERC1 near the edge of the protruding edge of migrating cells may facilitate the removal of PMAP components like Liprin-α1 away from the cell edge where this protein promotes and stabilized protrusion19. The altered properties of ERC1ΔN condensates may cause defective recycling of PMAPs components, as indicated by the differential effects on the distribution of endogenous Liprin-α1 between wildtype and truncated ERC1 (Fig. 6h, i). Therefore these results are consistent with a role of ERC1 in organizing physiological condensates needed to add/remove components from the cell edge to favor their turnover and the recycling of molecular components.
Deletion of the N-terminal region of ERC1 inhibits the motility of tumor cells
Our goal is to interfere with the formation of ERC1 condensates without interfering with the interaction of ERC1 with its PMAP partners, to address the role of LLPS in PMAPs function during cell motility. Based on previous studies on ELKS-1 LLPS in C. elegans21 and on in silico analysis with the FuzDrop method predicting sequence-based condensate-promoting polypeptide regions by LLPS42, several regions of ERC1 may be implicated in driving LLPS (Supplementary Fig. S4). Therefore it is difficult to obtain ERC1 constructs unable to form condensates by LLPS, but still able to interact with its PMAP partners. We hence focused on the analysis of ERC1ΔN that maintains the binding capacity to PMAP partners, to address the effects of altering the properties of ERC1 condensates on PMAPs function in cell motility. Indeed, our results indicate that ERC1ΔN: (a) can still generate condensates although with altered dynamic properties (Fig. 3); (b) can form homo-oligomers (Fig. 4); (c) and retains its binding ability to LL5β and Liprin-α1 (Figs. 4 and 5). Therefore, ERC1ΔN represents a good candidate to address how altering the dynamic properties of the ERC1 condensates may influence the function of the PMAP network during cell motility. We tested the effects of the deletion of the N-terminal IDR of ERC1 on different aspects of cell motility and invasion.
Matrigel invasion assays were used to test the invasive ability of MDA-MB-231 cells. This analysis showed that while overexpression of the full-length ERC1 protein did not affect invasion in vitro, expression of ERC1ΔN significantly reduced transwell invasion (Fig. 7a). Invasion by tumor cells requires both protrusive activity and the ability of cells to degrade the extracellular matrix. Therefore defective invasion by cells expressing ERC1ΔN could be due to impaired cell migration and/or to defective extracellular matrix degradation.
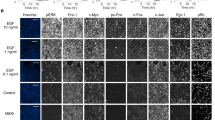
a MDA-MB- 231 cells expressing the indicated GFP-tagged proteins were used for the Matrigel Invasion assay (n = 9 wells from 3 independent experiments). Transfected cell counts were normalized to controls (GFP-transfected cells). Means ± SEM; n = 9 transwells from 3 experiments. One-way ANOVA, Kruskal Wallis and Dunn’s post hoc. Scale bar, 100 μm. b Effects of GFP-ERC1 and GFP-ERC1ΔN on two-dimensional random migration of MDA-MB-231 cells plated on fibronectin. Graph: means ± SEM of velocity (µm/min) of GFP–positive cells. c Plots with tracks of migrating GFP–positive cells (5 h). d Left: analysis of MSD from cell tracking data plotted as a function of time interval. Right: α values obtained by fitting MSD curves to least squares regression at 150 min (means ± SEM). In c–e, n = 253-280 cells from 3 experiments. One-way Anova, Kruskal-Wallis, Dunn’s posthoc. e MDA-MB-231 cells cotransfected with active SrcY527F (blue) and the indicated mCherry-tagged constructs (red). Transfected cells were plated on fluorescently labeled gelatin (green). Top panel: merged signals of mCherry (red), Src (blue), and FITC-gelatin (green). Lower panels show dark areas of gelatin degradation. Scale bar, 20 μm. Line profiles for ECM degradation in the selected cells are shown on the right. f Percentage of extracellular matrix–degrading MDA-MB-231 cells coexpressing active SrcY527F and the indicated mCherry-tagged protein. Mean ± SEM; n = 4 experiments. Significant differences were evaluated by Fisher’s χ2 test. g Percentage of the extracellular matrix degraded area normalized to the cell area. Means ± SEM; n = 190–244 cells from 4 experiments (2 data points are outside the y-axis limit). One-way Anova, Kruskal-Wallis, Dunn’s posthoc.
The analysis of free two-dimensional random migration on fibronectin of MDA-MB-231 cells showed no differences in cell speed between cells expressing either GFP-ERC1 or GFP-ERC1ΔN (Fig. 7b, c). Cell trajectories were used to evaluate the mean square displacement (MSD) that can be defined as a measure of the average distance a cell travels during a given time interval43. MSD curves were fitted by least-squares regression to clarify the directionality of cell migration by considering the α value44. By calculating the α values during the first 150 min of the random migration assays, we observed that both ERC1 and ERC1ΔN caused a depressed motion or sub-diffusion (α < 1) in comparison to random walk of GFP positive cells (α ≈ 1)43,44; in particular, ERC1ΔN positive cells displayed the lowest α values, suggestive of a compromised cell directionality (Fig. 7d).
