Abstract
Terminal cell differentiation, a process vital for tissue development and regeneration where progenitor cells acquire specialized functions and permanently exit the cell cycle, remains poorly understood at the molecular level. Using live-cell imaging and adipogenesis as a model, we show that the initial stage involves a variable number of cell divisions, driven by redundant CDK4/6 or CDK2 activation. Afterwards, a delayed decrease in cyclin D1 and an increase in p27 levels reduce CDK4/6 and CDK2 activity. This results in G1 lengthening and the induction of PPARG, the master regulator of adipogenesis. PPARG then induces p21, and later p18, ultimately causing irreversible inactivation of CDK4/6 and CDK2, and thus, permanent cell cycle exit. However, contrary to expectation, CDK inactivation alone is not sufficient to trigger differentiation commitment; ERK inactivation is also necessary. Our study reveals that the coordinated activation and subsequent delayed inactivation of CDK4/6, CDK2, and ERK are crucial for irreversible cell cycle exit and differentiation commitment in terminal cell differentiation.
Similar content being viewed by others
Introduction
The process of terminal cell differentiation produces specialized cell types, including neurons, muscle cells, and adipocytes (fat cells), which are essential for the proper functioning of tissues and organs1,2,3. This process generally begins with progenitor cell division, leading to an expansion of the progenitor pool, and culminates in permanent exit from the cell cycle and the development of specialized functions1,2,3. Failure of terminally differentiated cells to permanently exit the cell cycle is a defining feature of cancer2,4. Therefore, elucidating the mechanisms governing the timing of progenitor cell cycle exit and its connection to differentiation commitment is of fundamental importance.
When individual cells decide to stop proliferating and differentiate is highly variable and thus requires that the process be studied live in single cells5. We had previously developed such a method to investigate the terminal cell differentiation of adipocytes (fat cells)5,6. These studies revealed that preadipocytes commit to differentiate during the G1 phase of the cell cycle, but only if the expression of PPARG, the master transcriptional driver of adipogenesis, surpasses a critical threshold level (“differentiation commitment point”) at which multiple positive feedbacks keep PPARG levels persistently high7. While these studies demonstrated that permanent cell cycle exit and differentiation commitment both occur during G1 phase, it was not known which molecular components control progenitor exit from the cell cycle and, in particular, whether cell cycle exit precedes and is sufficient to trigger differentiation commitment.
In mammals, cell cycle entry and exit are controlled in G1 phase by the activity of two kinases, CDK4/6 and CDK28. However, the timing, mechanisms, and necessity of these kinase activities in controlling cell cycle exit during terminal cell differentiation had not been previously investigated. Moreover, the distinct roles of CDK4/6 and CDK2 in cell cycle entry and exit in other cell types8 raised the question of whether both kinase activities are needed to regulate cell cycle exit during terminal differentiation. Furthermore, genetic deletion and overexpression studies showed that cyclin D1 and the three CDK inhibitors, p21, p27, and p18, which regulate CDK4/6 and CDK2, also have critical roles in regulating adipogenesis and other differentiation processes9,10,11. However, if and how these critical CDK regulators inactivate CDK4/6 and CDK2 during terminal cell differentiation was also not known.
To understand how these critical kinases are regulated, we developed a live, single-cell method based on mosaic reporter transfection to track individual cells while measuring changes in critical signaling activities that govern both proliferation and differentiation. Specifically, we simultaneously measured the activity of the CDK4/6 and CDK2 kinases, which control cell cycle entry, along with the endogenous level of the master transcriptional regulator of adipogenesis, PPARG.
Markedly, we found that during terminal cell differentiation, CDK4/6 and CDK2 were inactivated after each mitosis, and then either jointly increased to start another cell cycle or jointly remained inactive. Using recently developed specific inhibitors for CDK2 and CDK4/612,13, we demonstrated that either CDK4/6 or CDK2 alone could trigger cell cycle entry, but both must be inactivated to increase PPARG levels to the threshold required for differentiation commitment. We identified a sequential order for CDK4/6 and CDK2 inactivation preceding differentiation commitment: the process begins with a decrease in cyclin D1 and an increase in cyclin-CDK inhibitor p27, which together reduce CDK4/6 and CDK2 activity to lengthen G1 phase. The lengthening of G1 phase provides sufficient time for PPARG levels to reach the differentiation commitment threshold and drive expression of the cyclin-CDK inhibitor p21, and later the CDK4/6 inhibitor p18, to permanently suppress CDK2 and CDK4/6 activity. Unexpectedly, inactivation of both CDK4/6 and CDK2 was insufficient for increasing PPARG levels and a subsequent inactivation of ERK kinase was also needed. Together, our study reveals an ordered signaling process that ends a variable cell division period and drives progenitor differentiation based on the inactivation of CDK4/6 and CDK2, followed by the inactivation of ERK.
Results
Either CDK4/6 or CDK2 must be active in G1 to drive adipocyte progenitor proliferation
We investigated the relationship between proliferation and terminal cell differentiation in OP9 preadipocytes in which CRISPR-mediated genome editing had been used to tag endogenous PPARG with mCitrine(YFP) and a FUCCI cell cycle phase reporter (APC/C-mCherry) had been co-expressed5,6 (Fig. 1a–d). First, we confirmed that adding an adipogenic stimulus (DMI) causes PPARG levels to increase, after an initial, variable delay (Fig. 1a, b)5,6. The DMI cocktail consists of the glucocorticoid analog dexamethasone, the phosphodiesterase inhibitor IBMX (which increases cAMP), and insulin.
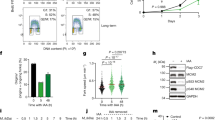
a OP9 cells expressing mCitrine-PPARG and a nuclear marker (H2B-mTurquoise2) were differentiated over 4 days with the standard adipogenic DMI cocktail (Dexamethasone—1 μM, IBMX—250 μM, and Insulin—1.75 nM). Scale bar —50 μm. b Representative single-cell time courses of mCitrine-PPARG from a typical experiment. In the standard DMI-induced differentiation protocol, the DMI cocktail is added to cell culture media for 48 h (horizontal black bar), and the media is replaced after 48 h with fresh media containing just insulin for another 48 h (white horizontal bar). Fifty single-cell time courses are shown as examples. The differentiation commitment threshold is indicated by the dashed line. Cells go on to either differentiate by increasing PPARG levels above the threshold (green time courses) or remain undifferentiated with low PPARG levels (gray time courses). Distribution of PPARG levels at 96 h is shown on the right. Data shown is representative of >2000 cells tracked in the experiment and three biological replicates. c Schematic of APC/C cell cycle reporter construct and dynamics of the reporter’s nuclear fluorescence intensity within a cell during different phases of the cell cycle. d Dual reporter cells expressing mCitrine-PPARG and mCherry-APC/C-reporter were differentiated using the standard DMI protocol. Representative time courses from four single cells. The APC/C-reporter signal is low during the G1 phase, increases during the S/G2 phases, and drops off at mitosis. In differentiating cells, commitment to differentiation (yellow circle) occurs during the G1-phase, whereas undifferentiated cells continue to divide intermittently during the 96 h experimental window (last panel from the right). Data shown are representative of >2000 cells tracked in the experiment and three biological replicates. e CDK4/6 and CDK2 kinase-translocation reporter (KTR) constructs. NLS nuclear localization signal sequence, NES nuclear export signal sequence, S consensus serine phosphorylation site for CDKs. f Representative cells expressing both the CDK4/6 and CDK2 reporters. Increasing CDK activities during the cell cycle leads to the phosphorylation and translocation of the two reporters from the nucleus to the cytosol. Scale bar—10 μm. g Single-cell time courses from two proliferating OP9 progenitor cells expressing CDK4/6 and CDK2 reporters. CDK4/6 and CDK2 activities were measured as the ratio of cytosolic to nuclear signal intensity. ‘M’ marks mitosis events. ‘G1/S’ marks the transition from G1 to S-phase, defined as when CDK2 and CDK4/6 reporter activities reach a value of 0.8, as indicated by the dashed line. h Single-cell time courses of CDK4/6 (top) and CDK2 (bottom) activity from 30 OP9 progenitor cells that undergo mitosis (seen by the drop in CDK activities) within 6 h of starting the imaging. CDK activity bifurcates into high (orange and blue) and low states (gray) after mitosis. Representative of two biological replicates. i–l DMSO (control), CDK4/6 inhibitor (CDK4i, Palbociclib, 1 μM), CDK2 inhibitor (CDK2i, Tagtociclib, 1 μM), or both CDK4/6 and CDK2 inhibitors (Dual CDKi, 1 μM of each) were added to the culture media of proliferating OP9 progenitor cells 24 h after the start of imaging (dashed vertical line). Plots show single-cell time courses from 30 cells that had undergone at least one mitosis event (CDK2 activity >1) since the start of imaging and were in G1-phase when drugs were added. Datapoints represent independent wells from the same experiment, with over 100 cells analyzed per well. m Percent of cells for data represented in i–l that entered S phase, as defined by the CDK2 signal increasing above a value of 0.8 within 16 h of drug addition (see the “Methods” section). One-way ANOVA with Dunnett’s multiple comparisons test, P = 9.16 × 10−8 (Control vs. CDK4/6i), P = 0.0271 (Control vs. CDK2i), P = 4.75 × 10−12 (Control vs. Dual CDKi).
PPARG expression has been shown to be bimodal, meaning cells are either undifferentiated (low PPARG) or differentiated (high PPARG) at 96 h after inducing differentiation7,14 (Fig. 1b). The same studies also showed that the PPARG level in undifferentiated cells must increase to a specific threshold level in order to switch to the differentiated state (Fig. 1b, threshold level is marked with a dotted line). Once this PPARG threshold level is reached, the cell becomes irreversibly differentiated and PPARG stays high. This bistability is a consequence of multiple positive feedback loops, which ensure that PPARG levels remain permanently high, even if the differentiation stimulus is removed7,14. Therefore, we can precisely mark the time when each cell in a population irreversibly commits to differentiation by analyzing when the cell reaches the PPARG threshold5,6,15 (“differentiation commitment point”). Previous studies have shown that at this differentiation commitment point, a cell switches from low to high PPARG expression and simultaneously induces mature adipocyte markers, including adiponectin, FABP4, neutral lipids, and GLUT45,6.
