Abstract
Serving as critical chromatin-binding effector proteins, the heterochromatin protein 1 (HP1) family possesses distinct biological functions, of which the mechanisms are unclear. Liquid-liquid phase separation (LLPS) of the HP1 family and their interaction with nucleosomes are crucial mechanisms governing chromatin activities. Here, we first investigate phosphorylated HP1α (pHP1α) LLPS by NMR spectroscopy, confocal microscopy, and molecular dynamics simulations. This reveals that the phosphorylated N-terminal extension (NTE) and hinge region are key determinants of pHP1α LLPS. We next explore the interactions between pHP1α and telomeric nucleosome arrays and a relatively more open Widom 601 nucleosome array under LLPS conditions. The results suggest that the interaction of pHP1α and nucleosomes is driven by the self-association of pHP1α mediated by intermolecular interaction between the NTE and hinge regions and the specific binding of the pHP1α chromo domain (CD) to the N-terminal histone tail of H3K9me3 in the nucleosomes. In addition, experiments elucidate that DNA can mediate the pHP1α LLPS by competing with the NTE to interact with hinge regions. Together, our findings provide atomic-level insights into the determinants of the molecular features driving the self-association of the pHP1α and the interactions with DNA and nucleosomes.
Similar content being viewed by others
Introduction
Chromatin-mediated gene activities are regulated by factors such as effector proteins and post-translational modifications (PTMs)1,2,3,4,5,6. The heterochromatin protein 1 (HP1) family are effector proteins that serve as a crucial component of heterochromatin, possessing diverse functions in gene regulation, DNA damage response, and cell differentiation in eukaryotic cells7,8,9,10,11. In humans, the HP1 family consists of three highly conserved proteins: HP1α, HP1β, and HP1γ. All three HP1 family members share a similar modular architecture, comprising two conserved domains, the N-terminal chromo domain (CD) and the C-terminal chromoshadow domain (CSD), which are flanked by three disordered regions, including the N-terminal extension (NTE), the hinge region, and the C-terminal extension (CTE). Among them, the hinge region contains patches of positively charged residues capable of binding DNA12. HP1α is widely distributed at heterochromatin regions, with its CD domain specifically recognizing histone H3 lysine 9 tri-methylation (H3K9me3), an epigenetic mark frequently associated with maintaining higher-order chromatin and locally silencing gene expression6,13. In-vivo investigations previously have demonstrated that the recruitment of HP1α and HP1β to an ectopic locus can trigger extensive gene silencing, spanning several kilobases of DNA in an active state14. The CSD domain mediates homodimerization and interactions with other proteins via a PXVXL motif, with dissociation constant values revealed at the nanomolar level for the CSD dimer of both HP1α and HP1β15,16,17,18. The interaction of HP1β with chromatin harboring H3K9me3 also requires dimerization, appropriate spacing of the CD, as well as general stabilization of the N-terminal region19,20. Notably, HP1β plays roles in both gene silencing and gene activation21,22. Previous investigations have also established that HP1β phosphorylation at Threonine-51 serves a critical function in the initiation phase of DNA damage response pathways, as inhibition of casein kinase 2 activity results in compromised HP1β recruitment to DNA lesions and subsequent impairment of downstream repair mechanisms23. In addition, HP1γ is distributed in both euchromatin and heterochromatin24,25. Phosphorylation of HP1γ at Serine-83 has been identified as a distinct euchromatic marker that facilitates interaction with the regulatory factor Ku70; this phosphorylation of HP1γ exhibits attenuated transcriptional repression capabilities and preferentially associates with regions undergoing active transcriptional elongation26. Although many studies have focused on the HP1 family, particularly HP1α, the distinct biological roles and related molecular mechanisms of the phosphorylated HP1α remain underexplored.
Chromatin exhibits a unique dynamic feature at the mesoscale, namely, undergoing liquid-liquid phase separation (LLPS) with or without binding proteins27,28,29. This phenomenon has been extensively studied over the past five years and is believed to provide dynamic, isolated compartments for gene activities30,31. Studies have unveiled that reconstituted chromatin can form liquid-like condensates under physiological salt conditions, driven by electrostatic interactions and a crucial role of histone tails32. These droplets exhibit tunable properties modulated by nucleosome plasticity33, linker DNA length and patterning, the presence of linker histone H1, and PTMs, ionic conditions, and chromatin fiber characteristics (such as fiber length and nucleosome repeat length)29,32,33,34,35,36,37,38. Intriguingly, an increasing number of chromatin-binding proteins like HP1α and methyl-CpG-binding protein 2 (MeCP2), have been recognized for their ability to undergo LLPS within heterochromatin regions39,40,41, a phenomenon implicated in the regulation of various gene processes. MeCP2 plays a pivotal role in heterochromatin organization through an oligomerization-based phase separation mechanism and can undergo the LLPS process that is facilitated by crowded environments and the presence of DNA39,42. LLPS has been demonstrated to drive the compartmentalization of heterochromatin, such as the formation of pericentric heterochromatin foci, which can be mediated by HP1α40,43. Previous studies have demonstrated that HP1α can undergo LLPS modulated by post-translational modification, specifically phosphorylation of its NTE, or the presence of DNA and nucleosomes44,45. In human and mouse cells, N-terminal serine residues (S11–14) of HP1α is also phosphorylated, modulating the protein function and localization, and phosphorylation of the N-terminal serine residues confers a stronger binding affinity for H3K9me3, leading to enhanced chromatin binding46. A nuclear magnetic resonance (NMR) study of a truncated HP1α peptide, which includes just the N-terminal region, revealed that phosphorylation does not perturb the overall tertiary structure; however, it appears to diminish the conformational dynamics of the NTE47. The phosphorylation of HP1α can affect its phase separation behavior by modulating its conformational dynamics and the charge distribution, which is the main factor driving its LLPS.