Given the inhibitory effects of ERC1ΔN expression on invasion, we tested the effects of the N-terminal deletion on the ability of tumor cells to degrade the extracellular matrix. ERC1 and other PMAP proteins are found near invadosomes in Src-transformed cells and in MDA-MB-231 cells expressing the constitutively active Src–Y527F mutant45. GFP-ERC1ΔN, but not GFP-ERC1, inhibited extracellular matrix degradation by MDA-MB-231 expressing SrcY527F (Fig. 7e). Both the percentage of extracellular matrix-degrading cells and the area of extracellular matrix degradation were inhibited with respect to control cells expressing GFP by expression of the deletion mutant, but not by full length ERC1 (Fig. 7f, g). Our data indicate that the expression of ERC1ΔN negatively affects the invasive capacity of tumor cells by inhibiting extracellular matrix degradation.
Discussion
The data presented here demonstrate that altering the biophysical properties of the supramolecular assemblies driven by ERC1/ELKS has consequences on cell motility. The N-terminal region ERC1(1-244), including an extended IDR and short coiled-coil regions, is sufficient for LLPS in vitro and in cells (Figs. 1 and 2). Interestingly, the deletion of this N-terminal region does not prevent the formation of cytoplasmic condensates by ERC1ΔN, but it significantly affects the morphology and liquid–like behavior of ERC1ΔN condensates (Fig. 3). Of note, the morphological and FRAP analyses presented in this study suggest that the two complementary portions of the ERC1 protein produce condensates that behave very differently. In contrast to the condensates formed by the full length ERC1 protein, the condensates formed by the N-terminal fragment ERC1(1-244) show more spherical and regular morphologies, with limited sizes compared to the ERC1 condensates (Fig. 1a). Analysis in living cells also suggests a more rigid behavior of the ERC1(1-244) condensates, with very rare fusion events observed by time-lapse imaging (Supplementary Movie 3). On the other hand, the complementary region ERC1ΔN forms condensates that show a clear liquid-like behavior with frequent fusion events leading to very large cytoplasmic condensates (Fig. 3d and Supplementary Movie 4). Moreover, the condensates formed by ERC1ΔN are highly dynamic as indicated by the faster recovery of fluorescence compared to the full length ERC1 condensates (Fig. 3f, g and Supplementary Movie 6). Altogether these data suggest that the two complementary regions ERC1(1-244) and ERC1ΔN contribute differently to the material properties of the condensates formed by the full length ERC1 protein.
Although the homotypic and heterotypic intermolecular interactions of ERC1ΔN with its PMAP partners are not affected (Figs. 4 and 5), ERC1ΔN expression has consequences on cell spreading (Fig. 6) and on tumor cell invasion in vitro (Fig. 7). These findings indicate that PMAPs forming at the leading edge of migrating cells are influenced by specific alteration of the properties of the ERC1 condensates, independently from the scaffolding capacity of ERC1ΔN.
The presence of poorly structured flexible IDRs46 is a common feature of many proteins that undergo LLPS47. Indeed often IDRs are necessary drivers of LLPS48. Interestingly, the N-terminal region including the IDR of ERC1/ELKS, although sufficient for LLPS, is not necessary for the formation of either ERC1 condensates or PMAPs in migrating tumor cells (Fig. 5d), but influences the properties of the ERC1–driven condensates. It is expected that multivalent weak protein-protein interactions are the driving force behind the assembly of ERC1 condensates and PMAPs49,50. It is conceivable that partially disrupting specific transient interactions by removing the N-terminal region of ERC1/ELKS alters the properties and function of PMAPs.
The structure of ERC1/ELKS is poorly characterized. This protein is predicted to include an N-terminal IDR and extended coiled coils51, consistent with the low-resolution structure obtained by rotary shadowing electron microscopy, showing that ERC1/ELKS forms elongated parallel dimers, likely via the extensive coiled coils within ERC120. Coiled coils may enhance LLPS by increasing multivalency through dimerization. The N-terminal IDR per se is unable to form condensates20, but it is necessary for the formation of ERC1(1-244) condensates (Fig. 1): it includes predicted short coiled-coils also necessary for the formation of ERC1(1-244) condensates (Fig. 1a).
The observed distinct properties of the condensates formed by ERC1, ERC1(1-244), and ERC1ΔN (Figs. 1–3) suggest that ERC1(1-244) contributes to the characteristics of the condensates formed by the full length protein: removal of the N-terminal region leads to ERC1ΔN condensates with very different morphology/properties with respect to those formed by either full length ERC1 or ERC1(1-244). The different morphologies of condensates formed by the two complementary parts of the ERC1 protein may reflect differences in the surface tension and/or visco-elastic properties that depend on the forces of attraction between the polypeptides within the two types of condensates. Surface tension represents an important physical property of condensates as it can affect their formation and function52, and may help explaining the specific effects observed by cells expressing ERC1ΔN. In this direction, ERC1(1-244) and ERC1ΔN display different mean net charges (+0.029 and –0.037, respectively) (Supplementary Table 1). One hypothesis is that differences in the density of charged residues may influence the properties of condensates by intermolecular electrostatic interactions48,53, contributing to the different dynamics of condensates formed by ERC1(1-244) and ERC1ΔN, respectively.
ERC1 promotes focal adhesion turnover in migrating tumor cells14. One hypothesis is that ERC1 condensates are needed for adding/removing components at the cell front, to favor their turnover and the turnover of adhesions to support continuous protrusion and cell movement19. In this direction, overexpression of the full-length ERC1 protein displaces endogenous Liprin-α1 from the cell edge and from around focal adhesions, while ERC1ΔN is unable to do so (Fig. 6h). It is conceivable that ERC1ΔN enhances cell spreading as a consequence of the altered properties of the ERC1ΔN–driven condensates, which in turn may affect the turnover of PMAPs and cemm motility.