We next made use of the live PPARG level measurements together with the APC/C-mCherry cell cycle reporter. This latter reporter is rapidly degraded at the end of mitosis, stays low in G1 phase, and then increases at the start of the G1/S phase to again gradually increase until mitosis5,16. G1-phase can thus be defined as the low period right after the sharp drop in the reporter fluorescence (Fig. 1c). We confirmed that cells only reached the threshold to commit to differentiation during G1-phase5 (Fig. 1d). Furthermore, we observed that addition of the differentiation stimulus resulted in a variable number of cell divisions that only continued if PPARG levels remained below the threshold5. Upon reaching the threshold, cells ceased cycling and exhibited a further increase in PPARG levels as they differentiated into mature adipocytes (Fig. 1d)5. In contrast, cells that maintained PPARG levels below the threshold continued to divide, indicating their undifferentiated progenitor state and preservation of their proliferative potential5 (Fig. 1d).
To elucidate the mechanisms controlling cell cycle exit during G1 phase, we focused on the two primary kinases regulating this phase: CDK4/6 and CDK28. We engineered a triple-reporter cell line by stably introducing fluorescence reporters for CDK4/6 and CDK2/1 activities into OP9 preadipocyte cells expressing mCitrine(YFP)-PPARG (Fig. 1e). These reporters undergo selective phosphorylation by CDK4/6 and CDK2/1, resulting in increased nuclear export as their respective CDK activities rise during G1 phase17,18 (Fig. 1f). Consequently, CDK4/6 and CDK2/1 activities can be quantified by measuring the ratio of nuclear to cytoplasmic localization. Here, we refer to the CDK2/1 reporter as a CDK2 reporter, as CDK1 is generally inactive during the G1 phase. Finally, we employed mosaic mixtures of OP9 preadipocytes with and without reporters to be able to effectively monitor individual fluorescently labeled preadipocytes over several days (Fig. 1f, also see Supplementary Fig. 1 and see the “Methods” section).
First, we measured CDK4/6 and CDK2 activities in cycling preadipocytes. Previous studies in epithelial cells had shown differences in the time courses of the two kinase activities, with CDK4/6 activity often staying high during and after mitosis despite CDK2 activity being low19. In contrast, we found that in preadipocytes, the CDK4/6 and CDK2 activities were closely correlated (Fig. 1g). Following a drop in both activities after mitosis, both activities then jointly increase after variable delays, or both stay permanently low (Fig. 1g-h). In most cell types, CDK4/6 activity is necessary for proliferation8,20, and addition of the specific CDK4/6 inhibitor Palbociclib12 strongly suppresses proliferation, as marked by the suppression of CDK2 reporter activity19. Here in OP9 preadipocyte cells, we also observed that Palbociclib strongly suppressed CDK4/6 activity. However, in many cells, CDK2 remained active, and proliferation continued (Fig. 1i-j).
We next inhibited CDK2 activity using the recently developed selective small molecule inhibitor of CDK2, Tagtociclib (PF-07104091)13. We observed that cells still entered the cell cycle, but only after a delay, as shown by the slower increase in the CDK2 reporter signal in Tagtociclib-treated cells compared to control cells (Fig. 1l-k). It should be noted that we observed an increase in the CDK2 reporter signal despite the addition of Tagtociclib. This increase in CDK2/1 reporter signal reflects previously identified mechanisms where cells treated with CDK2 inhibitors can use alternate pathways to reactivate CDK2 or by increasing CDK1 activity after a delay, bypassing the initial inhibition of CDK2 activity, to start S-phase21. Consistent with CDK2/1 activation requiring prior CDK4/6 activation, adding Palbociclib and Tagtociclib together completely prevented an increase in the CDK2 reporter signal (Fig. 1l). A quantitative analysis further shows that the percent of cells that enter S phase (i.e., proliferating cells) is completely suppressed only when both CDK4/6 and CDK2 are inhibited (Fig. 1m).
We conclude that the undifferentiated preadipocytes can proliferate by activating either CDK4/6 or CDK2 during the G1 phase. Furthermore, they can only exit the cell cycle if both CDK4/6 and CDK2 are inactivated.
Progenitor cells permanently exit the cell cycle during terminal differentiation by inactivating CDK4/6 and CDK2 in G1 phase
Having established the roles of CDK4/6 and CDK2 in unstimulated progenitor cells, we next measured the activity changes of these kinases in progenitor cells induced to differentiate. Upon adding the DMI adipogenic stimulus to the triple-reporter cells, we observed that CDK4/6 and CDK2 activities both became inactive after each mitosis. Subsequently, they either both increased after variable delays or remained persistently low (Fig. 2a). Notably, when we added either a CDK4/6 or CDK2 inhibitor along with DMI, cells could still increase the CDK2 reporter signal after mitosis, albeit with a delay (Fig. 2b, c). Complete suppression of proliferation during terminal differentiation was only achieved by adding both inhibitors (Fig. 2d, e). Thus, consistent with our findings in unstimulated progenitor cells (Fig. 1l, m), both CDK4/6 and CDK2 must be inactivated to terminate the proliferative period during terminal differentiation (Fig. 2a–e).
a–d DMSO (control), CDK4/6 inhibitor (CDK4/6i, Palbociclib, 1 μM), CDK2 inhibitor (CDK2i, Tagtociclib, 1 μM), or both CDK4/6 and CDK2 inhibitors (Dual CDKi, 1 μM of each) were added into the culture media 24 h after cells were induced to differentiate with DMI (dashed vertical line). Plots show fifty single-cell time courses from cells that had undergone at least one mitosis event (CDK2 activity >1) 12 h prior to and were in G1-phase when drugs were added. e Percent of cells for data represented in a–e in which CDK2 (blue) activities cross a value of 0.8 within 16 h of drug addition, representing cells that would transition from G1 to S-phase. Datapoints represent independent wells from the same experiment, with over 200 single cells analyzed per well. f Single-cell time courses of CDK4/6 (top) and CDK2 (bottom) activities aligned by when CDK4/6 activity reaches its minimum after mitosis (vertical dashed line). Baseline value was subtracted from both CDK4/6 and CDK2 activity values to compare their activation kinetics. g Single-cell time courses of CDK4/6 (top) and CDK2 (bottom) activities aligned by when CDK4/6 activity increases to a value of 0.6 after the drop at mitosis. Baseline value was subtracted from both CDK4/6 and CDK2 activity values to compare their activation kinetics. After baseline correction, CDK activity of 0.6 represents the transition from G1 to S-phase. f, g Are representative of three biological replicates. h Distribution of difference in time for CDK4/6 activity versus CDK2 activity to reach a value of 0.6 in cells after mitosis in the presence of DMI. Blue bars represent cells where CDK2 activity rises before CDK4/6 activity to a value of 0.6, whereas orange bars represent cells where CDK4/6 activity rises earlier than CDK2 activity. f, g Show data from the same experiment. i Progenitor cell divisions are driven by alternate routes through redundant CDK4/6 and CDK2 activation.
When analyzing the relative kinetics of CDK activity during G1 phase, we found that CDK4/6 activity increases slightly before CDK2 activity in most differentiating cells. We demonstrated this delayed activation of CDK2 using two distinct methods: aligning single-cell time courses to the minimum CDK4/6 activity after mitosis (Fig. 2f) or to the timepoint at which CDK4/6 activity reaches a value of 0.6, representing the transition from G1 to S-phase (Fig. 2g). Additionally, histogram analysis confirmed that most cells activate CDK2 only after CDK4/6 is activated (Fig. 2h). Taken together, these results support that progenitor cells first activate CDK4/6, which then boosts CDK2 activity to drive the next cell cycle. Or if CDK2 is inhibited, CDK4/6 boosts CDK1 activity.
Initially, it was puzzling how cells lacking either CDK2 or CDK4/6 activity could enter S phase, as both kinase activities are often considered necessary for triggering cell cycle22. However, gene knockout studies of cyclins and CDKs in mice23,24,25 support that CDK2 and CDK4/6 can, at least in some cells, on their own, phosphorylate RB and activate the cell cycle transcription factor E2F (Fig. 2i). Our data show that both CDK4/6 and CDK2 can be independently activated in the progenitor cells when either is inactive. Furthermore, each can on its own activate E2F in the G1 phase since many cells still enter the cell cycle when either CDK was inhibited (Figs. 1j, k and 2b, c). Progenitors lacking CDK2 activity can still activate CDK2/1 activity after a delay and enter S phase data (Figs. 1k and 2c). This supports a previous interpretation that CDK4/6 activated E2F can synthesize cyclin A, which can then activate CDK1 and/or residual CDK2 to initiate S phase (as diagrammed in Fig. 2i), but only after an apparent delay21,26. CDK2 knockout data also suggest that CDK1 can compensate for CDK2 and CDK2 is not essential in all conditions27. We thus conclude that there are two critical cell cycle redundancies in progenitor cells: progenitors can activate E2F in G1 phase by activating either CDK4/6 or CDK2, and can then enter S phase by either activating CDK2 or CDK1.
