Abstract
Phosphatidylinositol-3-phosphate (PI3P) is the key mediator of resistance to artemisinin drugs (ARTs) and that phosphatidylinositol 3-kinase (PfPI3K) is a promising target for eliminating ARTs resistance. However, few studies investigated the PfPI3K inhibitors. Here we show combination treatment with a potential PfPI3K inhibitor (idelalisib, which effectively inhibit PfPI3K activity resulting the decline in PI3P level) and dihydroartemisinin (DHA) exerts synergistic antimalarial effects and reverse ARTs resistance. The decline in PI3P level inhibits reactive oxygen species (ROS) degradation (resulting in ROS accumulation, apoptosis of parasites and the accumulation of ubiquitinated proteins), downregulates the gene expression of molecular chaperones and increases eIF2α phosphorylation, which finally leads to a dysregulated antioxidant system and disrupted proteostasis in parasites, thereby increasing the sensitivity of the parasites to ARTs and reversing ARTs resistance. Our results indicated that targeting PfPI3K with idelalisib was a successful antimalarial strategy for further controlling the development of ART resistance.
Similar content being viewed by others
Introduction
Malaria is an infectious disease that is transmitted through the bite of a female Anopheles mosquitoes infected with Plasmodium parasites that seriously threatens human life and health. Nearly half of the world’s population is at risk of contracting malaria, with 263 million cases and 597,000 deaths in 2024, according to the World Health Organization (WHO)1. Malaria caused by Plasmodium falciparum (P. falciparum), the most dangerous species, accounts for ~98% of malaria cases. The persistence of the P. falciparum parasite has resulted in a global malaria disease burden. Currently, the treatment of P. falciparum malaria relies heavily on artemisinin drugs (ARTs), including dihydroartemisinin (DHA), artemether, artesunate, etc., and DHA is the active metabolite of these drugs2,3. ARTs have strong antimalarial activity, fast-acting effects, low toxicity and no cross-resistance with most other antimalarial drugs. Therefore, the WHO recommends the use of artemisinin-based combination therapies (ACTs) as first-line treatments for uncomplicated P. falciparum malaria in endemic countries worldwide, both to improve patient outcomes and delay the onset of resistance. Encouragingly, remarkable reductions (37% from 2000 to 2015) of the global burden have been achieved, and countless lives have been saved because of the wide use of ACTs4,5. However, parasites with partial resistance to ARTs have emerged6. Thus, the rate of reduction in the global burden has slowed since 20157, inevitably leading to clinical treatment failure (~50% failure to cure in some regions)8. The spread of ART-resistant parasites (first in southeast Asia and recently in East Africa, mainly Uganda and Rwanda) has disastrous consequences for global malaria control9,10. This threatens the progress made in malaria control and derails the malaria elimination campaign, emphasizing the necessity for new drugs that can be used in combination therapies11,12. To date, a mutation in the parasite kelch13 gene (Pfkelch13, PF3D7_1343700, encoding the PfKelch13 protein) has been identified as the primary and definite marker of ART resistance13. PfKelch13 mutations in P. falciparum reduce the binding of PfKelch13 and phosphatidylinositol 3-kinase (PfPI3K) and thereby inhibiting the polyubiquitination of PfPI3K, disrupting the proteolysis of PfPI3K, and increasing the levels of the kinase, as well as its lipid product phosphatidylinositol-3-phosphate (PI3P). Moreover, increased PI3P levels alone efficiently confers artemisinin (ART) resistance, and there is a positive correlation between the PI3P level and ART resistance13. These findings indicate that PI3P is the key mediator of ART resistance and that PfPI3K is a promising target for eliminating ART resistance. Therefore, screening for high-efficiency PfPI3K inhibitors to be used in combination with DHA for the treatment of ART-resistant malaria is urgently needed. However, few studies have investigated PfPI3K inhibitors, since the three-dimensional structure of PfPI3K has not been fully resolved. Fortunately, a previous study revealed that PfPI3K and human Class III PI3K are highly homologous (67% identity in the catalytic structural domain and 44% identity in the helical domain)14, and existing broad-spectrum PI3K inhibitors (wortmannin and LY294002) were found to reduce the PI3P level in parasite13. Therefore, we believe that some PI3K inhibitors can also inhibit PfPI3K activity, thus serving as candidates for screening high-efficiency PfPI3K inhibitors. Our previous research has revealed five potential PfPI3K inhibitors among five marketed PI3K inhibitors and twenty-two compounds with human Class III PI3K protein (VPS34) inhibitory functions via molecular docking and molecular dynamics simulations (which will soon be published by co-investigator, Rongrong Wang). The antimalarial effects of the five potential PfPI3K inhibitors were also confirmed via in vivo pharmacodynamic evaluation. Overall, the combination of ARTs and PfPI3K inhibitors is expected to combat ART-resistant parasites and may be a novel strategy for treating malaria caused by ART-resistant parasites.
Therefore, on the basis of this idea, we used DHA (common active metabolites of all major ARTs) as a model drug and constructed Pfkelch13 mutation- and PI3P-overexpressing transgenic parasites on the P. falciparum 3D7 genetic background. Taking in vitro antimalaria activity and the synergistic antimalarial effects of DHA and PI3P level as evaluation indicators, we first screened and confirmed a potential PfPI3K inhibitor, which significantly inhibited PfPI3K activity and reduced the PI3P level, increases sensitivity to ARTs and reverses ART resistance. Then, the further mechanisms were also investigated. Overall, we provide critical drug candidates for combination treatment with ARTs. We confirmed that this combination strategy could reverse ARTs resistance in ART-resistant P. falciparum. This combination treatment can combat ART resistance and contribute to the elimination of malaria.
Results
The transgenic 3D7-PfKelch13C580Y and 3D7VPS34 P. falciparum strains were resistant to DHA
According to previous studies, the C580Y is recognized as the most prevalent mutation in ART-resistant populations15. Therefore, we produced the pSLI-N-sandwich-loxP (K13C580Y) plasmid containing the C580Y mutation site. Through sequencing alignment, the mutation site was successfully introduced (Supplementary Fig. 1A). To increase PI3P levels in the absence of the PfKelch13 mutation, we constructed the plasmid pVps34(human)-mCherry, which contained human VPS34 and synthesized only PI3P. The successful insertion was verified by plasmid gene sequencing. Therefore, we transfected 3D7 with three plasmids to obtain wild-type kelch13 parasites (3D7-PfKelch13WT), kelch13 mutant parasites (3D7-PfKelch13C580Y) and PI3P-overexpression parasites (3D7VPS34). After 5–7 days of drug selection, all the parasites died. After electroporation, 3D7-PfKelch13WT, 3D7-PfKelch13C580Y, and 3D7VPS34 were observed via microscopy at day 22, day 35 and day 30, respectively.
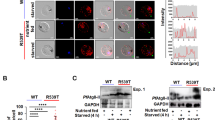
We confirmed the successful integration of GFP by integration-specific PCR (Supplementary Fig. 1B). In a Western blot, the anti-GFP antibody detected a major protein band of 165 kDa (Fig. 1A), which agreed with the predicted size of PfKelch13 fused with two FK506-binding protein (FKBP) domains and GFP. The anti-mCherry antibody detected a protein band of 130 kDa, which agreed with the predicted size of VPS34 fused with mCherry (Fig. 1B). The confocal images of 3D7-PfKelch13WT, 3D7-PfKelch13C580Y and 3D7VPS34 also indicated the successful construction of the transgenic strains (Fig. 1C).
The 20 µg of parasite protein samples were separated and transferred, which were then immunoblotted with anti-GFP antibody (A) and anti-mCherry antibody (B). A An anti-GFP antibody detected a major protein band of 165 kDa, which agreed with the predicted size of PfKelch13 fused with two FK506-binding protein (FKBP) domains and GFP. B An anti-mCherry antibody detected a protein band of 130 kDa, which agreed with the predicted size of VPS34 fused with mCherry. C Confocal images of untagged 3D7 (control), 3D7-PfKelch13WT, 3D7-PfKelch13C580Y, and 3D7VPS34. Tagged exogenous 3D7-PfKelch13WT and 3D7-PfKelch13C580Y are evident in the GFP panel, and VPS34 is evident in the red panel, while the nuclei are stained with DAPI, the parasites were visualized via brightfield microscopy, and images were merged for reference. D In vitro antimalarial activity of DHA against 3D7-PfKelch13WT, 3D7-PfKelch13C580Y, and 3D7VPS34. The IC50 values were calculated by GraphPad Prism 9.0 software (Analyse-Nonlinear regression curve fit-[Inhibitor] vs. normalized response—variable slope). The definitions of the maximum and minimum response values were 100% and 0%, respectively for the nonlinear regression curve fit. E The PI3P levels and the RSA values of 3D7 (control), 3D7-PfKelch13WT, 3D7-PfKelch13C580Y, and 3D7VPS34. For the PI3P level measurement, the parasites were not treated with any drugs. When the parasites reached the trophozoite stage, the red blood cells were lysed with saponin, and the lipids within parasites were extracted for PI3P detection. For the RSA value measurement, tightly synchronized early-ring parasites (0–3 h post-invasion, 2% hematocrit and 1% parasitemia) were exposed to 700 nM DHA combined (0.1% DMSO as control) for 6 h, washed thrice with Albumax-free RPMI and returned to culture for 66 h. Parasite survival rates (%RSAs) were determined by comparing the number of viable parasites between the drug-treated and untreated control and are expressed as percentages. Two-tailed unpaired Student’s t-test was employed. The mean (s.d.) are based on three replicates (the triplicate data points are shown). PI3P: **p < 0.01, ***p ≤ 0.001; RSA: ###p ≤ 0.001.
The Ring Stage Survival Assay (RSA) serves as a key in vitro indicator for determining the resistance of P. falciparum to ARTs. Meanwhile, given the established positive correlation between PI3P levels and ART resistance, we also quantified PI3P levels in these strains. In this study, the in vitro antimalarial activity (IC50) of DHA against 3D7, 3D7-PfKelch13WT, 3D7-PfKelch13C580Y, and 3D7VPS34 were 4.80 nM, 4.92 nM, 36.89 nM and 86.04 nM, respectively (Fig. 1D). The DHA IC50 value, RSA value and PI3P level of 3D7-PfKelch13WT was similar to 3D7 (control) (Fig. 1E). The RSA value of 3D7VPS34 (10.77 ± 0.77) % was significantly greater than that of 3D7-PfKelch13C580Y (5.79 ± 0.60) % (p = 0.001), and the RSA value of both strains was greater than 1.5%, which indicated that the introduction of the C580Y mutation or the insertion of human VPS34 led to parasite resistance to DHA. The PI3P level of 3D7VPS34 (52.74 ± 0.47) pmoles per 1 × 108 iRBCs was significantly greater than that of 3D7-PfKelch13C580Y (42.69 ± 0.76) pmoles per 1 × 108 iRBCs (p = 0.004), coinciding with the RSA results.