Our results support the hypothesis that ERC1 acts as a scaffold undergoing phase separation by a complex mechanism that includes a modulatory N-terminal region including an IDR. The distinct properties revealed by the analysis presented in this study, of the condensates produced by the complementary N-terminal ERC1(1-244) and ERC1ΔN, together with the functional consequences of altering the properties of condensates by the N-terminal ERC1ΔN deletion (including effects on cell spreading and Matrigel invasion), support the proposition that ERC1-driven PMAPs may be considered a potential target to interfere with the tumor metastatic process. The findings that removal of the N-terminal region affects ERC1 condensates and results in defects in cell invasion open to applications to interfere with tumor cell motility and metastasis. It is possible to hypothesize that interfering with PMAPs by agents such as small molecules, which can be intercalated within ERC1 condensates, may lead to alterations of the material properties of the PMAPs, with negative consequences on the invasive capacity of tumor cells. For this, more work is needed to characterize in more detail the intermolecular mechanisms that drive the formation of ERC1 condensates and of the PMAP protein network.
Methods
Plasmids and siRNAs
Plasmids for GFP-ERC1 (murine ELKSε, 1116 residues)54, FLAG-ERC1, Cherry-LL5β (murine isoform 3, 1206 residues)55, FLAG-Liprin-α1 (human), FLAG-Liprin-ΔEBR, FLAG-Liprin-EBR, GFP-ERC1-N, GFP-ERC1-C, GFP-ERC1Δ51, GFP-ERC1Δ147, and FLAG-βGalactosidase were described previously13,14,20,41,56. ERC1 fragments ERC1(1-420), ERC1(1-N), ERC1(1-244), ERC1(1-188, and ERC1ΔN were obtained by PCR from the plasmid pGFP-C1-ERC1, and subcloned into the pGFP-C1 or mCherry-C1 vector (Clontech Laboratories). For PCR, the following primers were used:
GFP-ERC1(1-420): forward 5′-GGAATTCCATGTATGGAAGTGCTCGATC-3′;
reverse 5′-GGAATTCTCACTCCCGCTCCTCACTAC-3′;
GFP-ERC1(1-244): forward 5′-GGAATTCCATGTATGGAAGTGCTCGATC-3’;
reverse 5′-GGAATTCTTACTGATTCAAGTCCCTCTG-3′;
GFP-ERC1(1-188): forward 5′-GGAATTCCATGTATGGAAGTGCTCGATC-3′;
reverse 5′-GGAATTCTTAGGTCTTGATACTATTCATTG-3’;
GFP-ERC1(60-420): forward 5′-GGAATTCCATGCAGTCCTTAAATGCTGCC-3′;
reverse 5’- GGAATTCTCACTCCCGCTCCTCACTAC-3’;
GFP-ERC1ΔN: forward 5′-GGAATTCCATGCTCACAGAGGAGAACTTCCAG-3′;
reverse 5′-GGAATTCTCAGGAGGACTCTTCCAG-3′;
GFP-ERC1Δ375: forward 5′-GTCGACTGCAGAATTCGAAGCTTGAGC-3′;
reverse 5’-ACAAAAGCCTTGCAAACTGTTATTGAAATG-3′;
monomeric GFP(A206K): forward 5′-TACCTGAGCACCCAGTCCAAACTGAGCAAAGACCCCAAC-3′; reverse 5′-GTTGGGGTCTTTGCTCAGTTTGGACTGGGTGCTCAGGTA-3′.
His-GFP-ERC1(1-244)pFastBac for the purification of GFP-ERC1(1-244) was obtained by Thermo Fisher Scientific. Liprin-α1, ERC1, and LL5α and β were silenced by transfecting validated siRNAs as described in ref. 14.
Antibodies and other reagents
Anti-ERC1 [ELKS-30] against residues 21-40 of ERC1a from Abcam, ab50312, mouse mAb, WB 1:1000; anti-ERC1 from Sigma-Aldrich, HPA019513, rabbit pAb, IF 1:150. Anti-FLAG mouse mAb, clone M2 from Sigma-Aldrich, WB 1:1000; IF 1:500. Anti-GFP rabbit pAb from Invitrogen, A11122, WB 1:2000. Anti-GFP chicken pAb from Abcam, ab13970; IF 1:1000. Anti-Liprin-α1 rabbit pAb from Proteintech, 14175-1-AP; WB 1:500, IF 1:150. Anti-paxillin mouse mAb (clone 349) from BD Biosciences; WB 1:2000; IF 1:150. Anti-Src (clone 327) was kindly provided by Dr. Sara Courtneidge (Oregon Health and Science University); IF 1:50. Hamster mAb for LL5α57 was kindly provided by Dr. Yuko Mimori-Kiyosue (RIKEN Center for Biosystems Dynamics Research, Kobe, Japan). Rabbit pAb anti-fibronectin was from Sigma-Aldrich, and rabbit pAb anti-DS-RED was from Clontech. Anti-LL5α/β (clone 1H12) was as described55. Oregon green 488-gelatin, Phalloidin Alexa-568S, and secondary Abs Alexa-488, Alexa-568, Alexa-546 and Alexa-647, and Alexa-647 (1:200) were from Life Technologies. HRP-conjugated anti-rabbit and anti-mouse secondary Abs (1:3000–1:5000) were from Jackson and Amersham Biosciences. Fibronectin was from Corning. 1,6-hexanediol was from Sigma.