PPARG induction requires that CDK4/6 and CDK2 are inactivated in G1 phase
We next used our triple-reporter cells to understand the relationship between CDK4/6 and CDK2 activities and the levels of PPARG during terminal differentiation. Figure 3a shows time courses of PPARG levels and CDK4/6 and CDK2 activities in cells that went on to differentiate in response to an adipogenic stimulus applied for the first 72 h. We found that CDK4/6 and CDK2 were inactivated at ~18–36 h, about the same time as PPARG reached the differentiation commitment threshold (Fig. 3a, dashed line). We note that the differentiation protocol includes withdrawal of DMI after 3 days and replenishment of fresh insulin-containing media, which resulted in a small increase of CDK4/6 and CDK2 activities at 72 h that did not trigger cell cycle entry (Fig. 3a). Such a reversible partial CDK activation without cells starting the cell cycle has previously been characterized in other cells in response to weak mitogenic stimuli26.
a Triple reporter cells expressing endogenous mCitrine-PPARG, CDK4/6 and CDK2 reporters were induced to differentiate with a DMI stimulus for 72 h (black horizontal bar), imaged and tracked over 5 days. Single-cell time courses of mCitrine-PPARG (left), CDK4/6 (middle) and CDK2 (right) activities from cells that increase PPARG levels beyond the commitment threshold. Representative of >10,000 cells tracked in the experiment. b Representative single cell time courses of mCitrine-PPARG (green) and CDK4/6 (Orange), and CDK2 (blue) reporters from differentiating cells. The yellow circle marks the time when cells reach the PPARG threshold (differentiation commitment timepoint). The blue circle marks the end of mitosis, as defined by when the CDK2 activity has dropped by more than half its peak level before mitosis. CDK4/6 activity is also seen to drop closely with CDK2 activity. c Histogram of the time between the end of mitosis (when CDK2 is inactivated) and the differentiation commitment time for all cells that differentiated. d, e 30 single cell time courses of CDK2 activity (top) and mCitrine-PPARG (bottom) from cells that have suppressed (d) or those that continue to increase CDK activity (e) between 12 and 36 h after DMI addition. Dashed line indicates the PPARG commitment threshold. Data for a–e are from the same experiment and are representative of three biological replicates. f Time course of total number of cells that were tracked in control and RB/p21 double knockdown conditions. Plot shows mean (line) ± SEM (shaded regions) from 5 to 6 replicate wells. g mCitrine-PPARG cells were treated with control or RB and p21 siRNA and imaged live during a standard 96 h DMI-induced differentiation protocol. Percent of cells with PPARG levels higher than the different commitment thresholds at 96 h was calculated (see Fig. 1b and “Methods” section) for control and RB/p21 knockdown conditions. Bars show the mean and individual datapoints from 5 to 6 replicate wells in the experiment. Representative of three biological replicates. Unpaired two-tailed t-test used to compare means of control and RB + p21 knockdown conditions, P = 1.66 × 10−9. h Heatmap plots from three hundred control and RB + p21 knockdown cells, showing the times at which cells increase PPARG and commit to differentiation (seen as the deep blue regions on the plots). Representative of three biological replicates. i Time course of mCitrine-PPARG levels and CDK2 activity from triple reporter cells that start with high CDK2 activity (>1) and eventually increase PPARG levels above the commitment threshold from control and RB + p21 knockdown conditions. Lines show the median, and the shaded regions represent 95% confidence intervals around the median. (Ctrl siRNA—680 cells, RB + p21 siRNA—124 cells). Representative of two biological replicates.
Figure 3b shows representative time courses from individual cells in which the drop in CDK activity after mitosis is marked with a blue dot and the PPARG threshold is marked with a yellow dot. These time courses, together with a histogram analysis in Fig. 3c, support that the inactivation of CDK4/6 and CDK2 precedes the increase in PPARG. Nearly all cells increase PPARG after CDK2 inactivation (Fig. 3d), but markedly, the PPARG increase happens with variable delays that can last for days. In cells in which the CDKs were inactivated early after adipogenic stimulation (<12 h, Fig. 3d), PPARG generally increased early, while PPARG stayed low in cells that inactivated CDKs late (Fig. 3e), further suggesting that inactivation of the CDKs is required for PPARG levels to increase.
To determine whether CDK4/6 and CDK2 inactivation is required before cells can increase PPARG and initiate differentiation, we used siRNA to deplete p21 and RB, two critical suppressors of CDK4/6 and CDK28, and thereby force cells to keep proliferating despite the presence of the differentiation stimulus. Indeed, increasing CDK4/6 and CDK2 activities during adipogenesis by depletion of p21 and RB resulted in increased proliferation, as evidenced by the strong increase in total cell numbers (Fig. 3f), while at the same time greatly reduced the fraction of cells that reached the PPARG threshold (Fig. 3g). Moreover, the few cells that eventually committed to differentiate only did so after a long delay (Fig. 3h), corresponding to the time when CDK2 activity started to become inhibited (Fig. 3i).
We conclude that differentiating progenitor cells decide in each G1 phase whether to enter an additional cell cycle by activating CDK4/6 and/or CDK2. However, they only undergo a limited number of divisions (from 1 to about 4, shown earlier in Fig. 1c) before invariably exiting the cell cycle, which requires that they permanently inactivate both CDK4/6 and CDK2 in G1 phase. Furthermore, active CDK4/6 and CDK2 suppress the PPARG increase and must be inactivated for progenitors to increase PPARG and commit to differentiation.
PPARG-induced p21 and p18 enforce the cyclin D1 and p27-regulated CDK4/6 and CDK2 inactivation
Our results, illustrated in Fig. 3a–d, demonstrated that the inactivation of CDK4/6 and CDK2 occurs before cells commit to differentiation. We observed that PPARG levels only rise after cells have remained in G1 phase for an extended period, devoid of CDK4/6 and CDK2 activity. Expression of the CDK inhibitors, p21, p27, and p18, was observed to coincide with growth arrest and differentiation during adipogenesis11. Furthermore, prior studies established that PPARG directly controls p21 and p18 expression transcriptionally5,11, which clarifies how cells can permanently exit the cell cycle once PPARG increases to the critical threshold for differentiation commitment. However, the specific regulators that inactivate CDK4/6 and CDK2 early in terminal differentiation, when PPARG levels are still low and have not yet reached this threshold, remained unclear. To address this, we investigated the changes in cyclin D1 (CCND1), p21, p27, and p18, the four primary regulators of CDK4/6 and CDK2 activity in G1 phase, throughout the time course of adipogenesis. We compared their levels during the G1-phase in individual cells, categorizing them by their PPARG levels (low or high).
When we measured the nuclear levels of cyclin D1 and p27 in individual G1 cells as a function of time, we found a gradual reduction of cyclin D1 and an increase in p27 in both PPARG-high and PPARG-low cells, arguing that the expression of these two regulators is not dependent on PPARG levels (Fig. 4a, b, see also Supplementary Fig. 2). Markedly, cyclin D1 levels decreased and p27 levels increased already at 24 h, before progenitor cells typically increase PPARG levels with the DMI stimulus. The decrease in cyclin D1 can explain the lower CDK4/6 activity, but the increase in p27 is surprising since p27 has been proposed in some studies to activate CDK4/6 activity28. However, one recent study suggests that localization is critical, and the nuclear level of p27 (which we measure here) is primarily inhibiting both cyclin D-CDK4/6 and cyclin E-CDK2 activity29. Thus, the combined decrease in nuclear cyclin D1 and increase in p27 can explain the inhibition of CDK4/6 and CDK2 activity and lengthening of the G1 phase before PPARG increases.
a–d OP9 cells expressing endogenous mCitrine-PPARG were plated across multiple 96-well plates in parallel, stimulated with DMI to induce differentiation and a plate was fixed at each of the indicated timepoints for single-cell immunofluorescence analysis. Immunofluorescence measurements of nuclear cyclin D1 (a), nuclear p27 (b), nuclear p21 (c), and nuclear p18 (d) levels from G1-phase cells that have PPARG levels higher (green) or lower (gray) than the differentiation commitment threshold at every timepoint. Plots show overall means and the individual means of 3 replicate wells in the experiment, wherein at least 1000 cells were analyzed per well. Representative of two biological replicates (also see Supplementary Figs. 2 and 3). e–g Scatter plot of nuclear cyclin D1 (e), p27 (f) and p21 (g) levels versus local cell density from single unstimulated OP9 cells in culture, measured 24 h after they were plated at different densities. Line on each plot shows a non-linear least squares regression fit to the datapoints. Pearson’s correlation co-efficient (‘r’) for the fitted line is indicated on each plot. Representative of two biological replicates. h As proliferating preadipocytes undergo differentiation, the increasing cell density causes cyclin D1 and p21 levels to decrease and p27 levels to increase independently of PPARG expression, thereby suppressing CDK activation across all cells. Additionally, in cells that increase PPARG levels in the presence of an adipogenic stimulus, PPARG drives the expression of p21 and p18 to maintain permanent cell cycle arrest. i Cell density causes a reduction in cyclin D1 and an increase in p27 levels to suppress CDK4/6 and CDK2 activity as progenitors proliferate. Increasing PPARG expression in the differentiation-committed state permanently shuts off CDK4/6 and CDK2 activities through expression of p21 and p18.
Our experiments revealed a clear divergence in p21 and p27 behavior during early adipogenesis. They also highlighted the heterogeneity among cells that commit to differentiation and those that do not. While p27 levels consistently rose in both PPARG-high and PPARG-low cells by 24 h, p21 levels diverged: they decreased in progenitors that did not show an increase in PPARG but rose significantly in PPARG-high cells. This aligns with prior research indicating that PPARG directly induces p21 expression5,11. However, the delayed increase of p21, occurring exclusively in PPARG-high cells, suggests that p21 is not the primary driver for the initial inactivation of CDK4/6 and CDK2 or for stopping the cell cycle early in adipogenesis. This early arrest is crucial to allow PPARG to reach the necessary threshold for differentiation. Furthermore, p18 also showed a significant increase with PPARG, but even later in the adipogenesis timeline (Fig. 4d). This indicates that p18, much like p21, contributes to permanent cell cycle arrest only after PPARG has reached its threshold and differentiation commitment has already taken place.