Screening of PfPI3K inhibitors by evaluating their in vitro individual antimalarial activity and synergistic antimalarial effects with DHA
Confirming that elevated PI3P levels were associated with DHA resistance, we screened for potent PfPI3K inhibitors that can efficiently inhibit the production of PI3P. Since the structure of PfPI3K has not been clearly resolved and researchers have also failed to elucidate the full-length PfPI3K protein14, further studies of PfPI3K inhibitors are difficult. Therefore, we have previously screened out five potential PfPI3K inhibitors through homology modeling, molecular dynamics simulation and in vivo pharmacodynamics (Supplementary Tables 1 and 2). The five potential PfPI3K inhibitors were idelalisib, umbralisib, taselisib, AZD6482, and copanlisib, meanwhile, the chemical structures were shown in Supplementary Fig. 2.
In this study, the IC50 values of idelalisib, umbralisib, taselisib, AZD6482 and copanlisib were 2.49 μM, 1.92 μM, 2.25 μM, 6.68 μM and 94.72 nM, respectively (Fig. 2A). This indicated that all the five inhibitors have a certain degree of killing activity against P. falciparum. The IC50 of copanlisib (94.72 nM) was the lowest among the five candidates, which indicated that copanlisib seemed to be more active against P. falciparum in vitro. However, the copanlisib has poor solubility and needs to be dissolved in hydrochloric acid.
The in vitro individual antimalarial activity was determined by standard 72-h susceptibility assays by the measurement of SYBR Green I fluorescence. Synchronized 2- to 4 h post-invasion ring-stage parasites were exposed to a serial dilution of idelalisib (A, ⅰ), umbralisib (A, ⅱ), taselisib (A, iii), AZD6482 (A, ⅳ) and copanlisib (A, ⅴ). The means (s.d.) were based on four replicates. The IC50 values were calculated by GraphPad Prism 9.0 software (Analyse-Nonlinear regression curve fit-[Inhibitor] vs. normalized response—variable slope). The definitions of the maximum and minimum response values were 100 and 0%, respectively, for the nonlinear regression curve fit. The antimalarial activity of the combination of DHA and PI3K inhibitors was determined by two coculture methods: (1) The synchronized 2- to 4 h post-invasion ring-stage parasites were cocultured with DHA at the IC50 and gradient concentration (from 1/4 × IC50 to 4 × IC50) of idelalisib (B, ⅰ), umbralisib (B, ⅱ), taselisib (B, ⅲ), AZD6482 (B, ⅳ) and copanlisib (B, ⅴ); 2) The synchronized 2- to 4 h post-invasion ring-stage parasites were cocultured with gradient concentrations of DHA (from 1/4 × IC50 to 4 × IC50) and with or without idelalisib (C, ⅰ), umbralisib (C, ⅱ), taselisib (C, ⅲ), AZD6482 (C, ⅳ) or copanlisib (C, ⅴ) at their IC50 values for 24 h. After treatment with the two compounds, the culture medium was replaced, and the parasite cultures were further incubated for another 48 h. The parasitemia rates (survival rates) were monitored by counting infected erythrocytes in Giemsa-stained thin blood smears using light microscopy. In B, the relative survival rate of no drug exposure was defined as 100% (first column), and the values of the other columns (parasitemia) were compared with the mean value of the first column. In C, the relative survival rate of no drug exposure was defined as 100% (first column), and the value of the other column (parasitemia) were compared with the mean value of the first column. The means (s.d.) are based on three replicates (the triplicate data points are shown). One-way ANOVA and LSD-T post hoc test comparisons between treatment groups were conducted. *p < 0.05, **p < 0.01, ***p ≤ 0.001.
A previous study revealed synergy between DHA and the PI3K inhibitor NVP-BGT22616. This confirms the feasibility of the combined use of the two drugs in enhancing the antimalarial effect. In this study, the five inhibitors exhibited substantially weaker antimalarial activity compared to DHA (IC50 of DHA: 4.80 nM). Consequently, in combination treatments, the antimalarial efficacy primarily relied on DHA. Building on the prior evidence of drug synergy, we further evaluated the impact of these five inhibitors on DHA’s antimalarial activity with two approaches. First, the ring-stage parasites were cocultured with DHA at the IC50 values and gradient concentrations (from 1/4 × IC50 to 4 × IC50) of idelalisib (Fig. 2B, i), umbralisib (Fig. 2B, ⅱ), taselisib (Fig. 2B, ⅲ), AZD6482 (Fig. 2B, ⅳ), and copanlisib (Fig. 2B, ⅴ). The results revealed that various concentrations of idelalisib, umbralisib and taselisib decreased the survival rates of P. falciparum 3D7. With increasing concentrations of the inhibitors, the killing effect of DHA on P. falciparum 3D7 became more obvious. In particular, idelalisib helped reduce the survival rate by approximate 50%, even at the 1/4 × IC50 concentration (Fig. 2B, ⅰ). Second, the ring-stage parasites were cocultured with gradient concentrations of DHA and idelalisib (Fig. 2C, i), umbralisib (Fig. 2C, ⅱ), taselisib (Fig. 2C, iii), AZD6482 (Fig. 2C, iv), or copanlisib (Fig. 2C, v) at their IC50 values, respectively. The results indicated that the combination of DHA and idelalisib decreased the survival rate of 3D7 at each concentration combination, especially when the concentration of DHA was less than or equal to the IC50 value (p < 0.05) (Fig. 2C, i). For the other four inhibitors, not all concentration combinations further reduced the parasitemia (Fig. 2C, ii–v). Although idelalisib has similar in vitro antimalarial activity to umbralisib and taselisib, its combination with DHA at various concentrations resulted in more parasites death than did DHA treatment alone. Therefore, we chose idelalisib for the following experiments.
Idelalisib decreased PI3P levels and reversed ART resistance by inhibiting PfPI3K activity
In this study, we determined the PfPI3K activity, PI3P level, RSA level and IC50 value to explore the effects of idelalisib combined with DHA. The IC50 of idelalisib against 3D7, 3D7-PfKelch13WT, 3D7-PfKelch13C580Y, and 3D7VPS34 was shown in Supplementary Fig. 3. We found that idelalisib decreased the IC50 value of DHA from 4.92 to 2.75 nM (Fig. 3A) in 3D7-PfKelch13WT and from 36.89 to 6.63 nM in 3D7-PfKelch13C580Y (Fig. 3B). However, the DHA IC50 value of 3D7VPS34 did not change significantly (from 86.04 to 73.43 nM, Supplementary Fig. 4). These findings indicated that with the addition of idelalisib, only a lower concentration of DHA was required to achieve the effect of high concentrations of DHA alone in 3D7-PfKelch13WT and 3D7-PfKelch13C580Y.
The idelalisib at the IC50 value decreased the DHA IC50 value in 3D7-PfKelch13WT (A) and 3D7-PfKelch13C580Y (B). The IC50 values were determined by standard 72-h susceptibility assays via the measurement of SYBR Green I fluorescence. The IC50 values were calculated by GraphPad Prism 9.0 software (Analyse-Nonlinear regression curve fit-[Inhibitor] vs. normalized response—variable slope). The definitions of the maximum and minimum response values were 100 and 0%, respectively for the nonlinear regression curve fit. For PfPI3K activity (C) and PI3P level detection (D), tightly synchronized ring-stage parasite cultures were exposed to 0.1% DMSO (control), DHA at the IC50, idelalisib at the IC50 or the combination of DHA and idelalisib (each at the corresponding IC50) for 24 h until the parasites reached the late-trophozoite stage. Briefly, for PfPI3K activity (C), the kinase assay of PfPI3K bound to the immunoprecipitated beads was measured using a Class III PI3-kinase kit from Echelon Biosciences (K-3000) following the manufacturer’s instructions. For PI3P level detection (E), tightly synchronized ring-stage parasite cultures were exposed to 0.1% DMSO (control) or different concentrations of idelalisib for 24 h until the parasites reached the late-trophozoite stage. Briefly for PI3P level detection (D, E), lipids were extracted from the parasite pellet using methanol, chloroform and hydrochloric acid. The PI3P level was assessed (in triplicate) using an Echelon PI3P mass ELISA Kit (K-3300) following the manufacturer’s instructions. In C–E, one-way ANOVA and LSD-T post hoc test comparisons between treatment groups were employed. The means (s.d.) are based on three replicates (the triplicate data points are shown). In F, tightly synchronized early ring parasites (0–3 h post-invasion, 2% hematocrit and 1% parasitemia) were exposed to 700 nM DHA combined with or without 5 μM idelalisib (0.1% DMSO as a control) for 6 h, washed thrice with Albumax-free RPMI and returned to culture for 66 h. Parasite survival rates (%RSAs) were determined by comparing the number of viable parasites between the drug-treated and untreated controls and are expressed as percentages. Two-tailed unpaired Student’s t-test was employed. The means (s.d.) are based on three replicates (the triplicate data points are shown). *p < 0.05, **p < 0.01, ***p ≤ 0.001.
Since no commercially available antibody against PfPI3K existed, we had a custom antibody developed. Prior to subsequent experiments, the accuracy of the custom anti-PfPI3K antibody was confirmed via serum ELISA (Supplementary Table 3) and Western blotting (Supplementary Fig. 5). The PfPI3K activity and PI3P levels of the transgenic parasites were significantly lower after combination treatment with DHA and idelalisib than control or DHA treatment alone (Fig. 3C, D). In particular, in 3D7-PfKelch13C580Y, the PI3P level was reduced to the normal level in 3D7-PfKelch13WT. Moreover, different concentrations of idelalisib significantly decreased the PI3P levels in 3D7-PfKelch13WT and 3D7-PfKelch13C580Y (Fig. 3E). As the idelalisib concentration increased, the reduction in the PI3P level became more pronounced. However, the reduction in PI3P levels in 3D7VPS34 was not obvious. The decreasing trend of PI3P levels was consistent with the decreasing trend of RSA values in 3D7-PfKelch13C580Y, which decreased from 5.79 to 1.47% (below 1.5%) (Fig. 3F), although the difference was not statistically significant. Together, idelalisib effectively inhibited the PfPI3K activity, resulting in a decrease in PI3P levels, resulting in sensitization ARTs and reversing ARTs resistance.