Cell culture and transfection
COS7 cells were cultured in DMEM with 10% fetal clone III (Hyclone). MDA-MB-231 human breast adenocarcinoma cells were grown in DMEM:F12 1:1 with 10% fetal bovine serum. NIH-3T3 cells were cultured in DMEM with 10% fetal bovine serum. Cells on plastic or on round 24 mm diameter glass coverslips were transfected with Lipofectamine-2000 (Life Technologies). For each 3.5 cm diameter plate 1–4 μg of plasmid DNA was used in Opti-MEM transfection medium. After 3.5–4 h the medium was replaced with complete medium; cells were processed after 24–72 h. All cells were regularly checked for mycoplasma contamination. Cells transfected for 24 h were incubated for 2′ at 37 °C with either 1.5% or 3% 1,6-hexanediol dissolved in culture medium58 before fixation for immunofluorescence.
Immunoprecipitation and immunoblotting
Cells were washed twice with ice-cold TBS (150 mM NaCl, 20 mM Tris-HCl pH 7.5), lysed with 50–150 μl of lysis buffer: 0.5% Triton X-100, 150 mM NaCl, 20 mM Tris-Cl pH 7.5, 1 mM NaV, 10 mM NaF, 1 mM DTT, anti-proteases Complete 1× (Roche), 0.5 mM PMSF (Sigma-Aldrich). After 15 min at 4 °C the insoluble material was removed by centrifugation at 16000 RCF for 15 min at 4 °C. Protein concentration was determined by Bradford protein assay (Bio-Rad). Denatured lysates were separated by SDS-PAGE and transferred to 0.45 μm nitrocellulose membranes (GVS). Membranes were incubated with primary antibodies, HRP-conjugated secondary antibodies, and revealed by Clarity with ChemiDoc MP Imaging System (Bio-Rad). Quantification of protein levels was done with ImageLab software (Bio-Rad). For reprobing, membranes were stripped by 5–10 min incubation at RT with 0.2 M glycine, 0.1% SDS, 1% Tween-20, pH 2.2, and washed at neutral pH before reprobing with the indicated antibodies. For immunoprecipitation cell lysates were incubated with either Protein-A–Sepharose beads (Cytiva), Pierce Protein G Agarose (Thermo Scientific) conjugated to antibodies, GFP-Trap® (Chromotek), or anti-FLAG-M2 Affinity Gel (Sigma-Aldrich) before processing for SDS-PAGE and immunoblotting (Supplementary Figs. 5 and 6 include uncropped blot images presented in Figs. 4 and 6, respectively).
In vitro LLPS assay
Purification of His6X-Strep(2X)-GFP-ERC1(1-244). Trichoplusia ni codon optimized ORF coding for His6X-Strep(2X)-GFP-ERC1(1-244) in pFastBac1 vector was obtained from Geneart (Thermo Fisher) and transposed into a baculovirus genome-containing BAC with the BAC-to-BAC methodology (Thermo Fisher). Infectious recombinant baculovirus was reconstituted in the Sf9 cell line grown in Sf-900 II medium (Thermo Fisher), according to manufacturer guidelines. A high titer viral stock was used to infect the High Five cells in the Insect-XPRESS culture medium (Lonza) for 72 h. Infected cells were lysed in 20 mM Tris pH 8.5, 150 mM KCl, 1 M urea, and the His6X-Strep(2X)-GFP-ERC1(1-244) fusion protein was purified by tandem affinity chromatography on Talon resin (Thermo Fisher) followed by a Strep Tactin column (IBA Lifescience). His6X-Strep(2X)-GFP-ERC1(1-244) was passed through a Superdex 200 size exclusion column equilibrated in 20 mM Tris pH 8.5, 150 mM KCl, 0.5 M L-arginine. Protein concentration was determined by UV280 and Bradford assay.
For in vitro LLPS, purified His6X-Strep(2X)-GFP-ERC1(1-244) was diluted in LLPS buffer (25 mM HEPES 7.5, 100 mM KCl) at different concentrations (5–20 µM) ± 10% PEG 3350. Reactions were incubated 5′ at RT. Samples were placed in home-made imaging chambers26, and observed at Leica SP5 confocal microscope with 63x oil lens (Zeiss).
Immunofluorescence and image analysis
Cells grown on fibronectin-coated coverslips were washed and fixed in 3% paraformaldehyde, permeabilized f with 0.1% Triton X-100, and incubated for 2 h with primary antibodies, then for 45 min with secondary antibodies, and mounted in ProLong Gold Antifade Reagent (Life Technologies). Wide field images were acquired with Zeiss Axio Observer Z.1 with Hamamatsu EMCCD 9100–02 camera equipped with Plan-Apochromat 63x (NA 1.4), 40x or 20x lenses. Confocal images were acquired with Leica TCS SP8 SMD FLIM laser scanning confocal microscope with HC PLAPO CS2 63× lens (NA 1.4), or with Leica TCS SP5 laser scanning confocal microscope with HCX PLAPO λ blue 63x lens (NA 1.4). Alternatively, images were taken at an Olympus FluoVIEW FV3000RS laser scanning confocal microscope equipped with UPLXAPO 60x (NA 1.42).
Colocalization of proteins with GFP-ERC1 in condensates was evaluated by the Pearson’s correlation coefficient on confocal images59. The coefficient (1 = perfect correlation, 0 = no correlation, −1 = perfect anti-correlation) was calculated using the plugin Colocalization Finder of ImageJ. For quantification, 3–4 GFP-ERC1–positive condensates were randomly picked from 6–10 cells per condition per experiment.