Our research established that p27 expression during adipogenesis is consistent with its function as the cyclin-dependent kinase (CDK) inhibitor responsible for inactivating CDK4/6 and CDK2 in the initial stages of adipogenesis, prior to the commitment to differentiation. Consequently, we sought to elucidate the mechanism by which p27 levels increase early in the adipogenic process. Studies in other cellular contexts have demonstrated that elevated cell density leads to a reduction in cyclin D1 levels and a corresponding increase in p27 levels29. We investigated whether this phenomenon is also evident during terminal cell differentiation. Such a mechanism appeared plausible given that progenitor cell density has been identified as a critical parameter for enhancing adipogenesis. Indeed, an increase in local progenitor cell density resulted in a reduction of nuclear cyclin D1 and a concomitant increase in p27 levels (Fig. 4e, f). This observation provides a mechanistic explanation for how heightened progenitor cell density during differentiation can lead to the inactivation of CDK4/6 and CDK2. In contrast, p21 levels were observed to decrease rather than increase at higher progenitor density (Fig. 4g). This finding further substantiates the argument that the initial inhibition of CDK4/6 and CDK2 activity during differentiation is primarily mediated by the reduction of cyclin D1 and the elevation of p27, rather than alterations in p21.
We conclude that adipogenic stimulation orchestrates a two-step cell cycle regulation process that promotes terminal differentiation: Initially, adipogenic cues promote progenitor cell expansion, thus causing a cell division-mediated increase in progenitor cell density (Fig. 4h, top box). This increase in cell density progressively reduces nuclear cyclin D1 levels and increases nuclear p27 levels in all the progenitor cells, independently of PPARG expression (Fig. 4h, top box). The cell-density-mediated changes in cyclin D1 and p27 levels lead to the inactivation of CDK4/6 and CDK2, thereby resulting in reversible cell cycle arrest and a lengthening of the G1 phase. Prior studies showed that the proliferation and differentiation processes compete to suppress each other at the end of every mitosis in the early phase of terminal cell differentiation5. For proliferating preadipocytes to commit to differentiation, they must lengthen their G1 phase after mitosis5. This extended G1 period is crucial as it provides sufficient time for PPARG levels to increase to the differentiation commitment point5. Once PPARG reaches this critical threshold, its levels remain irreversibly high due to positive feedback mechanisms7,14. This sustained elevation of PPARG then triggers the permanent expression of p21 and, subsequently, p185 (Fig. 4h, i). These proteins collectively act to permanently suppress CDK4/6 and CDK2 activity, ultimately establishing and maintaining the terminally differentiated state (Fig. 4h, i).
Delayed cell density and DMI-mediated ERK inactivation is also required for PPARG to increase
Since CDK2 and CDK4/6 inactivation is necessary for differentiation commitment, we next tested whether their inactivation is also sufficient to induce differentiation. To do so, we determined whether premature inhibition of CDK4/6 and CDK2 promotes differentiation. Control experiments confirmed that combined CDK4/6 and CDK2 inhibition lowered the percent of cells that proliferated, as measured by the presence of APC/C reporter fluorescence (Fig. 5a, Supplementary Fig. 4a) and by the presence of phosphorylated Rb (Fig. 5b, Supplementary Fig. 4b). Unexpectedly, when we inhibited CDK4/6 and CDK2 (dual CDKi) after 24 h, there was only a small increase in the percentage of differentiated cells (Fig. 5c), indicating that CDK4/6 and CDK2 inactivation is not the main factor restricting differentiation commitment.
a, b Proliferating OP9 preadipocytes expressing APC/C-reporter cell cycle reporter were treated with a combination of Palbociclib and CDK2i (1 μM each) for 24 h, fixed and immunostained for phosphorylated RB protein levels. Plots show the mean and the individual datapoints from 3 to 5 replicate wells in the experiment. Representative of two biological replicates (also see Supplementary Fig. 4). a Percent of proliferating cells (i.e., cells in S/G2/M phases) defined by the number of cells positive for nuclear APC/C reporter fluorescence (also see Fig. 1c and d). b Immunofluorescence staining was used to measure the percentage of cells positive for phosphorylated RB (pRB). c mCitrine-PPARG cells were imaged live during a standard 96 h DMI-induced differentiation protocol, and CDK inhibitors (or vehicle) were added and kept in starting 24 h after the DMI stimulus (experimental scheme, left panel). Dual CDKi refers to co-inhibition with Palbociclib and Tagtociclib, 1 μM each. Percent of cells with PPARG levels higher than the different commitment thresholds at 96 h was calculated (see Fig. 1b and “Methods” section) for control and Dual CDKi-treated conditions (right panel). Plots show the mean and individual datapoints points from 6 replicate wells in the experiment. Representative of three biological replicates. a–c Unpaired two-tailed t-test was used to compare sample means, a ****P = 1.59 × 10−5, b ****P = 7.5 × 10−5, c ****P = 7.75 × 10−8. d Western blot analysis for phospho-ERK (pERK) and total ERK and PPARG levels during adipogenesis. Beta-actin was used as a loading control. Data is representative of two biological replicates. e Scatter plot of pERK levels (nuclear + cytosolic) versus local cell density from single unstimulated OP9 cells in culture, measured 24 h after they were plated at different densities. Line on each plot shows a non-linear least-squares regression fit to the datapoints. Representative of two biological replicates. f Distribution of pERK from single OP9 cells at similar densities (cell density index of 30–45) before, at 24 and 48 h of DMI stimulation. One-way ANOVA with Tukey’s multiple comparisons was used to compare the differences between conditions. ****P = 3.27 × 10−11 (pre-stimulus vs. DMI 24 h), ****P = 3.27 × 10−11 (pre-stimulus vs. DMI 48 h), ****P = 7.58 × 10−6 (DMI 24 h vs. DMI 48 h). More than 500 cells were analyzed per condition. g mCitrine-PPARG cells were imaged live during a standard 96 h DMI-induced differentiation protocol, and MEK inhibitor (PD0325901—100 nM) or vehicle was added and kept for the first 24 h after the DMI stimulus. Percent of cells with PPARG levels higher than the different commitment thresholds at 96 h was calculated (see Fig. 1b and “Methods” section) for control and MEKi-treated conditions. Plots show the mean and individual datapoints points from 4 replicate wells in the experiment. Representative of three biological replicates. Unpaired two-tailed t-test was used to compare differences between sample means. ****P = 1.04 × 10−5 (Control vs. MEKi). h Scheme for the doxycycline-inducible MEK-CA construct (top left). Immunofluorescence images (bottom left) and distribution of HA-tagged MEKCA expression (right) in control and doxycycline-induced cells after 24 h of induction. Scale bar—25 μm. i–k mCitrine-PPARG cells were treated with doxycycline (2 μg/mL) to induce the expression of MEK-CA (or vehicle) during a standard 96 h DMI-induced differentiation protocol, DMI stimulation, and imaged over 96 h. i Representative single cell time courses of PPARG and APC/C cell cycle reporter from control and MEK-CA overexpressing cells. Yellow circle shows the differentiation commitment point. j Percent of cells with PPARG levels higher than the different commitment thresholds at 96 h was calculated (see Fig. 1b and “Methods” section) for control and MEK-CA overexpression conditions. Plot shows means and individual datapoints from 6 replicate wells in the experiment. Representative of two biological replicates. Unpaired two-tailed t-test was used to compare differences between sample means. ****P = 1.36 × 10−11. k MEK-ERK signaling may suppress the rise in PPARG both through promoting CDK activity and by directly phosphorylating and suppressing PPARG transcriptional activity, and preventing any positive feedback on its expression levels. l Besides doxycycline for MEK-CA expression, cells were also treated with vehicle, Dual CDKi (Palbociclib—4 μM + CDK2i—1 μM) or MEKi (PD0325901—100 nM) or Dual CDKi + MEKi. Inhibitors (or vehicle) were added and kept in starting 24 h after the DMI stimulus. Percent of cells with PPARG levels higher than the different commitment thresholds at 96 h was calculated (see Fig. 1b and “Methods” section) for control and inhibitor(s)-treated conditions. Plot shows means and individual datapoints from 6 replicate wells in the experiment. Representative of two biological replicates. One-way ANOVA with Tukey’s multiple comparisons test was used to compare differences between conditions. ****P = 3.81 × 10−13 (Control vs. MEK-CA), ****P = 2.77 × 10-10 (Control vs. MEK-CA + Dual CDKi), ns—not significant, P = 0.2590 (Control vs. MEK-CA + MEKi). m Representative single cell time courses of PPARG and APC/C cell cycle reporter from control and MEK-CA cells treated with Dual CDKi (Palbociclib—4 μM + CDK2i—1 μM) as described for l, showing variability in the timing of commitment to differentiation (yellow circle) after completion of mitosis (dashed line). n Distribution of the time taken after mitosis to undergo differentiation commitment from single differentiating cells for the experiment represented in (m). Horizontal line shows the median of each distribution. Mann–Whitney test used to compare medians of control and MEK-CA + CDKi conditions, ****P < 0.0001 (approximate), control—770 cells, MEK-CA + CDKi—655 cells. Representative of two biological replicates. Example traces in i and l from the same experimental dataset.
We hypothesized that this unknown suppressive factor might be MEK/ERK kinase activity since ERK can phosphorylate PPARG to suppress the transcriptional activity of PPARG30. However, there have been conflicting reports on whether ERK promotes or inhibits adipogenesis29,30,31, leaving its role unresolved. We considered that these contradictory results might be explained by the timing, when MEK/ERK is active during differentiation. We hypothesized that MEK/ERK signaling may be needed early in differentiation to promote proliferation, but must then be inactivated to allow for PPARG to increase the PPARG levels. This dual role seemed plausible, since MEK/ERK activity also promotes proliferation by inducing cyclin D1 expression32. Indeed, ERK activity was progressively reduced during differentiation as evidenced by the reduction of phospho-ERK levels by western blot analysis (Fig. 5d).