Then we explored the effect of idelalisib in combination with DHA by combination index (CI) method. The results obtained by the two dilution methods were consistent. Specifically, all the concentration combinations inhibited the 3D7-PfKelch13WT (Table 1 and Supplementary Fig. 6A) and 3D7-PfKelch13C580Y (Table 2 and Supplementary Fig. 6B) parasite survival rate more effectively than DHA or idelalisib treatment alone. According to CI, each concentration combination showed a certain degree of synergistic or additive effects in 3D7-PfKelch13WT and a greater degree of synergistic or additive effects in 3D7-PfKelch13C580Y. This indicated that the idelalisib increased the antimalarial activity of DHA across the tested dose range and the lethal effect was more notable in 3D7-PfKelch13C580Y strains.
The combination of DHA and idelalisib led to the ROS accumulation without promoting the DHA activation in 3D7-PfKelch13WT and 3D7-PfKelch13C580Y
We wondered whether the stronger antimalarial activity was due to more DHA activation. Therefore, we examined the total amount of ROS and the endocytosis, transport and digestion of hemoglobin to explore the DHA activation level when it was combined with idelalisib. The results of ROS levels demonstrated that in 3D7-PfKelch13WT and 3D7-PfKelch13C580Y, DHA, idelalisib and their combination all increased ROS levels, compared with control (Fig. 4A and Supplementary Table 4). Compared with DHA treatment alone, the combination of DHA and idelalisib significantly increased the ROS level (p < 0.01) in 3D7-PfKelch13WT and 3D7-PfKelch13C580Y by 21.57 ± 9.86% and 20.65 ± 5.93%, respectively.
The ROS levels were determined using a dihydroethidium fluorescent probe assay (A). Briefly, tightly synchronized ring-stage parasite cultures were exposed to 0.1% DMSO (control), DHA at the IC50, idelalisib at the IC50 or the combination of DHA and idelalisib (each at the corresponding IC50) for 24 h until the parasites reached the late-trophozoite stage. Then, the dihydroethidium fluorescent probe was added to the cultures for another 30 min of incubation at 37 °C in the dark. After that, the cultures were washed with PBS three times to remove additional probe. The ROS levels were measured at 488 nm (excitation wavelength) and 610 nm (emission wavelength). The endocytosis of Hb was determined by a horseradish peroxidase (HRP) endocytosis assay (B). Briefly, enriched schizont-stage parasites were allowed to invade erythrocytes that had been preloaded with HRP. When the parasites were reached the late ring/early trophozoite stage, the culture was adjusted into 24-well plates for 14 h of incubation with 0.1% DMSO (control), DHA at the IC50, idelalisib at the IC50 or the combination of DHA and idelalisib (each at the corresponding IC50). Parasite-associated HRP from parasite pellets was determined using a colorimetric HRP substrate, and color development was measured by absorbance at 450 nm. The absorbance at 450 nm was read to measure the formation of the colored HRP product. The OD450 of the first column (control) was defined as 100%, and the other values were normalized to those of the control. Hb fractionation assays (C, D, E) were performed according to previous studies. Tightly synchronized ring-stage parasite cultures were exposed to 0.1% DMSO (control), DHA at the IC50, idelalisib at the IC50 or the combination of DHA and idelalisib (each at the corresponding IC50) for 24 h until the parasites reached the late-trophozoite stage. The Hb fractionation assay was subsequently performed. C Proportion of total cellular iron made up of cytosolic hemoglobin (n = 4). D Proportion of total cellular iron made up of digestive vacuole-associated hemoglobin and free heme (n = 4). E Proportion of total cellular iron made up of hemozoin (n = 4). One-way ANOVA and LSD-T post hoc test comparisons between treatment groups were employed. The means (s.d.) are based on three replicates (A, B) or four replicates (C–E) (the triplicate or quadruplicate data points are shown). **p < 0.01, ***p ≤ 0.001.
We performed a horseradish peroxidase endocytosis assay to evaluate the endocytosis of Hb (Fig. 4B). The Hb level in the 3D7-PfKelch13C580Y mutant was 56.3% lower than that in the 3D7-PfKelch13WT control. Treatment with DHA had little effect on the Hb endocytosis, while treatment with idelalisib reduced the Hb endocytosis by 46.02% and 57.71% in 3D7-PfKelch13WT and 3D7-PfKelch13C580Y, respectively. Intriguingly, the combination of the two drugs restored Hb endocytosis.
To determine whether the combination of DHA and idelalisib potentially plays a role in the Hb trafficking pathway, we performed Hb fractionation assays by examining the proportion of heme found as cytosolic Hb (Fig. 4C), digested vacuole (DV)-associated free heme/Hb (Fig. 4D) and hemozoin (Fig. 4E). Compared with the control, the combination of DHA and idelalisib significantly decreased the cytosolic Hb in 3D7-PfKelch13WT (p = 0.001), while compared with DHA treatment alone, the combination of DHA and idelalisib significantly increased the cytosolic Hb (p = 0.006). This indicated that the idelalisib treatment decreased the transport of Hb to DV caused by DHA. In 3D7-PfKelch13C580Y, the combination of DHA and idelalisib significantly increased the level of cytosolic Hb compared with that in the control and DHA treatment alone groups (p < 0.05). This finding indicated that the combination treatment decreased the transport of Hb to DV in 3D7-PfKelch13C580Y. Moreover, in 3D7-PfKelch13C580Y, idelalisib treatment significantly increased the cytosolic Hb level (p < 0.001). These findings are consistent with those of a previous study indicating that PI3P serves as a transport vesicle in the parasite14. However, compared with the DHA treatment alone, the combination of DHA and idelalisib did not change the amount of free heme/Hb in the digestion vacuole in 3D7-PfKelch13WT and 3D7-PfKelch13C580Y. Finally, the combination of DHA and idelalisib reduced the percentage of total cellular iron present in the hemozoin fraction, although the difference was not statistically significant.
These data suggested that idelalisib inhibited the endocytosis of Hb and the trafficking of Hb to the digestive vacuole. The combination of DHA and idelalisib restored the Hb endocytosis in the two transgenic lines, but there was still a disruption in the delivery of Hb vesicles to the DV. However, this disruption did not significantly influence the level of heme, which is responsible for ART activation. This finding emphasized that idelalisib did not increase the activation of DHA and that the increasing level of ROS was not due to the increased DHA activation.
The addition of idelalisib attenuated the ROS scavenging capacity of 3D7-PfKelch13WT and 3D7-PfKelch13C580Y
P. falciparum relies on two antioxidant systems for ROS scavenging (Trx- and GSH-dependent systems). Therefore, we quantified the gene expression (transcriptional profiles) of several antioxidants. In this study, 3D7-PfKelch13C580Y had significantly upregulated expression of Fe-SOD, TrxR, nPrx, and GST than its sensitive counterpart did at the basal level (p values all < 0.01) (Fig. 5A). There was no significant difference in the expression of GR, γ-GCS, or GS. In 3D7-PfKelch13WT, DHA exposure did not significantly affect the gene expression of these antioxidants except for nPrx (p = 0.007) (Fig. 5B). Moreover, compared with DHA treatment alone, the combination of DHA and idelalisib significantly downregulated the expression of Fe-SOD and nPrx. In 3D7-PfKelch13C580Y, the combination of DHA and idelalisib significantly downregulated the expression of Fe-SOD, TrxR, nPrx, and GST (p values all < 0.001) (Fig. 5B).
Tightly synchronized ring-stage P. falciparum 3D7 cultures were exposed to 0.1% DMSO (control), DHA at the IC50, idelalisib at the IC50 or the combination of DHA and idelalisib (each at the corresponding IC50) for 24 h until the parasites reached the late-trophozoite stage. After incubation, the erythrocytes were lysed with saponin, and the parasite pellets were washed three times with PBS. Each treatment group had six replicates, with three replicates used for RNA extraction and another three replicates used for protein extraction. The RNA was extracted to determining the relative gene expression of the parasites. The graph also shows the relative gene expression of antioxidants in 3D7-PfKelch13WT (A) and 3D7-PfKelch13C580Y(B) strains. The expression levels were normalized to the level of the housekeeping gene seryl-tRNA synthetase. Relative expression (fold change) was calculated from the expression levels of the control of 3D7-PfKelch13WT. Values greater than 1 (dashed line) indicate upregulation. The protein was extracted to determine the antioxidant activity of the parasites. The total antioxidant capacity (C), activity of total SOD (D), GR (E) and TrxR (F), total glutathione content (tGSH) (G) and the ratio of reduced glutathione to oxidized glutathione (H) of 3D7-PfKelch13WT and 3D7-PfKelch13C580Y are shown in the graph. *p < 0.05, **p < 0.01, ***p ≤ 0.001.
Although enzyme synthesis is regulated by genes, it can also be induced or repressed by the action of specific substrates. We subsequently determined the total antioxidant capacity (Fig. 5C and Supplementary Table 5), total SOD activity (Fig. 5D and Supplementary Table 6), GR activity (Fig. 5E and Supplementary Table 7), TrxR activity (Fig. 5F and Supplementary Table 8), and GSH content (Fig. 5G, H; Supplementary Tables 9 and 10). In 3D7-PfKelch13WT, the total antioxidant capacity and GR activity were essentially unchanged after drugs exposure and compared with control and DHA alone, the combination of DHA and idelalisib significantly decreased the activity of total SOD and TrxR. Compared with 3D7-PfKelch13WT, 3D7-PfKelch13C580Y presented increases in total antioxidant capacity (171.78 ± 13.39%), total SOD activity (150.85 ± 8.06%), GR activity (171.38 ± 4.45%) and TrxR activity (14.80 ± 3.75%). However, the combination of DHA and idelalisib significantly attenuated the total antioxidant capacity by 39.93 ± 0.66% and decreased the activity of antioxidant enzymes by 81.34 ± 1.70%, 9.23 ± 5.65% and 29.90 ± 5.28% for total SOD, GR and TrxR, respectively. The amount of total glutathione (tGSH) and the ratio of reduced glutathione to oxidized glutathione (GSH/GSSH) were shown in Fig. 5G, H. Compared with 3D7-PfKelch13WT, 3D7-PfKelch13C580Y presented a 22.01 ± 3.36% increase in tGSH level at the basal level. Compared with the corresponding control, the combination of DHA and idelalisib significantly decreased the tGSH level in both transgenic lines. Moreover, compared with the DHA treatment alone, the combination of two drugs significantly reduced tGSH by 42.19 ± 5.38% in 3D7-PfKelch13C580Y (p < 0.001) while reduced tGSH by 7.72 ± 6.03% (p = 0.093) in 3D7-PfKelch13WT. 3D7-PfKelch13C580Y presented a (55.44 ± 15.77) % increase in the GSH/GSSH ratio compared with 3D7-PfKelch13WT at the basal level. Compared with the corresponding control, the combination of DHA and idelalisib significantly decreased the GSH/GSSH ratio in both transgenic lines. Compared with DHA treatment alone, the combination of two drugs significantly reduced the GSH/GSSH ratio by 72.13 ± 12.10% in 3D7-PfKelch13C580Y (p < 0.001) while reduced the GSH/GSSH ratio by 28.70 ± 18.17% (p = 0.097) in 3D7-PfKelch13WT. These results indicated that 3D7-PfKelch13C580Y possessed a stronger antioxidant capacity. The combination treatment effectively attenuated the antioxidant capacity of 3D7-PfKelch13WT and 3D7-PfKelch13C580Y, resulting in an attenuated ROS-scavenging capacity and a dysregulated antioxidant system.