The aspect ratio (A.R.) of intracellular condensates was measured with ImageJ. An ellipse was overlaid onto individual condensates, and the ratio between major and minor axis of the ellipse was measured. Condensates larger than 16 μm² were excluded because their shape could be influenced by other cytoplasmic structures (e.g., organelles), while condensates smaller than 0.5 μm² were excluded for the difficulty to obtain reliable A.R. values.
For the localization of endogenous Liprin-α1 near focal adhesions, cells immunostained for GFP, endogenous paxillin, and endogenous Liprin-α1 were analyzed as follows: 2–3 internal areas per cell (edge excluded), each including 3–10 paxillin-positive focal adhesions, were selected. Within each area, we measured the area of focal adhesions and the signal of endogenous Liprin-α1 (integrated density of the Liprin-α1 signal above a fixed threshold) by Image J software. Data were presented as Liprin-α1 signal/focal adhesion area.
FRAP analysis
FRAP experiments were performed as described20. Briefly, COS7 cells were plated on 3.5 cm diameter glass-bottom MatTek dishes (MatTek Corporation, Ashland, MA) pre-coated for 1 h at 37 °C with 2.5 μg/ml fibronectin, transfected for 24 h, and imaged at the Leica TCS SP8 SMD FLIM confocal microscope with adaptive focus control, Oko-Lab stage incubator (T, CO2), and FRAP module with LasX software (Leica). Cells were moved to imaging medium without phenol red for imaging. For full droplet bleaching FRAP experiments, a 2.5–5 μm diameter circular region of interest (ROI) was bleached with 1 pulse (0.2 s) of 100% laser power at 488 nm (Argon laser line). After bleaching, images were taken every 0.25 s for 5–10 min to monitor fluorescence recovery. The recovery was measured with ImageJ by calculating the fluorescent intensity at each time point as follows: R = [F(t)-F(0)]/[F(pre-bleach)-F(0)], where F(t) is the intensity of fluorescence at time t: F(t) = [F(ROI(t))-F(bcg(t))]/[F(ctrl(t))-F(bcg(t))], where “bcg” stands for background fluorescence outside the cell, and “ctrl” stands for control area in the cytosol for full droplet bleaching60. For FRAP analysis after spot bleaching, a 2 μm diameter ROI was bleached with 1 pulse (0.2 s) of 100% laser power at 488 nm (Argon laser line). After bleaching, images were taken every 0.2 s for 5 min; control for spot bleaching was on a nearby ERC1-positive condensate area. Curve fitting was performed with GraphPad Prism® (version 9.0), fitting the data to a non-linear regression double exponential equation61.
Condensate formation
The formation of condensates was followed in COS7 cells plated for one day on fibronectin–coated dishes (2.5 μg/ml for 1 h at 37 °C), and then transfected to express either GFP-ERC1 or GFP-ERC1ΔN. Right after transfection, the cells were imaged for 16-18 h (one frame every 5 min) with Live-Cell Imaging System equipped with 20x lens (Essen BioScience, Ann Arbor, MI) to follow the formation of the cytoplasmic condensates. For the graphs in Fig. 3e, the fluorescence intensity in each transfected cell was measured every hour for 11 h, starting from the beginning of detectable GFP-ERC1 or GFP-ERC1ΔN expression, by using the Image J software20.
Cell motility and live cell imaging
For cell spreading, MDA-MB-231 cells were re-plated 24–48 h after transfection on coverslips coated with 10 μg/mlc fibronectin. After 3 h or 18 h cells were processed for immunofluorescence. COS7 cells were re-plated 24 after transfection on coverslips coated with 10 μg/ml fibronectin. After 1 h cells were processed for immunofluorescence and images recorded at a Zeiss Axio Observer.Z1 inverted microscope with 40x or 63x lens equipped with Hamamatsu 9100 - 02 EM CCD Camera and Plan-Apochromat 63x (NA 1.4) lens. The projected cell area was quantified by ImageJ software (NIH, Bethesda, MD).
For two-dimensional random migration assays, MDA-MB-231 cells were replated 24 h after transfection on fibronectin–coated 6-well plates (2.5 μg/ml, overnight at 4 °C). After overnight culture, cells were washed with phosphate buffer saline and supplied with fresh medium. Cells were imaged for 5 h (one frame every 10 min) with IncuCyte Live-Cell Imaging System equipped with 10x lens (Sartorius). The pathway of transfected cells in two-dimensional migration assays was tracked and analyzed with Image J (Manual tracking and Chemotaxis plugins). About 70–100 cells/conditions were analyzed in each experiment.
Cell trajectories (X, Y) were used to calculate the MSD as follows: Dx = X(ti+1:end) − X(1:end-ti); Dy = Y(ti+1:end) − Y(1:end-ti); disp_sq = Dx2 + Dy2; MSD(ti) = mean(disp_sq); MSD_err(ti) = std(disp_sq)/sqrt(numel(disp_sq)); where D = displacement; ti = initial time; disp_sq = square displacement; MSD_err(ti) = MSD SEM; std(disp_sq) = standard deviation of square displacement43. The α values were extrapolated by fitting MSD data to a linear regression model (MSD(t)∝tα)44; where t = time. MSD and α values were calculated with MatLab®R2022b.