We identified two contributing factors that inactivate ERK during adipogenesis: First, the early period of differentiation is paralleled by increasing progenitor density due to the ongoing cell divisions. Analysis of ERK activity as a function of the local progenitor density showed that increasing the local density suppresses ERK activity (Fig. 5e). An analogous cell density-mediated ERK inactivation has been characterized in other cell types29. Second, when we analyzed ERK phosphorylation as a function of time after adipogenic stimulation by selecting progenitors with the same local density, we found that ERK is further inhibited by the adipogenic stimulus independent of local progenitor density (Fig. 5f). This inhibition of ERK can potentially be explained by the upregulation of the ERK phosphatase DUSP1 by glucocorticoids, since a glucocorticoid is part of the differentiation stimulus33. Crucially, when we treated preadipocytes with MEK inhibitor (MEKi) 24 h after DMI stimulus was added, a higher percentage of cells increased their PPARG levels after MEK inhibition (Fig. 5g), demonstrating that ERK inactivation promotes the increase in PPARG and differentiation commitment.
To directly determine whether ERK inactivation is necessary for the PPARG increase and commitment to differentiate, we generated a Doxycycline (Dox)-inducible CA-MEK construct to prolong ERK activity during adipogenesis29. We stably introduced this construct into OP9 preadipocyte cells expressing mCitrine(YFP)-PPARG (Fig. 5h, see also Supplementary Fig. 3). Control experiments showed that the CA-MEK induction prolonged cell proliferation (Fig. 5i).
Markedly, the addition of Dox to induce CA-MEK and persistently activate ERK strongly inhibited the PPARG increase (Fig. 5i, j). Since CDK4/6 activation also inhibits differentiation, this raises the question of whether ERK inhibits PPARG indirectly through activating CDK4/6 (Fig. 5k, pathway 1) or directly through inactivating PPARG (Fig. 5k, pathway 2). To test which of these mechanisms is critical, we inhibited CDK4/6 and CDK2 in CA-MEK expressing cells, which only partially restored the percentage of differentiating cells. However, additional MEK inhibition restored the fraction of differentiation cells seen in the control conditions (Fig. 5l). In further support that ERK activity has a direct repressive role independent of CDK4/6 and CDK2, differentiation commitment in all MEK-CA cells with inhibited CDK4/6 and CDK2 was greatly delayed (Fig. 5m, n).
We conclude that ERK must be inactivated to allow progenitors to increase PPARG levels and commit to differentiation. Inactivation of ERK is redundantly controlled by the adipogenic stimulus and by increasing local progenitor density through the ongoing cell divisions in the proliferative period. ERK activity inhibits differentiation commitment both indirectly by activating CDK4/6 and directly by preventing PPARG levels from increasing.
Sequential inactivation of CDK4/6-CDK2 and ERK separates the reversible proliferative period from the commitment to differentiate
Combined inhibition of CDK2, CDK4/6 and MEK 24 h after adipogenic stimulation had an additive effect compared to the inhibition of the two CDKs or MEK alone (Fig. 6a), suggesting that different cells in the population may be relying on prolonged ERK or CDK activation to suppress PPARG and differentiation. To directly determine whether inactivation of the CDKs or inactivation of ERK is the rate-limiting step for increasing PPARG, we monitored the time course of the PPARG increase after adding MEK or CDK inhibitors 24 hours after the DMI stimulus. Markedly, MEK-inhibited cells rapidly increased PPARG levels compared to control cells (Fig. 6b), while inhibition of the CDKs only increased PPARG after a delay. Moreover, combined CDK and MEK inhibition increased PPARG levels much more rapidly than CDK inhibition alone, implying that inhibition of ERK activity is generally the rate-limiting step acutely increasing PPARG (Fig. 6c). This acceleration of the PPARG increase by MEK inhibition is more clearly shown when comparing the averaged signals in Fig. 6d, e. The accelerated increase in PPARG by MEK inhibition can also be shown in individual cells in a heat map analysis by sorting 500 progenitors by the time when PPARG increases and comparing them under MEK and CDK-inhibited conditions (Fig. 6f, g). We conclude that ERK inactivation is an acute trigger increasing PPARG levels in cells that have already inactivated CDKs.
a mCitrine-PPARG cells were imaged live during a standard 96 h DMI-induced differentiation protocol, and inhibitors, Dual CDKi (Palbociclib—4 μM + CDK2i—1 μM), MEKi (PD0325901—100 nM), Dual CDKi + MEKi or vehicle were added and kept in starting 24 h after the DMI stimulus. Percent of cells with PPARG levels higher than the different commitment threshold at 96 h was calculated (see Fig. 1b and “Methods” section) was calculated for control and inhibitor(s)-treated conditions. Plot shows the means and individual datapoints from 4 replicate wells in the experiment. One-way ANOVA with Dunnett’s multiple comparisons test was used to compare differences between conditions, **P = 0.0085 (Control vs. Dual CDKi), **P = 0.0018 (Control vs. MEKi), ****P = 2.71 × 10−6 (Control vs. Dual CDKi + MEKi). b 50 single cell time courses of mCitrine-PPARG from cells that eventually commit to differentiate across the control and MEKi (b) and, Dual CDKi and Dual CDKi + MEKi (c) treatment conditions. The bold line shows the mean PPARG levels. Representative of >500 cells tracked per condition in the experiment. Shaded regions highlight the differences seen within 24 h of drug addition. d, e Mean mCitrine-PPARG levels for control and MEKi (d) and Dual CDKi and Dual CDKi + MEKi (e) conditions 4 h prior to and for 24 h (shaded regions in b, c) after the drugs were added. f, g Heatmap plots from control and MEKi (f) and Dual CDKi and Dual CDKi + MEKi (g) treatment conditions, showing the times at which cells increase PPARG and commit to differentiation (seen as the deep blue regions on the plots). Data in a–g are from the same experiment and are representative of three biological replicates. h Progenitor cells exposed to differentiation stimulus undergo divisions as long as CDK4/6 and CDK2 activity increase during the G1 phase. Active MEK-ERK signaling in G1-phase drives cyclin D1 expression and CDK activity. ERK also phosphorylates and suppresses the transcriptional activity of any PPARG expressed in cells, preventing any downstream positive feedback that increases PPARG expression. As progenitors proliferate, increasing cell density and reduction in ERK activity cause a decrease in cyclin D levels and an increase in p27 levels to suppress CDK4/6 and CDK2 activation in the G1 phase. This induces a reversible state of cell cycle arrest until further ERK inactivation increases PPARG transcriptional activity, which both induces differentiation commitment and increases p21 and p18 levels to enforce permanent cell cycle arrest. Thus, inactivation of both CDK and ERK activities in the G1-phase is required for cells to commit to differentiate and establish a post-mitotic state. Extending activation of either CDK or ERK signaling delays or prevents differentiation.
Discussion
Our study investigated how cell cycle exit is coordinated with the commitment to terminal differentiation. We employed live single-cell reporter analysis to directly elucidate the relationship between PPARG, the primary transcriptional regulator of adipogenesis commitment, and the two critical G1-phase kinases, CDK4/6 and CDK2, which control cell cycle entry and exit. We demonstrated that redundant activation of CDK4/6 and CDK2 in G1 phase drives the proliferative phase of adipogenesis, allowing for a variable but limited number of cell divisions (Fig. 6h). Following each mitosis, CDK4/6 and CDK2 activities decrease, providing cells the opportunity to decide between proceeding to the next cell cycle or exiting. Only cells that maintain CDK4/6 and CDK2 inactivity for sufficient durations can increase PPARG to the threshold where progenitors commit to terminally differentiate.
We identified two sequential steps leading to the permanent inactivation of CDK4/6 and CDK2 during adipogenesis. Initially, increased local progenitor density, resulting from ongoing proliferation, transiently inhibits CDK4/6 and CDK2 by reducing cyclin D1 and increasing p27 levels. Subsequently, PPARG-mediated induction of p21 and p18 permanently inhibits CDK4/6 and CDK2 after cells commit to differentiation. Surprisingly, inactivation of CDK4/6 and CDK2 alone was insufficient to induce the PPARG increase and commitment to differentiation. Cells could only increase PPARG and commit to differentiation if they also inactivated ERK, which we identified as the rate-limiting step initiating differentiation commitment.
Our study showed that inhibiting CDK2/4/6 or MEK led to only a modest rise in PPARG+ cells (Fig. 5c, g). This implies that ERK and CDK inhibition may not directly induce PPARG expression, but instead creates a permissive state for quiescence, which then facilitates differentiation. This is supported by Fig. 5i, where constitutively active MEK suppresses PPARG expression, likely indirectly by promoting proliferation, since this is a well-established role of MEK-ERK signaling. As shown in Fig. 5l, both MEK-ERK and CDK must be inhibited to create this permissive state, since as long as MEK is active, dual inhibition of CDK2 and CDK4/6 only modestly induces differentiation. Previous studies demonstrated that increased cell density results in both ERK and CDK inhibition29, supporting that the increasing cell density as progenitor cells expand during the early proliferative phase of terminal cell differentiation helps to create this permissive state for differentiation.
Our findings expand upon previous research demonstrating that progenitor cells permanently exit the cell cycle during terminal differentiation and that proliferation and differentiation mutually suppress each other1,2,3. However, due to significant variability in the timing of individual cell cycle exit and differentiation initiation5, a mechanistic understanding of how these processes are coordinated has remained elusive without live-cell analysis. For example, earlier studies provided evidence for a positive regulation of PPARG by Rb-E2F29, but our more detailed timing analysis now shows that CDK4/6 and CDK2 activity, which activate E2F, are suppressing PPARG. Our adipogenesis cell model, incorporating endogenously tagged PPARG and mosaic time-course analysis, enabled us to precisely determine the differentiation commitment point while simultaneously monitoring the activity of both CDK4/6 and CDK2 relative to differentiation commitment during the 4-to-5-day differentiation process.