ROS accumulation caused greater mitochondrial membrane potential loss and DNA breakage in 3D7-PfKelch13WT and 3D7-PfKelch13C580Y
Since mitochondrial membrane potential depolarization is an early marker of apoptosis, while DNA fragmentation represents its final stage, we therefore assessed the effects of accumulated ROS on both mitochondrial membrane potential (Fig. 6A) and DNA fragmentation (Fig. 6B–D). The results demonstrated that, compared with DHA treatment alone, the combination of DHA and idelalisib significantly increased the degree of mitochondrial membrane potential loss by 27.89 ± 1.43% (3D7-PfKelch13WT) and 29.81 ± 3.61% (3D7-PfKelch13C580Y) (p < 0.001). Compared with DHA treatment alone, the combination of DHA and idelalisib significantly increased the percentage of TUNEL-positive cells by 2.51 ± 0.18% (3D7-PfKelch13WT) and 6.19 ± 0.13% (3D7-PfKelch13C580Y) (p < 0.001). This result indicated that the mitochondrial structure and the DNA was obviously damaged by the combination of the drugs in both strains.
Tightly synchronized ring-stage P. falciparum 3D7 cultures were exposed to 0.1% DMSO (control), DHA at the IC50, idelalisib at the IC50 or the combination of DHA and idelalisib (each at the corresponding IC50) for 24 h until the parasites reached the late-trophozoite stage. After incubation, the parasite pellets were collected. The degree of mitochondrial membrane potential depolarization was subsequently determined using a TMRE fluorescent probe assay (A) by measuring the optical density at 550 nm (excitation wavelength) and 575 nm (emission wavelength). DNA fragmentation was determined using a TUNEL kit (B–D). CytoFLEX flow cytometry was used to analyse and calculate the percentage of TUNEL-positive cells, which indicated the level of DNA fragmentation. The combination of DHA and idelalisib induced more DNA fragmentation in 3D7-PfKelch13C580Y (D) than in 3D7-PfKelch13WT (C). One-way ANOVA and LSD-T post hoc test comparisons between treatment groups were employed. The means (s.d.) are based on three replicates (A, B) or four replicates (C, D) (the data points are shown). ***p ≤ 0.001.
The combination of DHA and idelalisib caused disruptions in proteostasis
As the accumulation of ubiquitinated proteins is toxic to parasites, we next examined the consequences of drugs exposure on global ubiquitination (Fig. 7A, B). Compared with DHA treatment alone, the combination of DHA and idelalisib increased the amount of ubiquitinated proteins by 20.72% and 10.16% in 3D7-PfKelch13WT and 3D7-PfKelch13C580Y, respectively, although the difference was not statistically significant. The accumulation of damaged proteins further induces endoplasmic reticulum (ER) stress. Therefore, we examined the ER stress response hallmark eIF2α phosphorylation (Fig. 7C, D). Compared with DHA treatment alone, the combination of DHA and idelalisib induced 17.69% and 10.73% increases in eIF2α phosphorylation in 3D7-PfKelch13WT and 3D7-PfKelch13C580Y, respectively, although the difference was not statistically significant.
In addition, idelalisib upregulated the expression of genes in the UPR pathway in 3D7-PfKelch13WT (E) but downregulated them in 3D7-PfKelch13C580Y (F). Tightly synchronized ring-stage P. falciparum 3D7 cultures were exposed to 0.1% DMSO (control), DHA at the IC50, idelalisib at the IC50 or the combination of DHA and idelalisib (each at the corresponding IC50) for 24 h until the parasites reached the late-trophozoite stage. After incubation, the erythrocytes were lysed with saponin and the parasite pellets were washed three times with PBS. Each treatment group had six replicates, with three replicates used for RNA extraction and another three replicates used for protein extraction. For Western blot analysis, 20 µg of parasite protein samples was separated and transferred which was then immunoblotted with anti-ubiquitin (A, B), anti-eIF2α (C, D) or phospho-eIF2α (C, D) antibodies. For RT-qPCR, the RNA was extracted. The expression levels were normalized to the level of the housekeeping gene seryl-tRNA synthetase. Relative expression (fold change) was calculated from the expression levels of the control of 3D7-PfKelch13WT. Values greater than 1 (dashed line) indicate upregulation. One-way ANOVA and LSD-T post hoc test comparisons between treatment groups were employed. The means (s.d.) are based on three replicates (the triplicate data points are shown). p < 0.05, **p < 0.01.
Then, we examined the genes expression (transcriptional profiles) of unfolded protein response (UPR) pathway (Fig. 7E, F). 3D7-PfKelch13C580Y had, at the basal level, significantly upregulated expression of Bip, PDI, ERC, SDF, CYPB, TCP1, and prefoldin than its sensitive counterpart did (p < 0.05), with the exception of TCP1-b. In 3D7-PfKelch13WT, the combination of DHA and idelalisib significantly upregulated the expression of all the test genes (Fig. 7E). However, in 3D7-PfKelch13C580Y, the combination of DHA and idelalisib significantly downregulated the expression of the test genes to 3D7-PfKelch13WT basal levels (Fig. 7F).
The above results indicated that the accumulation of ROS induced by idelalisib resulted in increased ubiquitinated proteins, resulting in the persistent ER stress in 3D7-PfKelch13WT and 3D7-PfKelch13C580Y. Moreover, in 3D7-PfKelch13C580Y, idelalisib inhibited the degradation of damaged proteins. Together, these effects together led to disrupted proteostasis in the parasites.
Discussion
Mutations in Pfkelch13, resulting in increased levels of PfPI3K, as well as its lipid product PI3P, are the primary and definite molecular mechanism of ARTs resistance. PfPI3K was determined to be a promising target for eliminating ART resistance; thus, novel combination antimalarials that specifically target PfPI3K are needed to counter the resistant parasites that are rapidly emerging and spreading. Therefore, in this study, we aimed to screen high-efficiency PfPI3K inhibitors to accelerate the progress in the control of ART resistance and the elimination of malaria.
To better explore the effects of PfPI3K inhibitors, the construction of transgenic parasite lines with simple and comparable genetic backgrounds was needed. Therefore, we introduced the C580Y mutation site on the 3D7 genetic background using the SLI approach, which allowed us to compare the wild-type strains with the isogenic mutants. SLI was first reported by Tobias Spielmann17. This method can inactivate native target proteins, accompanied by the introduction of exogenous target proteins, and allowed us to explore the functions of the target protein and the pathway that it regulates accurately. A previous study showed that the PfPI3K was closest to the class III PI3Ks VPS34, by comparing its amino acid sequence and domain organization14. Therefore, we inserted exogenous VPS34, which expresses PI3P only, into the genome of PI3P-overexpressing transgenic parasites. The transgenic parasites were subsequently used throughout the study.
Prior to this study, our group used the predicted 3D structure of PfPI3K published by Mbengue et al.13 as the modeled protein and selected five marketed PI3K inhibitors and twenty-two compounds with human class III PI3K protein inhibitory functions as docking ligands for molecular docking. We subsequently verified the stability of the binding between the model protein and the ligands via molecular dynamics simulation, resulting in five PfPI3K inhibitor candidates. On the basis of the above work, we further identified idelalisib as the most effective PfPI3K inhibitor. Idelalisib is a PI3K inhibitor and a second-line drug for the treatment of certain hematological malignancies18. Being an anticancer drug, idelalisib has shown good safety index for host cells19. Meanwhile, it is known that PfPI3K phosphorylates phosphatidylinositol to generate PI3P, which promotes ART resistance. Our results showed that idelalisib could decrease the PI3P level by approximately 50% in both ART-sensitive and ART-resistant strains. Idelalisib decreased the IC50 value of DHA and the RSA value in the resistant strain. This result was consistent with our hypothesis and emphasized that idelalisib efficiently inhibited PfPI3K, significantly increased the antimalarial activity of ARTs and even reversed ART resistance. Thus, idelalisib is a promising DHA combination drug that needs to be further explored.
Although the exact action mode of ARTs is not fully understood, a popular theory proposes that the cleavage of the endoperoxide bridge in ARTs, which relies on free heme liberated from digested hemoglobin, is a prerequisite for ART activation. Once ARTs are activated, the resulting high levels of ROS are deleterious, attacking and damaging biomolecules ranging from nucleic acids to proteins that eventually lead to cell death20,21,22,23. Therefore, we examined the total amount of ROS and the endocytosis, transport and digestion of Hb to explore the DHA activation level when it was combined with idelalisib. We found that the combination of DHA and idelalisib significantly increased the ROS level, while the amount of the byproduct of Hb digestion, heme, did not increase. Since the ARTs are activated in a heme-dependent manner, resulting in the generation of ROS, this result may seem paradoxical. Thus, we believe that the accumulation of ROS was not due to increased production of ROS but rather to decreased ROS degradation. In addition, we found that idelalisib exposure alone inhibited the endocytosis and transport of hemoglobin. This finding was consistent with those of previous studies14.