Time-lapse imaging to assess PMAPs in living cells was performed on MDA-MB-231 cells co-transfected overnight with either GFP, GFP-ERC1, or GFP-ERC1ΔN together with mCherry-Zyxin. Cells were recorded after replating for 18 h on 35 mm diameter fibronectin-coated (2.5 μg/mL o.n. at 4 °C) glass bottom dishes (MatTek Corporation). For imaging cells were supplied with phenol red-free DMEM medium with 10% FBS (Thermo Scientific), and recorded at 37 °C with 5% CO2. Images were acquired for 50 min at 1 frame/min with a Nikon CSU-X1 Spinning Disk Nikon TE2 inverted microscope with 100x oil lens (1.4 NA) and an ORCA-Flash 4.0 camera (Hamamatsu) with NIS Elements acquisition software (Nikon)15.
Matrigel invasion
Transfected MDA-MB-231 cells were for 24 h were seeded (100,000 cells in 100 μl) on Matrigel-coated 8 μm-pore polycarbonate membrane transwells (Corning) in DMEM/F12 medium with 0.1% bovine serum albumin (Sigma-Aldrich). NIH-3T3-conditioned medium placed in the lower chamber was used as chemoattractant. After 5 h at 37 °C non-invading cells were removed from the upper side of the filter with a cotton swab; cells invading and crossing the membrane were fixed and stained with DAPI and anti-GFP Ab.
Extracellular matrix degradation
Gelatin degradation assays were performed as published45,62. Briefly, glass coverslips coated for 1 h at RT with 0.5 mg/ml poly-L-lysine were quenched 15 min at 4 °C with 0.5% glutaraldehyde in phosphate buffer saline and coated for 10 min at RT with Oregon–green–conjugated gelatin (Life Technologies) diluted 1:4 in 0.2% gelatin in phosphate buffer saline. The coverslips were finally coated with 10 μg/ml fibronectin in phosphate buffer saline for 1 h at 37 °C. Cells transfected for 24 h were re-plated on gelatin-coated coverslips and cultured for 6 h at 37 °C before processing for immunofluorescence. Dark areas of gelatin degradation and projected cell areas were quantified by ImageJ on thresholded images. Invadopodia were identified by staining for phalloidin. Thirty–50 cells/condition/experiment were analyzed.
Bioinformatics
The average net charge at pH 7 of the complementary ERC1 regions, ERC1(1-244) and of ERC1ΔN, was determined as the difference between positively charged amino acids (Lys and Arg) and negatively charged amino acids (Glu and Asp). The mean net charge was obtained by dividing the total net charge by the number of residues of the respective polypeptide63. The total hydrophobicity was calculated using the Kyte and Doolittle scale considering 5-residue intervals64. The mean hydrophobicity was obtained by dividing the total hydrophobicity by the number of residues of the respective polypeptide. The polypeptide sequences were analyzed with ProtParam and ProtScale65. Structural prediction by Alphafold235 was obtained using the ColabFold platform66.
Statistics and reproducibility
Statistical analysis was performed with GraphPad Prism 9.0. Datasets were tested for normality with Shapiro-Wilk test. For datasets with normal distribution, the statistical significance was calculated by the unpaired two-tailed Student’s t-test or one-way ANOVA with Dunnett’s or Tukey’s posthoc. For datasets with non-normal distribution statistical significance was calculated using Kruskal-Wallis test with Dunn’s posthoc. Data are presented as means ± SEM of at least three experiments
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data associated to the results presented in this study will be available in the San Raffaele Open Research Data Repository (https://ordr.hsr.it/research-data/), https://doi.org/10.17632/tsv85hkcw3.1.
References
Ridley, A. J. et al. Cell migration: integrating signals from front to back. Science 302, 1704–1709 (2003).
Duffy, M. J., McGowan, P. M. & Gallagher, W. M. Cancer invasion and metastasis: changing views. J. Pathol. 214, 283–293 (2008).
Parsons, J. T., Horwitz, A. R. & Schwartz, M. A. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 11, 633–643 (2010).
Lappalainen, P., Kotila, T., Jégou, A. & Romet-Lemonne, G. Biochemical and mechanical regulation of actin dynamics. Nat. Rev. Mol. Cell Biol. 23, 836–852 (2022).
Elosegui-Artola, A., Trepat, X. & Roca-Cusachs, P. Control of mechanotransduction by molecular clutch dynamics. Trends Cell Biol. 28, 356–367 (2018).
Hida, Y. & Ohtsuka, T. CAST and ELKS proteins: structural and functional determinants of the presynaptic active zone. J. Biochem. 148, 131–137 (2010).
Pehkonen, H., de Curtis, I. & Monni, O. Liprins in oncogenic signaling and cancer cell adhesion. Oncogene 40, 6406–6416 (2021).
Paranavitane, V., Coadwell, W. J., Eguinoa, A., Hawkins, P. T. & Stephens, L. LL5beta is a phosphatidylinositol (3,4,5)-trisphosphate sensor that can bind the cytoskeletal adaptor, gamma-filamin. J. Biol. Chem. 278, 1328–1235 (2003).
Ribolla, L. M. et al. Interfering with the ERC1-LL5β interaction disrupts plasma membrane-associated platforms and affects tumor cell motility. PLoS ONE 18, e0287670 (2023).
Lansbergen, G. et al. CLASPs attach microtubule plus ends to the cell cortex through a complex with LL5β. Dev. Cell 11, 21–32 (2006).
Grigoriev, I. et al. Rab6 regulates transport and targeting of exocytotic carriers. Dev. Cell 13, 305–314 (2007).