Previous studies have reported conflicting roles of ERK activity in promoting or suppressing adipogenesis. A study by Farmer’s group concluded that ERK activity is required for adipogenesis, while Spiegelman’s group demonstrated that ERK activity directly phosphorylates PPARG and inhibits its transcriptional activity11,30. It was also plausible that MEK-ERK activity might indirectly suppress PPARG by activating CDK4/6, since MEK-ERK activity has been shown to induce Cyclin D expression32. Furthermore, we show that the delayed inactivation of ERK arises both from the increase in local progenitor density due to the cell division early in adipogenesis and from the adipogenic stimulation itself. ERK inactivation is likely controlled downstream of the adipogenic stimulus by the induction of DUSP phosphatases, which can dephosphorylate ERK and the levels of which have been seen to increase during adipogenesis previously34. Expression of DUSP1 has also been shown to be responsive to glucocorticoids, an important component of the adipogenic stimulus33.
Our study supports that ERK may be both promoting and suppressing adipogenesis, but just at different times in the process: First, ERK activity, by inducing Cyclin D expression32, activates CDK4/6 and drives cell cycle progression early in differentiation. Second, we show that ERK must be inhibited late in differentiation, not only to inhibit CDK4/6 but also to allow PPARG to increase independently of CDK4/6 and CDK2 (summarized in Fig. 6h). Furthermore, we show that the delayed inactivation of ERK arises both from the increase in local progenitor density due to the cell division early in adipogenesis and from the adipogenic stimulation itself. Thus, ERK activity plays a critical role in promoting proliferation early in adipogenesis but must subsequently be inhibited to both inactivate CDK4/6 and allow PPARG to increase.
In line with an early and late role for ERK activity during adipogenesis, ERK has been shown to phosphorylate CEBPB, an early transcriptional driver of PPARG expression35,36,37. CEBPB and PPARG levels are low in undifferentiated progenitor cells38,39. In response to a differentiation stimulus, C/EBPβ levels increase rapidly in all progenitor cells, which in turn drives expression of PPARG38,39. However, live-cell imaging studies showed that while all progenitor cells increase CEBPB expression in response to an adipogenic stimulus, only a fraction of these cells subsequently increase PPARG to the threshold level required for differentiation commitment. These findings collectively suggest that increased C/EBPβ expression and ERK phosphorylation are critical for initiating the differentiation process. However, ERK phosphorylation of CEBPB early in adipogenesis contrasts with ERK phosphorylation of PPARG, which occurs days later during the differentiation commitment phase. Whereas early ERK phosphorylation of CEBPB is important for initiating the differentiation process in all cells6,35,36,37, inhibiting ERK later in adipogenesis is critical to prevent phosphorylation of PPARG, which results in degradation of PPARG30. Only by inhibiting ERK and preventing phosphorylation/degradation of PPARG are preadipocytes able to increase PPARG to the threshold for differentiation commitment. Taken together, these prior studies and our current study highlight the distinct temporal roles of CEBPB and its ERK phosphorylation versus PPARG and its ERK phosphorylation in the initiation and commitment phases of terminal cell differentiation, respectively.
Previous studies have indicated the role of p21, p27, and p57 in suppressing CDK activity during terminal differentiation5,40,41, supporting the role of these cell cycle inhibitors in promoting cell cycle exit across multiple differentiation contexts. Specific to adipogenesis, a prior study demonstrated that knocking out the cell cycle inhibitors p21 and p27 in mice led to a significant increase in the number of differentiated cells (hyperplasia) and resulted in obesity10. Our findings suggest that this observed adipose tissue hyperplasia stems from an extended proliferative phase early in the terminal differentiation process. This prolonged proliferation, occurring due to the absence of p21 and p27, ultimately facilitates the generation of a greater number of adipocytes per progenitor cell. Data available on the Human Protein Atlas database also shows that human adipose tissue samples have been seen to express significant levels of p21 and p18 RNA and protein, further emphasizing the importance of these regulators in maintaining CDK inhibition in the context of adipose tissue differentiation (Supplementary Fig. 7, proteinatlas.org42).
Our core findings that inhibiting CDK4/6, CDK2, and ERK is necessary for PPARG to increase and drive differentiation are supported by independent evidence from in vivo studies, as well as studies using other in vitro models of adipogenesis. In our study, we used the mouse OP9 adipocyte cell model, but studies in the mouse NIH3T3 and 3T3L1 cell models have shown evidence of a transient proliferative period early in adipogenesis that is influenced by CDK2 and ERK signaling30,43. In vivo studies in mice and humans have demonstrated that adipose progenitor cells undergo a transient proliferative period before terminal differentiation39,44 and that increased ERK activation impairs adipogenesis45. Furthermore, adipogenesis has been shown to be a protective mechanism in humans under obesogenic conditions46. By providing new mechanistic insights into how the synergistic inhibition of proliferation and ERK activity allows PPARG to increase and initiate terminal differentiation, our study bridges a critical gap. This expanded context demonstrates that our research on the molecular mechanisms governing these proliferation and differentiation decisions is applicable to diverse in vivo settings, making our findings highly relevant for developing new therapies to control adipogenesis in both mouse and human physiology.
The regulatory mechanisms governing adipogenesis may also explain how proliferation and differentiation are interconnected in other cellular systems, such as neurons and muscle. In these systems, progenitor cells initially undergo expansion through repeated cell divisions before daughter cells exit the cell cycle and terminally differentiate. Similar to PPARG inducing p21 in adipocytes, the main transcriptional drivers of differentiation, NeuroD and myoD, also induce p21 in neurons and muscle, respectively47,48. Given the general role of ERK, CDK4/6, and CDK2 in promoting proliferation, it is plausible that external differentiation stimuli and local cell density generally first activate these proteins to control the length of the proliferative period. This proliferative period may not only facilitate epigenetic changes needed for differentiation, but also regulate the extent of regenerative responses by controlling the number of differentiated cells produced from each progenitor cell.
Together, these results suggest the following framework for terminal cell differentiation: stimulated progenitors start with a variable proliferative period initiated by ERK, CDK4/6 and CDK2 activation, which must subsequently be terminated by reduced nuclear cyclin D1 and elevated p27 levels, leading to the inactivation of CDK4/6 and CDK2. In addition to this transient cell cycle arrest in G1, ERK activity must subsequently be inhibited to trigger differentiation commitment, which ultimately locks cells in permanent cell cycle arrest through the expression of p21, alongside other CDK inhibitors such as p18.
Methods
Cell lines and constructs
The CDK1/2 (pLV-EF1a-DHB-mTurquoise2) and CDK4/6 (pLV-EF1a-mCherry-KTR-RbCterm) reporter constructs were generated and characterized in Spencer et al. 2013 and Yang et al. 2020, respectively17,18. The MEK1-CA construct was generated as described in Fan et al. 202029. Third-generation lentiviral packaging system was used to generate lentiviruses for all the reporter constructs, as well as the fluorescently tagged nuclear reporter H2B constructs (H2B-mTurquoise2 and H2B-iRFP670) and used to infect and stably express these reporters in the OP9 preadipocyte cell line with endogenously tagged citrine-PPARG2. The dual-reporter cell line with endogenously tagged citrine-PPARG2 and stably expressed mCherry-Geminin-degron reporter was generated and characterized in Zhao et al 20205. For all cell lines, either FACS was used to select and enrich for reporter-positive cells or cells were subjected to antibiotic selection post-lentiviral infection.
Cell culture and differentiation
OP9 cell lines were cultured as previously published7,49. Briefly, the cells were cultured in phenol red-containing MEM-α media (Thermo Fisher Scientific) supplemented with 20% FBS, 100 units/mL of Penicillin, 100 µg/mL Streptomycin and 292 µg/mL L-glutamine. Cells were induced to differentiate in the same basal media but supplemented with 10% FBS. In most experiments, a commonly-used DMI protocol was used to induce adipogenesis, in which an adipogenic cocktail (DMI) consisting of dexamethasone (1 µM, Sigma-Aldrich), IBMX (125 µM, Sigma-Aldrich), and insulin (1.75 nM, Sigma-Aldrich) was added to the cell culture media for 48 h, then aspirated away and replaced with fresh media containing 1.75 nM insulin for another 48 h. Whenever experiments were performed with the triple reporter cell line expressing endogenous mCitrine-PPARG, CDK4/6 and CDK2 reporters, DMI-containing media were kept on for 72 h instead of the usual 48 h. This extended stimulation time was required to obtain a larger fraction of cells that go on to increase their PPARG levels beyond the differentiation threshold. For all live-cell microscopy experiments, cells were differentiated in the same media as described above, but without phenol red.
Inhibitors
The final concentrations for the small molecule chemical inhibitors used in this study were: CDK4/6 inhibitor Palbociclib (PD-0332991)—1 or 4 µM as indicated. CDK2 inhibitor Tagtociclib (PF-07104091)—1 µM, MEK inhibitor Mirdametinib (PD-0325901)—100 nM. 4 µM Palbociclib was used in experiments where we drove the expression of constitutively active MEK and, therefore, expected basal CDK4/6 activity to be higher. For drug additions during adipogenesis, 40 μl of serum-free MEM-alpha media containing 5× concentration of the inhibitors was added to individual wells containing cells growing in 160 μl media, 24 h after the DMI stimulus. Equal volumes of DMSO were used as a vehicle control. Thereafter, media containing insulin and inhibitors, or a vehicle, were used to replace DMI-containing media for the last 2 days of the differentiation protocol. All inhibitors were purchased from Selleck Chemicals.