The primary radicals are handled more by Fe-SOD, which converts O2•− to H2O224,25. Parasites are highly dependent on peroxiredoxins to detoxify H2O2, which ultimately converts H2O2 to water26. Peroxiredoxins are kept in reduced forms by Trx- and GSH-dependent systems, which are the most important antioxidant systems in P. falciparum. These two systems play interconnecting roles in scavenging ROS and keeping biomolecules in reduced and functional states. GST plays an interconnecting role in the reductive functions of GSH and helps reduce of H2O2 via the use of GSH as a cofactor27. TrxR, with its reductive function, is required for the generation of several isoforms of Trx28, which are supportive of the GSH system and are directly involved in reducing oxidized biomolecules and ROS levels. The combination of DHA and idelalisib downregulated the gene expression of Fe-SOD, TrxR, nPrx, and GST; suppressed the activity of antioxidant enzymes; and reduced the amount of total GSH and the ratio of GSH and GSSH in the ART-resistant parasites. In addition, the expression of GR, γ-GCS, and GS in response to the combination of DHA and idelalisib did not significantly changed. GR was slightly downregulated, whereas γ-GCS and GS were slightly upregulated. This result was consistent with previous works reporting that GR is a repressible enzyme whose level decreases due to elevated expression of γ-GCS29. These findings indicate that idelalisib inhibits antioxidant function, resulting in the accumulation of toxic ROS.
In this study, idelalisib decreased in the mitochondrial membrane potential, increased of DNA fragmentation, and increased the amount of ubiquitinated proteins30,31,32. This indicated that the accumulation of toxic ROS in parasites caused increased and sustained damage. Parasite proteostasis mainly includes the production and accumulation of damaged proteins and their degradation and elimination33. The downregulation of a molecular chaperone participating in the UPR decreases the capacity of parasites to quickly repair or degrade damaged proteins34,35. A previous study utilized experimental and bioinformatics data to assemble a network of molecular chaperones consisting of two major protein complexes, the reactive oxidative stress complex (PROSC) and the TCP-1 ring complex (TRiC), in P. falciparum36,37. The two chaperonin complexes play pivotal roles in the UPR. In this study, idelalisib significantly downregulated the gene expression of PROSC. In addition, idelalisib did not significantly change the gene expression of TRiC, which was more strongly associated with the synthesis of novel proteins. We hypothesized that idelalisib had little effect on the new protein synthesis, but significantly inhibited the protein degradation. Moreover, the accumulation of ubiquitinated proteins also promoted eIF2α phosphorylation, which inhibited the folding of newly synthesized proteins38. Taken together, these results indicate that idelalisib leads to global perturbation of cell function and viability, resulting in disruptions in proteostasis.
In addition, the 24 h drug treatments were performed in the whole experiment for reasons of practicality and consistency with other studies. The effects observed from such prolonged drug treatments may reflect secondary changes associated with loss of parasite viability. Further work using shorter treatments13 (4 h or less) may be more informative and this need more experiments to clarify. Meanwhile, although our initial screening focused on anti-malarial activity, we acknowledge that further profiling against human PI3K and other related kinases is essential to evaluate selectivity and minimize potential off-target effects. And further study should be performed and these studies will be critical in advancing the most promising inhibitors toward further development.
Previous studies indicate that K13 mutations not only directly increase PI3K and PI3P levels, thereby mediating ART resistance in parasites, but also impair hemoglobin uptake in ART-resistant strains carrying PfKelch13 mutations. This results in reduced production of hemoglobin-derived heme and consequently decreased activation of ART. Recent research has further revealed the role of PfKelch13 in ART activation through its heme-binding capability39. Therefore, the mechanisms underlying reversion to ART-based drugs warrant more in-depth investigation.
In conclusion, our study demonstrated that idelalisib is a potential PfPI3K inhibitor with strong synergistic antimalarial activity with DHA and can reverse ARTs resistance. This feature of idelalisib was achieved by effectively inhibiting the PfPI3K activity, resulting in a reduction in PI3P level. The degradation of toxic ROS produced by DHA activation was subsequently suppressed, resulting in increased mitochondrial membrane potential loss, increased DNA fragmentation, and increased levels of ubiquitinated proteins. Moreover, the reduction in PI3P levels also suppressed the expression of genes encoding molecular chaperones that participate in the UPR pathway, which is responsible for the degradation of ubiquitinated proteins, resulting in the aggregation of ubiquitinated proteins. We found that ubiquitinated protein aggregation also promoted eIF2α phosphorylation, resulting a persistent ER stress, leading to a disrupted proteostasis in P. falciparum. The reduction in PI3P levels by the PfPI3K inhibitor increased the accumulation of ROS as well as the amount of ubiquitinated proteins, thereby increasing sensitivity to ARTs and reversing ARTs resistance (Fig. 8). Overall, we provide critical novel candidates for combination treatment with ARTs. We confirmed that this combination strategy could further sensitize ART-sensitive P. falciparum to ARTs and reverse ART resistance in ART-resistant P. falciparum. This combination treatment will help combat ART resistance and contribute to the elimination of malaria.
In both ART-sensitive and ART-resistant parasites, idelalisib effectively inhibited PfPI3K activity, resulting in reduced PI3P levels. The reduction in the PI3P level led to the accumulation of ROS produced by activated DHA in P. falciparum. The accumulation of ROS was due to the decreased ROS degradation. Toxic ROS accumulation induced more mitochondrial membrane potential loss, resulting in more DNA fragmentation. Moreover, the reduction in PI3P levels also suppressed the expression of genes encoding molecular chaperones that participate in the UPR pathway, which is responsible for the degradation of damaged proteins induced by ROS in ART-resistant transgenic strains, resulting in the accumulation of ubiquitinated proteins. The accumulation of ubiquitinated proteins also promoted eIF2α phosphorylation, resulting in the persistent of ER stress, leading to disrupted proteostasis in P. falciparum. Overall, the reduction in PI3P by the PfPI3K inhibitor increased the accumulation of ROS as well as the ubiquitinated proteins via the UPR pathway, thereby increasing sensitivity to ARTs and reversing ART resistance. The red arrows represent the mechanism of ART resistance. The green arrows represent the mechanism by which the PfPI3K inhibitor increases sensitivity to ARTs.
Methods
Cloning and generation of constructs for P. falciparum transfection
All the constructs used for generating transgenic P. falciparum 3D7 parasites were assembled into different plasmids and finally cloned and inserted into pSLI-N-sandwich-loxP (K13) or p1xNLS-FRB-mCherry-hsp86-BSD. These two plasmids were both gifts from Tobias Spielmann (Addgene plasmid # 85793 and Addgene plasmid # 85795, respectively)17.
The pSLI-N-sandwich-loxP (K13) plasmid contains a recoded Pfkelch13 gene with its N-terminus tagged with the green fluorescent protein (GFP) sandwiched between two FK506-binding protein (FKBP) sequences. This plasmid was constructed by the selection-linked integration (SLI) approach, which was previously reported. The homologous recombination method was used to introduce the C580Y point mutation into the recoded Pfkelch13 gene in this plasmid. Briefly, the plasmid was used as a template to amplify the PCR1 product (using PfKelch13AvrIIF and C580Yreverse) and the PCR2 product (using C580Yforward and PfKelch13KpnIR). These two PCR products were subsequently cloned to the AvrII/KpnI site of pSLI-N-sandwich-loxP (K13) to generate pSLI-N-sandwich-loxP (K13C580Y). The primers used are listed in Supplementary Table 11. The construction schematic diagram is shown in Supplementary Fig. 7.
Human VPS34, the mammalian orthologue of PfPI3K, synthesizes only PI3P. We used pCMV-Myc-VPS34 (CAT# P1799, MiaoLingBio, China) as a template to amplify human VPS34 using VPS34XhoIF and VPS34SpeIR. The PCR product was subsequently cloned and inserted into the XhoI/SpeI site of p1xNLS-FRB-mCherry-hsp86-BSD to generate pVps34(human)-mCherry. The primers used are listed in Supplementary Table 11. The construction schematic diagram is shown in Supplementary Fig. 8.
P. falciparum culture and synchronization
P. falciparum 3D7 (gift from Artemisinin Research Center, Institute of Chinese Materia Medica) and transgenic parasites were cultured with O+ human erythrocytes (3% to 4% hematocrit, purchased from Shanxi Province Blood Center, China) in RPMI medium (Sigma-Aldrich) supplemented with 0.5% Albumax II (Invitrogen), 0.2 mM hypoxanthine, 11 mM glucose, 0.17% NaHCO3, and 10 mg/mL gentamycin and incubated at 37 °C in an atmosphere of 90% N2, 5% O2, and 5% CO2 in a humidified incubator using standard methods40. Parasite growth was monitored by counting infected erythrocytes in Giemsa-stained thin blood smears using light microscopy.
Synchronization was performed at the ring stage by 5% D-sorbitol treatment41,42. Mature schizont parasites were isolated using the Percoll/Sorbitol enrichment method with a 40/70% discontinuous Percoll gradient43,44. Parasite growth was synchronized via successive rounds of treatment with Percoll and D-sorbitol to obtain populations of pure ring-stage, trophozoite-stage, and schizont-stage parasites.
P. falciparum transfection and selection to obtain transgenic P. falciparum strains with comparable genetic backgrounds
Synchronous P. falciparum 3D7 parasites in culture were transfected with the indicated plasmids (purified and diluted to 10 μg/μL) via a preloading approach first described by Deitsch et al., with some modifications45. Briefly, 150 μL of uninfected erythrocytes, 100 μg of each plasmid DNA and 1× cytomix were mixed with a final volume of 400 μL and transferred to a 0.2 cm cuvette. For electroporation, a Bio-Rad Gene Pulser Xcell was used at 310 V and 950 μF46,47. After electroporation, these erythrocytes (already preloaded with plasmid DNA) were washed with complete medium to remove broken cell contents. The in-tact, preloaded erythrocytes were then resuspended in 10 mL of complete medium and incubated with enriched schizonts.
After 2–3 complete growth cycles, the transfectants were selected either with 2.5 nM WR99210 (which selects for the human dihydrofolate reductase [hDHFR]) and 0.9 μM DSM1 (which is a dihydroorotate dehydrogenase inhibitor) or with 1 μg/ml blasticidin S hydrochloride until stable cell lines were obtained. Parasites were observed via microscopy throughout the whole drug selection period48,49. The parasites were then screened for monoclonal clones by limiting dilution. Briefly, infected red blood cells were loaded onto a hemocytometer for erythrocyte density quantification and parasitemia assessment. On the basis of erythrocyte counts and parasitemia, parasites were diluted with uninfected erythrocytes and complete medium to generate 0.5 parasites/200 µL working solution, which was homogenized and distributed into 96-well plates (200 µL/well). The complete medium containing the screening drugs in the 96-well plates was replaced every 48 h, and the parasites were observed until the single parasite populations were obtained.