Stehbens, S. J. et al. CLASPs link focal-adhesionassociated microtubule capture to localized exocytosis and adhesion site turnover. Nat. Cell Biol. 16, 561–573 (2014).
Astro, V., Chiaretti, S., Magistrati, E., Fivaz, M. & de Curtis, I. Liprin-alpha1, ERC1 and LL5 define polarized and dynamic structures that are implicated in cell migration. J. Cell Sci. 127, 3862–3876 (2014).
Astro, V. et al. Liprin-alpha1 and ERC1 control cell edge dynamics by promoting focal adhesion turnover. Sci. Rep. 6, 33653 (2016).
Ramella, M. et al. Dual specificity kinase DYRK3 regulates cell migration by influencing the stability of protrusions. iScience 27, 109440 (2024).
Hyman, A. A., Weber, C. A. & Jülicher, F. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 (2014).
Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017).
Astro, V. & de Curtis, I. Plasma membrane-associated platforms: dynamic scaffolds that organize membrane-associated events. Sci. Signal. 8, re1 (2015).
de Curtis, I. Biomolecular condensates at the front: cell migration meets phase separation. Trends Cell Biol. 31, 145–148 (2021).
Sala, K. et al. The ERC1 scaffold protein implicated in cell motility drives the assembly of a liquid phase. Sci. Rep. 9, 13530 (2019).
McDonald, N. A., Fetter, R. D. & Shen, K. Assembly of synaptic active zones requires phase separation of scaffold molecules. Nature 588, 454–458 (2020).
Liang, M. et al. Oligomerized Liprin-alpha promotes phase separation of ELKS for compartmentalization of presynaptic active zone proteins. Cell Rep. 34, 108901 (2021).
Liang, M. et al. Structural basis of the target-binding mode of the G protein-coupled receptor kinase-interacting protein in the regulation of focal adhesion dynamics. J. Biol. Chem. 294, 5827–5839 (2019).
Linding, R. et al. Protein disorder prediction: implications for structural proteomics. Structure 11, 1453–1459 (2003).
McDonnell, A. V., Jiang, T., Keating, A. E. & Berger, B. Paircoil2: Improved prediction of coiled coils from sequence. Bioinformatics 22, 356–358 (2006).
Alberti, S. et al. A user’s guide for phase separation assays with purified proteins. J. Mol. Biol. 430, 4806–4820 (2018).
Zacharias, D. A., Violin, J. D., Newton, A. C. & Tsien, R. Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 96, 913–916 (2002).
Brangwynne, C. P., Mitchison, T. J. & Hyman, A. A. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. USA 108, 4334–4339 (2011).
Tsumoto, K. et al. Role of arginine in protein refolding, solubilization, and purification. Biotechnol. Prog. 20, 1301–1308 (2004).
André, A. A. M. & Spruijt, E. Liquid-liquid phase separation in crowded environments. Int. J. Mol. Sci. 21, 5908 (2020).
André, A. A. M., Yewdall, N. A. & Spruijt, E. Crowding-induced phase separation and gelling by co-condensation of PEG in NPM1-rRNA condensates. Biophys. J. 122, 397–407 (2023).
Chiaretti, S., Astro, V., Chiricozzi, E. & de Curtis, I. Effects of the scaffold proteins liprin-α1, β1, and β2 on invasion by breast cancer cells. Biol. Cell 108, 65–75 (2016).
Nakata, T. et al. Fusion of a novel gene, ELKS, to RET due to translocation t(10;12)(q11;p13) in a papillary thyroid carcinoma. Genes Chromosom. Cancer 25, 97–103 (1999).
Ko, J., Na, M., Kim, S., Lee, J. R. & Kim, E. Interaction of the ERC family of RIM-binding proteins with the liprin-alpha family of multidomain proteins. J. Biol. Chem. 278, 42377–42385 (2003).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Südhof, T. C. The presynaptic active zone. Neuron 75, 11–25 (2012).
van der Vaart, B. et al. CFEOM1-associated kinesin KIF21A is a cortical microtubule growth inhibitor. Dev. Cell 27, 145–160 (2013).
Bernadzki, K. M. et al. Liprin-α-1 is a novel component of the murine neuromuscular junction and is involved in the organization of the postsynaptic machinery. Sci. Rep. 7, 9116 (2017).
Toretsky, J. A. & Wright, P. E. Assemblages: functional units formed by cellular phase separation. J. Cell Biol. 20, 579–588 (2014).
Astro, V., Asperti, C., Cangi, M. G., Doglioni, C. & de Curtis, I. Liprin-α1 regulates breast cancer cell invasion by affecting cell motility, invadopodia, and extracellular matrix degradation. Oncogene 30, 1841–1849 (2011).
Asperti, C., Astro, V., Totaro, A., Paris, S. & de Curtis, I. Liprin-alpha1 promotes cell spreading on the extracellular matrix by affecting the distribution of activated integrins. J. Cell Sci. 122, 3225–3232 (2009).
Hardenberg, M., Horvath, A., Ambrus, V., Fuxreiter, M. & Vendruscolo, M. Widespread occurrence of the droplet state of proteins in the human proteome. Proc. Natl. Acad. Sci. USA 117, 33254–33262 (2020).
Wang, Y., Jeong, Y., Jhiang, S. M., Yu, L. & Menq, C. H. Quantitative characterization of cell behaviors through cell cycle progression via automated cell tracking. PLoS ONE 9, e98762 (2014).