Fluorescent imaging
Cells were plated in full growth medium at a density of 20,000 cells/cm2 (for experiments at low density) 24 h prior to imaging onto either CellVis (P96-1.5H-N) or Ibidi μ-Plates (catalog no. 89626). OP9 cells have a fibroblast-like morphology and show significant migratory behavior in culture, often transiently moving very close to or over neighboring cells. Therefore, for live-cell imaging experiments, to overcome the challenge of reliably identifying and tracking the same cells over several hours to days as they reach very high cell densities during the differentiation protocol, the cell suspension used for plating included a mixture of cells that either stably expressed fluorescently tagged H2B or not such we had a mosaic of labeled and unlabeled cells in every field of view being imaged. Ratio of tagged to untagged cells used was 1:1 to 3:1 (more labeled cells) whenever we expected perturbations to reduce proliferation of cells, and 1:3 (more unlabeled cells) when the perturbations in the experiment were expected to increase proliferation. Before image acquisition, the full growth medium was aspirated and replaced with fresh MEMα without phenol red (R&D Biosystems, M34750) supplemented with 10% FBS containing the adipogenic stimulus wherever indicated. Live-cell imaging was performed on a Nikon ECLIPSE Ti2 inverted microscope using a ×10 or ×20 (for CDK reporter activity assays), Plan Apo 0.45-numerical aperture (NA) objective on a stage-top humidified 37 °C incubator maintained with 5% CO2. Images were acquired every 12–15 min for three or four fluorescent channels as the experiment required (CFP, YFP, RFP and iRFP). Total light exposure time was kept to <800 ms for each time point. Four to nine non-overlapping sites were imaged inside each well.
siRNA-mediated gene silencing
siRNA targeting p21, RB and the Negative Control siRNA were purchased from QIAGEN (FlexiTube siRNA). For siRNA knockdown in the live-cell imaging experiments, OP9 cells were transfected by reverse transfection using Lipofectamine RNAiMax (Invitrogen). Briefly, our reverse-transfection protocol was: 19 µL of OptiMEM was mixed with 0.5 µL of a 10 µM siRNA stock solution, and 0.5 µL of Lipofectamine RNAiMax. Transfection mix was incubated at room temperature for 10 min and 80 µL of culture media was added, which contained the desired number of cells per well. Then the entire (~100 µL) volume was plated into one well of a 96-well plate. The siRNA/RNAiMax mixture was left on the cells for 6–12 h before being aspirated away and replaced with 100 µL fresh culture media. DMI was added 24 h after siRNA transfection. The siRNA sequences are shown in Table 1.
Immunofluorescence (IF) staining
All cultured cells were fixed with 4% PFA in PBS for 15 min at room temperature, followed by three washes with PBS using an automated plate washer (Biotek). Cells were then pretreated with ice-cold methanol for 10 min, washed thrice with PBS and blocked for 1 h in PBS containing 5% FBS and 0.3% Triton X-100. Cells were then incubated overnight with primary antibodies diluted in PBS containing 1% BSA and 0.3% Triton X-100 at 4 °C. Primary antibodies used in this study were: mouse anti-PPARγ (Santa Cruz Biotech, sc-7273, 1:1000), mouse anti-p21 (Santa Cruz Biotech, sc-6246, 1:100), rabbit anti-p21 (Abcam, ab-188224, 1:2000), mouse anti-p27 (Abcam, ab-193379, 1:500), rabbit anti-p27(Abcam, ab-32034, 1:500), cyclin D1 (Abcam, ab-16663, 1:500), rabbit anti-p18 (Abcam, ab192239, 1:500), rabbit anti-HA (Cell Signaling Technology, 3274, 1:1000) and rabbit anti-phospho ERK1/2 (T202/Y204) (Cell Signaling Technology, 4370, 1:250). Following primary antibody incubation, cells were washed with PBS (3×), incubated with Alexa fluor-conjugated anti-rabbit or anti-mouse secondary antibodies (1:1000, Thermo Fisher Scientific) in 1 PBS containing 1% BSA and 0.3% Triton X-100 for 1.5 h, followed by incubation with Hoechst (1:10,000 in PBS containing 1% BSA and 0.3% Triton X-100) for 30 min at room temperature. Cells were then washed three times with PBS using the automated plate washer prior to imaging.
Image data processing and analysis
Data processing and analysis of fluorescent images were performed in MATLAB R2020a (MathWorks). Unless indicated otherwise, fluorescent images were processed, and intensity data were extracted by automated image segmentation, tracking and measurement using custom-written MACKtrack cell tracking software previously described in Kovary et al. 2018 and Zhao et al. 20205,50. PPARG levels were quantified from fixed samples based on median fluorescence signal within the nuclei of individual cells. Cells were scored as ‘PPARG high’ if the marker expression level was above a cut-off determined from the bimodal distribution of PPARG expression at the end of the experiment.
For live imaging data acquired from OP9 cells, the CFP channel capturing H2B-mTurquoise2 or the far-red channel capturing H2B-iRFP670 fluorescence was used for nuclear segmentation and cell tracking. Obtained single-cell traces were filtered to remove incomplete or mis-tracked traces according to the following criteria: cells or their daughter cells that were absent at any point during the entire time lapse experiment, cells that had a large increase or decrease in PPARG intensity normalized to the previous timepoint and cells where there are many and large fluctuations in H2B signal.
The percentage of PPARG high cells was calculated by counting cells in which the PPARG levels were above the threshold (calculated as described in the next section) at that time point as a fraction of the total number of cells being tracked at that time point.
To measure CDK4/6 and CDK2 activities, the median fluorescence in the reporter channel was calculated for both the nuclear and cytosolic regions around the nucleus. Nuclear fluorescence was calculated by directly using the H2B segmentation mask. For cytosolic fluorescence measurement, an annular region of interest around the nucleus was generated by isometrically expanding the nuclear segmentation mask by 4 pixels, deleting the nuclear mask from it and thereafter calculating the median reporter fluorescence within the annulus. Finally, the cellular CDK activity was calculated as the ratio of cytosolic to nuclear reporter fluorescence intensity for each cell at every timepoint.
Previous studies have shown that cells enter S-phase whenever the activity of CDK2, as measured by the respective reporters, reaches a value between 0.8 and 1. We use the CDK2 activity value of 0.8 as a cut-off to identify cells that have entered S-phase17,18,26.
Local cell density measurements were performed by a method similar to that described in Fan et al. 202029. Briefly, a nuclear marker channel (DAPI or H2B) was used to segment the nuclei. Next, a centroid was estimated for every nucleus in the image. All nuclei (i.e., their centroids) that were <~200 μm away from the edge of the image were excluded from further analysis. Finally, the number of neighboring centroids that were within ~100 μm of every nucleus in all directions was counted as a measure of local cell density index.
Estimating a differentiation commitment point (i.e., time at which cells increase PPARG above a threshold)
PPARG values at the end of a differentiation experiment typically exhibit a bimodal distribution when the differentiation stimulus is removed after 2 days5,6,14. Under such conditions, a cut-off PPARG value (commitment threshold) was determined by scanning the entire PPARG time series data to estimate a PPARG value that identified the most number cells that would satisfy all of these three conditions: (1) PPARG levels at the start of the experiment was below threshold, (2) PPARG levels just prior to removal of adipogenic stimulus was above the threshold and (3) PPARG at the end of the experiment (96 or 120 h) was above the threshold. Differentiation commitment time was then estimated for every cell that retained PPARG levels at or higher than this threshold at the end of the experiment by identifying when PPARG levels first crossed this threshold during the experiment.
Western blotting
OP9 cells were plated in parallel on multiple 60 mm cell culture dishes in growth media at a density of ~20,000 cells/cm2 and stimulated with an adipogenic stimulus starting 48 h after plating. At each time point indicated, cells on a single plate were washed twice with ice-cold PBS, scraped off and lysed in ice-cold RIPA lysis buffer (Millipore Sigma, 20-188) containing a cocktail of protease and phosphatase inhibitors (Thermo Fisher Scientific, 78444). The lysates were incubated on ice for 20 min and spun down at 13,000×g for 10 min. The supernatant was collected, and total protein amounts for each sample were determined using 5 µL with a standard Bradford assay. 35–40 µg protein was used per sample for SDS-PAGE and western blotting. Blots were blocked in TBS containing 0.1% Tween-20 (TBST) and 10% dry milk and incubated overnight with primary antibodies diluted in TBST with 5% BSA. The following primary antibodies were used: rabbit anti-PPARG (Cell Signaling Technology, 2443, 1:1000), rabbit anti-phospho ERK1/2 (T202/Y204) (Cell Signaling Technology, 4370, 1:2000), mouse anti-ERK1/2 (Cell Signaling Technology, 4696, 1:2000) and mouse HRP-tagged anti-beta-actin (Santa Cruz technology, sc-47778, 1:4000). Following primary antibody incubation, blots were washed (3×) in TBST for a total of 30 min and incubated with fluorescently labeled antibodies diluted in TBST with 5% BSA, for 1.5 h in the dark at room temperature. Secondary antibodies used were as follows: Alexa Fluor goat anti-rabbit 680 (Thermo Fisher Scientific, 1:10,000), IRDye 800 CW goat anti-mouse (LI-COR Biosciences, 1:10,000) and HRP-tagged goat anti-mouse (Cell Signaling Technology, 1:2000). Following secondary antibody incubation, blots were washed (3×) for a total of 20 min and imaged on a LI-COR Odyssey fluorescence and chemiluminescence imager.
Statistics
In each experiment, 2 or more replicate wells were included for every treatment condition. Experiments were also repeated across different days, indicated as biological replicates in the figure legends. Statistical tests used for comparisons are also indicated in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
References
Hardwick, L. J. A., Ali, F. R., Azzarelli, R. & Philpott, A. Cell cycle regulation of proliferation versus differentiation in the central nervous system. Cell Tissue Res. 359, 187–200 (2015).
Ruijtenberg, S. & van den Heuvel, S. Coordinating cell proliferation and differentiation: antagonism between cell cycle regulators and cell type-specific gene expression. Cell Cycle 15, 196–212 (2016).
Buttitta, L. A. & Edgar, B. A. Mechanisms controlling cell cycle exit upon terminal differentiation. Curr. Opin. Cell Biol. 19, 697–704 (2007).