A schematic diagram of the integration of the transgenic strains via SLI is shown in Supplementary Fig. 9. The well incubated transgenic parasites from the single parasite populations with mutant kelch13 were verified by PCR (the primer 1 and 2 are listed in Supplementary Table 12 and the legend of Supplementary Fig. 9). The PCR cycle was as follows: initial denaturation at 94 °C for 3 min; 30 cycles of denaturation at 94 °C for 30 s and annealing at 55 °C for 30 s and extension at 72 °C for 2 min; and a final extension at 72 °C for 5 min. Western blotting probed with anti-GFP antibody (Abclonal-AE078; 1∶1000) and confocal images in the GFP panel, and the nuclei were stained with DAPI. The transgenic parasites with VPS34 were verified by Western blotting probed with an anti-mCherry antibody (Abclonal-AE127; 1∶1000), and confocal images are shown in the red panel, while the nuclei were stained with DAPI.
Ring-stage survival assay (RSA) for determining the resistance of P. falciparum to ART
In vitro RSA was performed as previously described50,51. Briefly, tightly synchronized early ring parasites (0–3 h post-invasion, 2% hematocrit and 1% parasitemia) were exposed to 700 nM DHA combined with or without 5 μM idelalisib (0.1% DMSO as a control) for 6 h, washed thrice with Albumax-free RPMI and returned to culture for 66 h. Blood smears were prepared and stained with Giemsa. Each sample was analyzed in duplicate and 10,000 erythrocytes were assessed independently by light microscopy. Parasite survival rates (%RSA) were expressed as ratios by comparing the number of viable parasites between the drug-treated and untreated controls. The means (s.d.) of RSA value of three replicates was determined by two independent slide readers.
In vitro 72-h susceptibility assays for determining the antimalarial activity of DHA and PI3K inhibitors
Parasite susceptibility to DHA and PI3K inhibitors was evaluated as previously described52,53. Briefly, synchronized 2- to 4-h post-invasion ring-stage parasites were exposed to a 10-point serial dilution of DHA, idelalisib, umbralisib, taselisib, copanlisib or AZD6482 in a 96-well plate at 2% hematocrit and 1% parasitemia in a 200 μL final volume. After 72 h of incubation, the DNA was released by adding 100 μL of lysis buffer (30 ml of 1 M Tris-HCL, 15 ml of 0.5 M EDTA, 1.2 ml of 10% saponin, and 1.2 ml of Triton X-100) with 0.05% SYBR Green I (Solarbio). Parasite drug susceptibility was assessed by the measurement of SYBR Green I fluorescence. The definitions of the maximum and minimum response values were 100% and 0%, respectively for the nonlinear regression curve fit. The definitions of the maximum and minimum response values (100% and 0%, respectively).
Measurement of PI3P levels in standard and transgenic P. falciparum
Tightly synchronized ring-stage parasite cultures were exposed to 0.1% DMSO (control), DHA at the IC50, idelalisib at the IC50 or the combination of DHA and idelalisib (each at the corresponding IC50) for 24 h until the parasites reached the late-trophozoite stage. After that, infected red blood cells (iRBCs), with or without drug treatment were lysed with saponin to remove hemoglobin (Hb). The resulting pellets (corresponding to 6 × 107 parasites) were washed with cold PBS. Lipids were extracted using methanol, chloroform, and hydrochloric acid. The PI3P level was assessed (in triplicate) using an Echelon PI3P Mass ELISA Kit (K-3300)13 following the manufacturer’s instructions.
Generation of anti-PfPI3K antibody and PI3-kinase assay for determining the kinase activity
Custom antibodies against P. falciparum PI3K (PfPI3K, PF3D7_0515300) were generated in rabbits against the C-terminal peptide CVLKVQEKFRLDLNDE by HuaBio, China. The reactivity of the custom antibody was confirmed by serum ELISA and Western blotting and was used for immunoprecipitation assays.
Tightly synchronized ring-stage parasite cultures were exposed to 0.1% DMSO (control), DHA at the IC50, idelalisib at the IC50 or the combination of DHA and idelalisib (each at the corresponding IC50) for 24 h until the parasites reached the late-trophozoite stage. After incubation, the parasite pellets were collected via lysis in saponin (0.25%) and washed three times with cold PBS.
PfPI3K was immunoprecipitated from the parasite pellet. Proteins were extracted for 1 h at 4 °C with 1× protease inhibitor cocktail (MCE). The supernatant was incubated with anti-PfPI3K antibodies for 12 h at 4 °C, and then the protein A/G agarose beads (preequilibrated in lysis buffer) were added for 3 h of additional incubation. PfPI3K-bound beads were washed three times and then used for kinase activity assay. The kinase assay of PfPI3K bound to the immunoprecipitated beads was measured using a Class III PI3-Kinase Kit from Echelon Biosciences (K-3000)13 following the manufacturer’s instructions.
The analysis of synergy between DHA and idelalisib
According to the IC50 of DHA and idelalisib obtained from experiment above, 9 concentration combinations of the two drugs (4 × IC50, 2 × IC50, IC50, 1/2 × IC50, 1/4 × IC50, 20% IC50, 40% IC50, 60% IC50, and 80% IC50) are used in pairs. The experiment was conducted as SYBR Green 1 fluorescence method. The he synergy analysis was performed by CompuSyn and the combination index method (the Chou-Talalay method; median-drug effect analysis) was used to explore whether there was a synergistic effect between DHA and idelalisib. This method was widely adopted pharmacodynamic approach employs the combination index (CI) to define the combination effect of two drugs. The synergistic efficacy of drug combinations was classified with the CI serving as the quantitative metric: strong synergy: CI < 0.1; high synergy: 0.1 ≤ CI < 0.3; moderate synergy: 0.3 ≤ CI < 0.7; low synergy: 0.7 ≤ CI < 0.85; marginal synergy: 0.85 ≤ CI < 0.9; additive effect: 0.9 ≤ CI < 1.1 and antagonism: CI ≥ 1.1.
Horseradish peroxidase (HRP) endocytosis assay and Hb fractionation assay for determining the degree of DHA activation
The endocytosis assay was performed as previously described54,55. The HRP content in the subsequent trophozoite-stage parasites was determined to represent the degree of Hb endocytosis. Briefly, erythrocytes were preloaded with HRP by hypotonic lysis (5 mM HEPES, 11 mM glucose, 2 mM MgCl2 and 2 mM Na2ATP) containing 200 mg/ml HRP. After incubation for 10 min at 30 °C, a hypertonic solution (280 mM NaCl, 40 mM KCl and 11 mM glucose) was added, and the mixture was then incubated for 5 min at 37 °C to reseal the erythrocytes. The enriched mature schizonts were added to 600 μL of HRP-loaded erythrocytes. When the parasites reached the late ring/early trophozoite stage, the culture was adjusted into 24-well plates for 14 h of incubations with the drug and DMSO controls. Then the erythrocytes were lysed with saponin and the pellets were washed three times with PBS. After that, the pellets were permeabilized in colorimetric HRP substrate buffer (0.05 M Na-phosphate, 0.05 M citric acid, pH 4.8) containing 1% Triton. Following centrifugation at 16,000 g, 100 μL of the supernatant was added to 100 μL of substrate buffer containing 3 mg/ml o-phenylenediamine and 1 ml/ml H2O2. The absorbance of the colored HRP product was measured at 450 nm.
The Hb fractionation assay was adapted from previous research using parasites stocked previously frozen at −80 °C56,57,58,59. Tightly synchronized ring-stage parasite cultures were exposed to 0.1% DMSO (control), DHA at the IC50, idelalisib at the IC50 or the combination of DHA and idelalisib (each at the corresponding IC50) for 24 h until the parasites reached the late-trophozoite stage. After incubation, the parasite pellets were collected by lysis with saponin (0.25%) and washed three times with cold PBS. The fractionation assay was performed using parasite pellets. For the cytosolic Hb fraction, the parasite pellets were resuspended in 50 μL ultrapure water and sonicated for 5 min in a water bath sonicator. After that, 50 μL of 0.2 M HEPES was added. The supernatant containing the cytosolic Hb fraction was carefully transferred to new tubes and 50 μL of 4% of SDS was added before the samples were incubated at 95 °C for 5 min. 200 μL samples which containing the cytosolic Hb fraction were transferred to a 96-well plate and the absorbance at 405 nm was read immediately.
The remaining pellets were resuspended in 50 μL of ultrapure water and 50 μL of 4% of SDS before being sonicated for 5 min and incubated at 95 °C for 5 min. After that, 50 μL of 0.2 M HEPES, 50 μL of 0.3 M NaCl and 50 μL of 25% (v/v) pyridine were added to the samples before they were centrifuged at 1500 × g for 20 min. A total of 200 μL of the supernatant, which contained the free heme/DV-associated Hb fraction was transferred to a 96-well plate, and the absorbance at 405 nm was read immediately.
The remaining pellets were solubilized with 50 μL ultrapure water and 50 μL 0.3 M NaOH and then vortexed for 10 s. After sonicating for 15 min, 50 μL 0.2 M HEPES, 50 μL 0.3 M HCl and 50 mL of 25% (v/v) pyridine were added to the samples. 200 μL samples which contained hemozoin fractions were transferred to a 96-well plate, and the absorbance at 405 nm was read immediately.
A standard curve of hematin (porcine) was prepared to quantify heme Fe. Briefly, serial dilutions of hematin (in 0.3 M NaOH) and the other components (0.2 M HEPES, 4% SDS, 0.3 M NaCl, 0.3 M HCl, 25% pyridine in 0.2 M HEPES and ultrapure water), which were added to form the heme-pyridine complex were mixed in a 96-well plate, and the absorbance was recorded at 405 nm. The amount of heme for each fraction was calculated by comparing the OD 405 nm to the standard curve and then dividing by the number of cells used in the assay.
ROS quantification assay, antioxidant capacity and antioxidant enzyme activity assays for determining the content and degradation of ROS
Tightly synchronized ring-stage parasite cultures were exposed to 0.1% DMSO (control), DHA at the IC50, idelalisib at the IC50 or the combination of DHA and idelalisib (each at the corresponding IC50) for 24 h until the parasites reached the late-trophozoite stage. After incubation, the parasite pellets were collected via lysis in saponin (0.25%) and washed three times with cold PBS. The ROS quantification assay, antioxidant capacity and antioxidant enzyme activity assays were performed using parasite pellets.