Ingram, K. et al. NKX2-1 controls lung cancer progression by inducing DUSP6 to dampen ERK activity. Oncogene 41, 293–300 (2022).
Sala, K., Raimondi, A., Tonoli, D., Tacchetti, C. & de Curtis, I. Identification of a membrane-less compartment regulating invadosome function and motility. Sci. Rep. 8, 1164 (2018).
Uversky, V. N. Intrinsically disordered proteins in overcrowded milieu: membraneless organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 44, 18–30 (2017).
Boeynaems, S. et al. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 28, 420–435 (2018).
Borcherds, W., Bremer, A., Borgia, M. B. & Mittag, T. How do intrinsically disordered protein regions encode a driving force for liquid-liquid phase separation?. Curr. Opin. Struct. Biol. 67, 41–50 (2021).
Fung, H. Y. J., Birol, M. & Rhoades, E. IDPs in macromolecular complexes: the roles of multivalent interactions in diverse assemblies. Curr. Opin. Struct. Biol. 49, 36–43 (2018).
Schmit, J. D., Feric, M. & Dundr, M. How hierarchical interactions make membraneless organelles tick like clockwork. Trends Biochem. Sci. 46, 525–534 (2021).
Held, R. G. & Kaeser, P. S. ELKS active zone proteins as multitasking scaffolds for secretion. Open Biol. 8, 170258 (2018).
Wang, H., Kelley, F. M., Milovanovic, D., Schuster, B. S. & Shi, Z. Surface tension and viscosity of protein condensates quantified by micropipette aspiration. Biophys. Rep. 1, 100011 (2021).
Schuster, B. S. et al. Identifying sequence perturbations to an intrinsically disordered protein that determine its phase-separation behavior. Proc. Natl. Acad. Sci. USA 117, 11421–11431 (2020).
Inoue, E. et al. ELKS, a protein structurally related to the active zone protein CAST, is involved in Ca2+-dependent exocytosis from PC12 cells. Genes Cells 11, 659–672 (2006).
Kishi, M., Kummer, T. T., Eglen, S. J. & Sanes, J. R. L. L. 5beta: a regulator of postsynaptic differentiation identified in a screen for synaptically enriched transcripts at the neuromuscular junction. J. Cell Biol. 169, 355–366 (2005).
Totaro, A., Paris, S., Asperti, C. & de Curtis, I. Identification of an intramolecular interaction important for the regulation of GIT1 functions. Mol. Biol. Cell 18, 5124–5138 (2007).
Hotta, A. et al. Laminin-based cell adhesion anchors microtubule plus ends to the epithelial cell basal cortex through LL5alpha/beta. J. Cell Biol. 189, 901–917 (2010).
Liu, X. et al. Time-dependent effect of 1,6-hexanediol on biomolecular condensates and 3D chromatin organization. Genome Biol. 22, 230 (2021).
Dunn, K. W., Kamocka, M. M. & McDonald, J. H. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell. Physiol. 300, C723–C742 (2011).
Day, C. A., Kraft, L. J., Kang, M. & Kenworthy, A. K. Analysis of protein and lipid dynamics using confocal fluorescence recovery after photobleaching (FRAP). Curr. Protoc. Cytom. Chapter 2, Unit2.19 (2012).
Firmino, J., Tinevez, J. Y. & Knust, E. Crumbs affects protein dynamics in anterior regions of the developing Drosophila embryo. PLoS ONE 8, e58839 (2013).
Artym, V. V., Zhang, Y., Seillier-Moiseiwitsch, F. O., Yamada, K. M. & Mueller, S. C. Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res. 66, 3034–3043 (2006).
Uversky, V. N., Gillespie, J. R. & Fink, A. L. Why are “natively unfolded” proteins unstructured under physiologic conditions?. Proteins 41, 415–427 (2000).
Kyte, J. & Doolittle, R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 (1982).
Gasteiger, E. et al. Protein Identification and Analysis Tools on the Expasy Server. In: John M. Walker, ed. The Proteomics Protocols Handbook, 571–607 (Humana Press, 2005).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Acknowledgements
Support to IdC by Fondazione AIRC (Grant IG 2023 ID 29097) and by Ministero dell’Università e della Ricerca (PRIN 2022 - Project 2022EMZJL4 - Supramolecular assemblies in cell invasion as targets for cancer therapy; supported by the European Union - Next Generation EU, Mission 4 Component 2 Investment 1.1, CUP D53D23007880008) and to LMR by a fellowship from FCSR-Fronzaroli are gratefully acknowledged. Work performed by LMR was in partial fulfillment of the requirements for obtaining the PhD degree at the Vita-Salute San Raffaele University, Milano, Italy. We thank Dr. Jean-Michel Cioni (San Raffaele Scientific Institute) for support with time-lapse confocal imaging and the personnel of the Advanced Light and Electron Microscopy BioImaging Center (ALEMBIC) of the San Raffaele Scientific Institute for technical support.
Author information
Authors and Affiliations
Contributions
LMR, MR and MP performed experimental studies. MD provided advice on experimental design. IdC designed the experiments. IdC and LMR analyzed the data. IdC wrote the manuscript with the help of LMR.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Dragomir Milovanovic, Paul Atherton, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Kaliya Georgieva. [A peer review file is available.]
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ribolla, L.M., Patrone, M., Degano, M. et al. Altering the biophysical properties of ERC1/ELKS–driven condensates interferes with cell motility. Commun Biol 8, 1045 (2025). https://doi.org/10.1038/s42003-025-08470-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08470-5