Ghaben, A. L. & Scherer, P. E. Adipogenesis and metabolic health. Nat. Rev. Mol. Cell Biol. 20, 242–258 (2019).
Zhao, M. L. et al. Molecular competition in G1 controls when cells simultaneously commit to terminally differentiate and exit the cell cycle. Cell Rep. 31, 107769 (2020).
Bahrami-Nejad, Z. et al. A transcriptional circuit filters oscillating circadian hormonal inputs to regulate fat cell differentiation. Cell Metab. 27, 854–868.e8 (2018).
Ahrends, R. et al. Controlling low rates of cell differentiation through noise and ultrahigh feedback. Science 344, 1384–1389 (2014).
Fassl, A., Geng, Y. & Sicinski, P. CDK4 and CDK6 kinases: from basic science to cancer therapy. Science 375, eabc1495 (2022).
Tokumoto, Y. M., Apperly, J. A., Gao, F.-B. & Raff, M. C. Posttranscriptional regulation of p18 and p27 Cdk inhibitor proteins and the timing of oligodendrocyte differentiation. Dev. Biol. 245, 224–234 (2002).
Naaz, A. et al. Loss of cyclin-dependent kinase inhibitors produces adipocyte hyperplasia and obesity. FASEB J. 18, 1925–1927 (2004).
Morrison, R. F. & Farmer, S. R. Role of PPARgamma in regulating a cascade expression of cyclin-dependent kinase inhibitors, p18(INK4c) and p21(Waf1/Cip1), during adipogenesis. J. Biol. Chem. 274, 17088–17097 (1999).
Finn, R. S. et al. Palbociclib and letrozole in advanced breast cancer. N. Engl. J. Med. 375, 1925–1936 (2016).
Yap, T. A. et al. First-in-human phase 1/2a study of a potent and novel CDK2-selective inhibitor PF-07104091 in patients (pts) with advanced solid tumors, enriched for CDK4/6 inhibitor resistant HR+/HER2− breast cancer. J. Clin. Oncol. 41, 3010–3010 (2023).
Park, B. O., Ahrends, R. & Teruel, M. N. Consecutive positive feedback loops create a bistable switch that controls preadipocyte-to-adipocyte conversion. Cell Rep. 2, 976–990 (2012).
Zhang, Z.-B., Sinha, J., Bahrami-Nejad, Z. & Teruel, M. N. The circadian clock mediates daily bursts of cell differentiation by periodically restricting cell-differentiation commitment. Proc. Natl Acad. Sci. USA 119, e2204470119 (2022).
Sakaue-Sawano, A. et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498 (2008).
Spencer, S. L. et al. The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell 155, 369–383 (2013).
Yang, H. W. et al. Stress-mediated exit to quiescence restricted by increasing persistence in CDK4/6 activation. eLife 9, e44571 (2020).
Chung, M. et al. Transient hysteresis in CDK4/6 activity underlies passage of the restriction point in G1. Mol. Cell 76, 562–573.e4 (2019).
Liu, C. et al. Altered G1 signaling order and commitment point in cells proliferating without CDK4/6 activity. Nat. Commun. 11, 5305 (2020).
Arora, M. et al. Rapid adaptation to CDK2 inhibition exposes intrinsic cell-cycle plasticity. Cell 186, 2628–2643.e21 (2023).
Harbour, J. W., Luo, R. X., Santi, A. D., Postigo, A. A. & Dean, D. C. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell 98, 859–869 (1999).
Malumbres, M. et al. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell 118, 493–504 (2004).
Kozar, K. et al. Mouse development and cell proliferation in the absence of D-cyclins. Cell 118, 477–491 (2004).
Berthet, C., Aleem, E., Coppola, V., Tessarollo, L. & Kaldis, P. Cdk2 knockout mice are viable. Curr. Biol. 13, 1775–1785 (2003).
Konagaya, Y., Rosenthal, D., Ratnayeke, N., Fan, Y. & Meyer, T. An intermediate Rb–E2F activity state safeguards proliferation commitment. Nature 631, 424–431 (2024).
Suski, J. M. et al. CDC7-independent G1/S transition revealed by targeted protein degradation. Nature 605, 357–365 (2022).
Guiley, K. Z. et al. p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition. Science 366, eaaw2106 (2019).
Fan, Y. & Meyer, T. Molecular control of cell density-mediated exit to quiescence. Cell Rep. 36, 109436 (2021).
Hu, E., Kim, J. B., Sarraf, P. & Spiegelman, B. M. Inhibition of adipogenesis through MAP kinase-mediated phosphorylation of PPARG. Science 274, 2100–2103 (1996).
Wiesner, M., Berberich, O., Hoefner, C. & Blunk, T. & Bauer-Kreisel, P. Gap junctional intercellular communication in adipose-derived stromal/stem cells is cell density-dependent and positively impacts adipogenic differentiation. J. Cell. Physiol. 233, 3315–3329 (2018).
Lavoie, J. N., L’Allemain, G., Brunet, A., Müller, R. & Pouysségur, J. Cyclin D1 expression is regulated positively by the p42/p44 and negatively by the p38/HOG pathway. J. Biol. Chem. 271, 20608–20616 (1996).
Kassel, O. Glucocorticoids inhibit MAP kinase via increased expression and decreased degradation of MKP-1. EMBO J. 20, 7108–7116 (2001).
Ferguson, B. S., Nam, H., Stephens, J. M. & Morrison, R. F. Mitogen-dependent regulation of DUSP1 governs ERK and p38 signaling during early 3T3-L1 adipocyte differentiation: DUSP1 regulates MAPKs in early adipogenesis. J. Cell. Physiol. 231, 1562–1574 (2016).
Hamm, J. K., Park, B. H. & Farmer, S. R. A role for C/EBPβ in regulating peroxisome proliferator-activated receptor γ activity during adipogenesis in 3T3-L1 preadipocytes. J. Biol. Chem. 276, 18464–18471 (2001).
Park, B.-H., Qiang, L. & Farmer, S. R. Phosphorylation of C/EBPβ at a consensus extracellular signal-regulated kinase/glycogen synthase kinase 3 site is required for the induction of adiponectin gene expression during the differentiation of mouse fibroblasts into adipocytes. Mol. Cell. Biol. 24, 8671–8680 (2004).
Prusty, D., Park, B.-H., Davis, K. E. & Farmer, S. R. Activation of MEK/ERK signaling promotes adipogenesis by enhancing peroxisome proliferator-activated receptor γ (PPARγ) and C/EBPα gene expression during the differentiation of 3T3-L1 preadipocytes. J. Biol. Chem. 277, 46226–46232 (2002).
Farmer, S. R. Transcriptional control of adipocyte formation. Cell Metab. 4, 263–273 (2006).
Rosen, E. D. & Spiegelman, B. M. What we talk about when we talk about fat. Cell 156, 20–44 (2014).
Zhang, M., Kim, S. & Yang, H. W. Non-canonical pathway for Rb inactivation and external signaling coordinate cell-cycle entry without CDK4/6 activity. Nat. Commun. 14, 7847 (2023).
Bahrami, A. R., Matin, M. M. & Andrews, P. W. The CDK inhibitor p27 enhances neural differentiation in pluripotent NTERA2 human EC cells, but does not permit differentiation of 2102Ep nullipotent human EC cells. Mech. Dev. 122, 1034–1042 (2005).
Uhlén, M. et al. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Tang, Q.-Q., Otto, T. C. & Lane, M. D. Mitotic clonal expansion: a synchronous process required for adipogenesis. Proc. Natl. Acad. Sci. USA 100, 44–49 (2003).
Jeffery, E., Church, C. D., Holtrup, B., Colman, L. & Rodeheffer, M. S. Rapid depot-specific activation of adipocyte precursor cells at the onset of obesity. Nat. Cell Biol. 17, 376–385 (2015).
Hosooka, T. et al. Dok1 mediates high-fat diet-induced adipocyte hypertrophy and obesity through modulation of PPAR-γ phosphorylation. Nat. Med. 14, 188–193 (2008).
Hammarstedt, A., Gogg, S., Hedjazifar, S., Nerstedt, A. & Smith, U. Impaired adipogenesis and dysfunctional adipose tissue in human hypertrophic obesity. Physiol. Rev. 98, 1911–1941 (2018).
Liu, Y., Encinas, M., Comella, J. X., Aldea, M. & Gallego, C. Basic helix-loop-helix proteins bind to TrkB and p21Cip1 promoters linking differentiation and cell cycle arrest in neuroblastoma cells. Mol. Cell. Biol. 24, 2662–2672 (2004).
Halevy, O. et al. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science 267, 1018–1021 (1995).
Wolins, N. E. et al. OP9 mouse stromal cells rapidly differentiate into adipocytes: characterization of a useful new model of adipogenesis. J. Lipid Res. 47, 450–460 (2006).
Kovary, K. M., Taylor, B., Zhao, M. L. & Teruel, M. N. Expression variation and covariation impair analog and enable binary signaling control. Mol. Syst. Biol. 14, e7997 (2018).
Acknowledgements
This work was supported by NIH RO1-DK131432-01A1 (M.N.T.) and startup funds from the Drukier Institute and Weill Cornell Medical School (M.N.T.). We thank all members of the Teruel lab and Meyer lab for helpful discussions.
Author information
Authors and Affiliations
Contributions
S.S. and M.N.T. designed the study; S.S. and H.E.B. performed experiments and collected and analyzed data. T.M. gave conceptual advice. M.N.T. provided supervision and funding acquisition. S.S. and M.N.T. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Stephen Farmer and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Kaliya Georgieva. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sharma, S., Berger, H., Meyer, T. et al. Inactivation of CDK4/6, CDK2, and ERK in G1-phase triggers differentiation commitment. Commun Biol 8, 1727 (2025). https://doi.org/10.1038/s42003-025-09038-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-09038-z