For overall ROS quantification, dihydroethidium (Elabscience) was used as a fluorescent probe. Dihydroethidium itself produces blue fluorescence, whereas its oxidation in cell results in red fluorescence. ROS levels were measured at 488 nm (excitation wavelength) and 610 nm (emission wavelength).
The total antioxidant capacity was determined via colorimetry at OD593 nm using an Elabscience T-AOC Colorimetric Assay kit (E-BC-K136-M) according to the manufacturer’s instructions. The T-AOC is quantified on the basis of the reduction of Fe3+-TPTZ to blue Fe2+-TPTZ by antioxidant substances under acidic conditions, which can be determined by measuring the absorbance at OD593 nm.
The total superoxide dismutase (T-SOD) activity was determined via colorimetry at OD550nm using a T-SOD Activity Assay Kit (hydroxylamine method) (E-BC-K019-S, Elabscience) according to the manufacturer’s instructions. The assay utilizes a xanthine/xanthine oxidase reaction system to generate superoxide anion radicals (O₂•−). These radicals oxidize hydroxylamine to form nitrite, which subsequently reacts with a chromogenic agent to produce a purple‒red compound. The introduction of SOD into the system catalyzes the dismutation of O₂•−, thereby reducing nitrite formation and attenuating purple‒red color reaction.
The glutathione reductase (GR) activity was determined following the manufacturer’s instructions for the GR Activity Assay Kit (E-BC-K099-M, Elabscience). The assay is assessed for its ability to catalyze the NADPH-dependent reduction of oxidized glutathione (GSSG) to reduced glutathione (GSH). The enzymatic activity was calculated on the basis of the rate of NADPH oxidation, as monitored by the decrease in absorbance values at 340 nm.
Thioredoxin reductase (TrxR) activity was determined following the manufacturer’s instructions for the TrxR Activity Assay Kit (E-BC-K548-M, Elabscience). The assay is determined through its catalytic reduction of a specific chromogenic substrate. The reaction product exhibited a characteristic absorption peak at 412 nm, with enzymatic activity quantified by the rate of absorbance change at this wavelength.
The quantification of total glutathione (T-GSH) and oxidized glutathione (GSSG) was performed following the manufacturer’s instructions for the T-GSH/GSSG Colorimetric Assay Kit (E-BC-K097-S, Elabscience). The total glutathione content (GSSG + GSH) was determined through a two-step enzymatic reaction. In the first step, oxidized glutathione (GSSG) is enzymatically reduced to reduced glutathione (GSH) by glutathione reductase (GR) in the presence of NADPH. The generated GSH then reacts with the chromogenic substrate DTNB, yielding a stoichiometric amount of yellow-colored TNB. The total glutathione concentration is directly proportional to TNB formation, which is quantified via spectrophotometry by measuring the absorbance at 412 nm. To specifically quantify GSSG, endogenous GSH in the sample was first eliminated. The remaining GSSG is subsequently reduced to GSH via the GR/NADPH system and subjected to the DTNB reaction as described above. The GSSG concentration was calculated using the measurement after background interference was corrected for.
Mitochondrial membrane potential and TUNEL assay for determining the effect of ROS on P. falciparum
Tightly synchronized ring-stage parasite cultures were exposed to 0.1% DMSO (control), DHA at the IC50, idelalisib at the IC50 or the combination of DHA and idelalisib (each at the corresponding IC50) for 24 h until the parasites reached the late-trophozoite stage. After incubation, the parasite pellets were collected via lysis with saponin (0.25%) and washed three times with cold PBS. The mitochondrial membrane potential and DNA fragmentation (TUNEL assay) were determined using parasite pellets. TMRE (tetramethylrhodamine, ethyl ester; Beyotime), which is a membrane-permeable orange‒red cationic fluorescent probe, was used to detect the mitochondrial membrane potential. It can accumulate in intact mitochondria, whereas depolarized or inactive mitochondria with reduced membrane potential lead to reduced TMRE accumulation. The relative fluorescence intensity was measured at 550 nm/575 nm. A TUNEL kit (Elabscience) was used to analyse DNA fragmentation by flow cytometry at 650 nm/670 nm. Beckman Navios flow cytometry and the FlowJo software were used to analyse and calculate the percentage of TUNEL-positive cells. The gating strategy for analyzing the DNA fragmentation was shown in Supplementary Fig. 10.
Quantitative real-time RT-PCR for determining the expression of antioxidants and genes encoding molecular chaperones involved in the UPR pathway
Tightly synchronized ring-stage parasite cultures were exposed to 0.1% DMSO (control), DHA at the IC50, idelalisib at the IC50 or the combination of DHA and idelalisib (each at the corresponding IC50) for 24 h until the parasites reached the late-trophozoite stage. After incubation, the parasite pellets were collected via lysis with saponin (0.25%) and washed three times with cold PBS. Total RNA was extracted from different parasite pellet samples by using RNA extraction reagent (TRIgent, Mei5bio). Subsequently, reverse transcription was conducted using the HiScript III RT SuperMix for qPCR (+gDNA wiper) Kit (Vazyme), and qPCR was performed using the ChamQ Universal SYBR qPCR Master Mix (Vazyme) with primers corresponding to the sequences of BiP (PF3D7_0917900), PDI (PF3D7_0827900), ERC (PF3D7_1108600), SDF (PF3D7_1010700), CYPB (PF3D7_1115600), TCP1-b (PF3D7_0306800), TCP1 (PF3D7_1357800), Prefoldin (PF3D7_0718500), GR (PF3D7_1419800), γ-GCS (PF3D7_0918900), GS (PF3D7_0512200), Fe-SOD (PF3D7_0814900), TrxR (PF3D7_0923800), nPrx (PF3D7_1027300), and GST (PF3D7_1419300). The 2−ΔΔCT method was employed to determine the expression of the target genes relative to that of the seryl-tRNA synthetase (PF07_0073). The primer sequences used for qRT-PCR are listed in Supplementary Table 13. The PCR cycle was as follows: initial denaturation at 95 °C for 3 min; 40 cycles of denaturation at 95 °C for 10 s and annealing/extension at 60 °C for 30 s; and a melting curve analysis step comprising denaturation at 95 °C for 15 s, annealing at 60 °C for 1 min, and final denaturation at 95 °C for 15 s.
Western blotting for verifying the transgenic parasites, determining the content of damaged proteins and the level of ER stress
Tightly synchronized ring-stage parasite cultures were exposed to 0.1% DMSO (control), DHA at the IC50, idelalisib at the IC50 or the combination of DHA and idelalisib (each at the corresponding IC50) for 24 h until the parasites reached the late-trophozoite stage. After incubation, the parasite pellets were collected via lysis with saponin (0.25%) and washed three times with cold PBS. The parasite pellets were solubilized in lysis solution, mixed with protein sample buffer, then 20 µg of parasite protein samples were separated on 6, 8, or 10% SDS‒PAGE gels and transferred to 0.22 μm PVDF membranes (Millipore). The membranes were blocked with 5% BSA for 2 h at room temperature and probed with primary antibody overnight at 4 °C, followed by incubation with secondary antibody for 2 h at room temperature. The primary antibodies used included rabbit polyclonal anti-β-actin (Abclonal-AC038; 1∶1000), rabbit anti-ubiquitin (Abclonal-A0162; 1∶1000), rabbit anti-phospho-eIF2α (Abclonal-AP0692; 1∶1000), mouse anti-eIF2α (Cell Signaling Technology-L57A5; 1∶1000) antibodies. All the primary antibodies were validated. Appropriate HRP-coupled secondary antibodies were used, and immunoblots were revealed by ECL. Chemiluminescence was detected using the Bio-Rad ChemiDocTM MP imaging system. Western blot band intensities were quantified using ImageJ software to determine grayscale values. Integrated density (IntDen) and mean gray values were measured, with background signal subtracted from corresponding band values to account for nonspecific noise. For normalization, target band intensities were expressed relative to a loading control (β-actin) using the formula: Normalized Intensity = (Target IntDen − Background IntDen)/(Control IntDen − Background IntDen).
Statistics and reproducibility
The means (s.d.) from three experimental replicates, each containing duplicate data points, are shown. GraphPad Prism 9.0 software was used for all the statistical analyses. The statistical significance was determined using one-way ANOVA, and least significant difference (LSD)-T tests were used for comparisons between treatment groups. One-way ANOVA was first performed to test the overall significance across all groups. Only when ANOVA indicated a statistically significant difference, we proceeded with post-hoc pairwise comparisons using the LSD-T test or two-tailed unpaired Student’s t tests were used for comparisons between two groups. The synergistic effects of DHA and the PI3K inhibitor were evaluated via CompuSyn software. A minimum of three biological replicates were performed for each experiment. To ensure analytical robustness, all the assays were performed in triplicate across independent biological replicates.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The authors declare that the source data supporting the findings of this study are available within the paper and the associated Supplementary Information files (Supplementary Materials and the Supplementary Data file).
References
Venkatesan P. WHO World Malaria Report 2024. Lancet Microbe 6, 101073 (2025).
White, N. J. et al. Malaria. Lancet 383, 723–35 (2014).
Bhatt, S. et al. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature 526, 207–211 (2015).
Ward, K. E., Fidock, D. A. & Bridgford, J. L. Plasmodium falciparum resistance to artemisinin-based combination therapies. Curr. Opin. Microbiol. 69, 102193 (2022).
Gething, P. W. et al. Mapping Plasmodium falciparum mortality in Africa between 1990 and 2015. N. Engl. J. Med. 375, 2435–2445 (2016).
Björkman, A., Gil, P. & Alifrangis, M. Alarming Plasmodium falciparum resistance to artemisinin-based combination therapy in Africa: the critical role of the partner drug. Lancet Infect. Dis. 24, e540–e541 (2024).
Siddiqui, F. A., Liang, X. & Cui, L. Plasmodium falciparum resistance to ACTs: emergence, mechanisms, and outlook. Int J. Parasitol. Drugs Drug Resist. 16, 102–118 (2021).
van der Pluijm, R. W. et al. Determinants of dihydroartemisinin-piperaquine treatment failure in Plasmodium falciparum malaria in Cambodia, Thailand, and Vietnam: a prospective clinical, pharmacological, and genetic study. Lancet Infect. Dis. 19, 952–961 (2019).
Spring, M. D. et al. Dihydroartemisinin-piperaquine failure associated with a triple mutant including kelch13 C580Y in Cambodia: an observational cohort study. Lancet Infect. Dis. 15, 683–91 (2015).
Amaratunga, C. et al. Dihydroartemisinin-piperaquine resistance in Plasmodium falciparum malaria in Cambodia: a multisite prospective cohort study. Lancet Infect. Dis. 16, 357–65 (2016).
Balikagala, B. et al. Evidence of artemisinin-resistant malaria in Africa. N. Engl. J. Med. 385, 1163–1171 (2021).
Haldar, K., Bhattacharjee, S. & Safeukui, I. Drug resistance in Plasmodium. Nat. Rev. Microbiol. 16, 156–170 (2018).
Mbengue, A. et al. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature 520, 683–7 (2015).
Vaid, A., Ranjan, R., Smythe, W. A., Hoppe, H. C. & Sharma, P. PfPI3K, a phosphatidylinositol-3 kinase from Plasmodium falciparum, is exported to the host erythrocyte and is involved in hemoglobin trafficking. Blood 115, 2500–7 (2010).
Ashley, E. A. et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 371, 411–23 (2014).
Mott, B. T. et al. High-throughput matrix screening identifies synergistic and antagonistic antimalarial drug combinations. Sci. Rep. 5, 13891 (2015).
Birnbaum, J. et al. A genetic system to study Plasmodium falciparum protein function. Nat. Methods 14, 450–456 (2017).
Yang, Q., Modi, P., Newcomb, T., Quéva, C. & Gandhi, V. Idelalisib: first-in-class PI3K delta inhibitor for the treatment of chronic lymphocytic leukemia, small lymphocytic leukemia, and follicular lymphoma. Clin. Cancer Res. 21, 1537–42 (2015).
Yadav, K. et al. Repurposing of existing therapeutics to combat drug-resistant malaria. Biomed. Pharmacother. 136, 111275 (2021).
Bridgford, J. L. et al. Artemisinin kills malaria parasites by damaging proteins and inhibiting the proteasome. Nat. Commun. 9, 3801 (2018).
Ismail, H. M. et al. Artemisinin activity-based probes identify multiple molecular targets within the asexual stage of the malaria parasites Plasmodium falciparum 3D7. Proc. Natl. Acad. Sci. USA 113, 2080–5 (2016).
Wang, J. et al. Haem-activated promiscuous targeting of artemisinin in Plasmodium falciparum. Nat. Commun. 6, 10111 (2015).
Gopalakrishnan, A. M. & Kumar, N. Antimalarial action of artesunate involves DNA damage mediated by reactive oxygen species. Antimicrob. Agents Chemother. 59, 317–25 (2015).
Soulère, L. et al. Screening of Plasmodium falciparum iron superoxide dismutase inhibitors and accuracy of the SOD-assays. Bioorg. Med. Chem. 11, 4941–4 (2003).
Akerman, S. E. & Müller, S. 2-Cys peroxiredoxin PfTrx-Px1 is involved in the antioxidant defence of Plasmodium falciparum. Mol. Biochem. Parasitol. 130, 75–81 (2003).
Kawazu, S., Komaki-Yasuda, K., Oku, H. & Kano, S. Peroxiredoxins in malaria parasites: parasitologic aspects. Parasitol. Int. 57, 1–7 (2008).
Müller, S. Redox and antioxidant systems of the malaria parasite Plasmodium falciparum. Mol. Microbiol. 53, 1291–305 (2004).
Hirt, R. P., Müller, S., Embley, T. M. & Coombs, G. H. The diversity and evolution of thioredoxin reductase: new perspectives. Trends Parasitol. 18, 302–8 (2002).
Lüersen, K., Walter, R. D. & Müller, S. Plasmodium falciparum-infected red blood cells depend on a functional glutathione de novo synthesis attributable to an enhanced loss of glutathione. Biochem. J. 346, 545–52 (2000).
Chen, M. Z. et al. A thiol probe for measuring unfolded protein load and proteostasis in cells. Nat. Commun. 8, 474 (2017).
Ismail, H. M. et al. A click chemistry-based proteomic approach reveals that 1,2,4-trioxolane and artemisinin antimalarials share a common protein alkylation profile. Angew. Chem. Int. Ed. Engl. 128, 6511–6515 (2016).
Tank, E. M. & True, H. L. Disease-associated mutant ubiquitin causes proteasomal impairment and enhances the toxicity of protein aggregates. PLoS Genet. 5, e1000382 (2009).
Hipp, M. S., Park, S. H. & Hartl, F. U. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 24, 506–14 (2014).
Ellgaard, L. & Helenius, A. Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4, 181–91 (2003).
Tongaonkar, P. & Selsted, M. E. SDF2L1, a component of the endoplasmic reticulum chaperone complex, differentially interacts with {alpha}-, {beta}-, and {theta}-defensin propeptides. J. Biol. Chem. 284, 5602–9 (2009).
LaCount, D. J. et al. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature 438, 103–7 (2005).
Pavithra, S. R., Kumar, R. & Tatu, U. Systems analysis of chaperone networks in the malarial parasite Plasmodium falciparum. PLoS Comput. Biol. 3, 1701–15 (2007).
Zhang, M. et al. Inhibiting the Plasmodium eIF2α kinase PK4 prevents artemisinin-induced latency. Cell Host Microbe. 22, 766–776.e4 (2017).
Rahman, A. et al. Artemisinin-resistant Plasmodium falciparum Kelch13 mutant proteins display reduced heme-binding affinity and decreased artemisinin activation. Commun. Biol. 7, 1499 (2024).
Jiang, L. et al. PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum. Nature 499, 223–7 (2013).
Lambros, C. & Vanderberg, J. P. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 65, 418–20 (1979).
Blackman, M. J. Purification of Plasmodium falciparum merozoites for analysis of the processing of merozoite surface protein-1. Methods Cell Biol. 45, 213–20 (1994).
Rivadeneira, E. M., Wasserman, M. & Espinal, C. T. Separation and concentration of schizonts of Plasmodium falciparum by Percoll gradients. J. Protozool. 30, 367–70 (1983).
Xiao, B. et al. Epigenetic editing by CRISPR/dCas9 in Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 116, 255–260 (2019).
Deitsch, K., Driskill, C. & Wellems, T. Transformation of malaria parasites by the spontaneous uptake and expression of DNA from human erythrocytes. Nucleic Acids Res. 29, 850–3 (2001).
Skinner-Adams, T. S., Lawrie, P. M., Hawthorne, P. L., Gardiner, D. L. & Trenholme, K. R. Comparison of Plasmodium falciparum transfection methods. Malar. J. 2, 19 (2003).
Hasenkamp, S., Merrick, C. J. & Horrocks, P. A quantitative analysis of Plasmodium falciparum transfection using DNA-loaded erythrocytes. Mol. Biochem. Parasitol. 187, 117–20 (2013).
Sherling, E. S. et al. The Plasmodium falciparum rhoptry protein RhopH3 plays essential roles in host cell invasion and nutrient uptake. Elife 6, e23239 (2017). pii.
Wilde, M. L. et al. Protein kinase A is essential for invasion of Plasmodium falciparum into human erythrocytes. mBio. 10, e01972-19 (2019).
Mukherjee, A. et al. Artemisinin resistance without pfkelch13 mutations in Plasmodium falciparum isolates from Cambodia. Malar. J. 16, 195 (2017).
Witkowski, B. et al. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in vitro and ex vivo drug-response studies. Lancet Infect. Dis. 13, 1043–9 (2013).
Mukherjee, A., Gagnon, D., Wirth, D. F. & Richard, D. Inactivation of Plasmepsins 2 and 3 sensitizes Plasmodium falciparum to the antimalarial drug piperaquine. Antimicrob. Agents Chemother. 62, e02309-17 (2018).
Smilkstein, M., Sriwilaijaroen, N., Kelly, J. X., Wilairat, P. & Riscoe, M. Simple and inexpensive fluorescence-based technique for high-throughput antimalarial drug screening. Antimicrob. Agents Chemother. 48, 1803–6 (2004).
Smythe, W. A., Joiner, K. A. & Hoppe, H. C. Actin is required for endocytic trafficking in the malaria parasite Plasmodium falciparum. Cell Microbiol. 10, 452–64 (2008).
Hoppe, H. C. et al. Antimalarial quinolines and artemisinin inhibit endocytosis in Plasmodium falciparum. Antimicrob. Agents Chemother. 48, 2370–8 (2004).
Birrell, G. W. et al. Multi-omic characterization of the mode of action of a potent new antimalarial compound, JPC-3210, against Plasmodium falciparum. Mol. Cell Proteom. 19, 308–325 (2020).
Combrinck, J. M. et al. Optimization of a multi-well colorimetric assay to determine haem species in Plasmodium falciparum in the presence of anti-malarials. Malar. J. 14, 253 (2015).
Combrinck, J. M. et al. Insights into the role of heme in the mechanism of action of antimalarials. ACS Chem. Biol. 8, 133–7 (2013).
Mukherjee, A. et al. A phosphoinositide-binding protein acts in the trafficking pathway of hemoglobin in the malaria parasite Plasmodium falciparum. mBio. 13, e0323921 (2022).
Acknowledgements
We thank all the members who contributed to this study. We thank Rongrong Wang for her permission for details of the unpublished work to be disclosed in this manuscript (including the Tables S1 and S2). This research was supported by the National Natural Science Foundation of China (grant number 82173767).
Author information
Authors and Affiliations
Contributions
Conceptualization Ideas: N.S., R.R.W., and S.Q.Z. Methodology: N.S. and Q.X.L. Validation: N.S., T.J.Y.Y., and J.Q.Y. Investigation: N.S. and R.R.W. Resources: Y.J.L. and S.Q.Z. Writing: N.S. Supervision: L.N.C., Y.J.L., and S.Q.Z. Project Administration: S.Q.Z. Funding acquisition: S.Q.Z.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Tawanda Zininga and the other anonymous reviewer(s) for their contribution to the peer review of this work. Primary handling editors: Nishith Gupta and Kaliya Georgieva. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shi, N., Wang, R., Yu, T. et al. PfPI3K inhibition reversed artemisinin resistance by increasing ROS and ubiquitinated protein levels in Plasmodium falciparum. Commun Biol 8, 1661 (2025). https://doi.org/10.1038/s42003-025-09057-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-09057-w