Furthermore, it has been reported that nucleosome arrays in conjunction with the phosphorylated HP1α (pHP1α), can give rise to a new liquid phase capable of recognizing methylated histone H3 tails45. HP1α can undergo phase separation with nucleosomes, and its N-terminal was hypothesized to be the key component driving the inter-molecular contacts44. However, questions remain regarding the key molecular features that contribute to the distinct LLPS behaviors of the pHP1α and the relevant interactions involved in phase separation with nucleosomes. Recent investigations have leveraged the power of solid-state and solution-state NMR spectroscopy to elucidate, at atomic resolution, the intricate interplay between nucleosome-protein interactions, nucleosome-DNA interactions, and the modulation of chromatin dynamics48,49,50,51,52,53,54,55,56.
The HP1α protein has been shown to be essential for normal telomere function in Drosophila, such as capping and control of telomere elongation57,58. The H3K9me3 mark is prevalent at telomeres59. However, information on the HP1α-nucleosome interaction in telomeric chromatin is almost completely lacking. Recently, a unique columnar structure of telomeric chromatin in the condensed, highly compact form of closely stacked nucleosomes was discovered60. We hypothesize that due to this compact form of telomeric DNA, its interaction with HP1α is likely different from that of canonical chromatin represented by nucleosome arrays with a Widom 601 DNA sequence, which adopts a zig-zag conformation in its condensed form.
In this work, we aim to explore the interaction between pHP1α and chromatin fibers with different compacting levels under LLPS conditions at the molecular level, as well as the contribution of the histone-free DNA in the LLPS condensation. Using NMR and molecular dynamics (MD) simulations, we first thoroughly characterize the structural features of full-length pHP1α corresponding to KCl-induced reversible phase separation of the protein (in the absence of chromatin), which revealed the interaction between the phosphorylated NTE and the hinge region. We then establish the formation of pHP1α LLPS in the presence of telomeric nucleosome arrays under conditions in which the CD region is exposed, and the protein displays increased interactions with nucleosomes, greatly promoted by the CD domain and the interaction between the phosphorylated NTE and the hinge region, which was further characterized by NMR. We compare with the NMR structural information of the pHP1α interaction with the canonical Widom 601 nucleosome arrays. We also elucidate the potential roles of pHP1α interaction with free DNA in regulating chromatin condensation. This study establishes comprehensive atomic-level features of pHP1α and the molecular interactions with nucleosomes governing LLPS, thus providing valuable clues into the formation and maintenance of heterochromatin and its diverse functional significance.
Results
Phosphorylation of HP1α does not change the protein structure
We co-expressed HP1α and CKII (Fig. S1), yielding the phosphorylation of the N-terminal serine residues (S11-14) (Fig. S1c) of the protein, and performed NMR experiments to obtain the pHP1α structure information. The full-length pHP1α (Fig. 1a, b) was observed in complementary triple-resonance NMR experiments, and partial 1H, 13C, and 15N resonance assignments were obtained (deposited in the biological magnetic resonance data bank under entry 52976). High-resolution solution-state NMR data were acquired, as exemplified by the spectrum in Fig. 1c. A suite of 2D 1H-15N HSQC, 3D HNCO, HN(CA)CO, HNCA, HN(CO)CA, and HNCACB spectra were collected to facilitate the backbone chemical shift assignments of pHP1α residues, with the representative of CCPNmr strip plots shown in Fig. S2. The exceptional quality of the NMR spectra enabled sequential resonance assignments for the complete NTE, CD domain, and CTE. However, most residues of the hinge region and CSD domain were absent in the NMR spectra due to the unfavorable relaxation properties of the pHP1α dimers. The overlapping peaks with a large dynamic range of peak intensities in the middle of the NMR spectrum presumably correspond to the variable and disordered segments of pHP1α.
With the resonance assignments, we obtained the secondary structure of pHP1α using TALOS-N61, and the result is shown in Fig. 1d. The N- and C-terminal regions (Y177-S191) exhibit randomly coiled structures, the CD (amino acids Y20-W41) adopts β-sheet structures, and the region proximal to the hinge region is predominantly α-helical. The hinge region (K71-D112) is characterized by a mixture of α-helical and random coil structures. Overall, the phosphorylation of HP1α does not alter its structure significantly, which is consistent with the secondary structure of truncated HP1α peptides without phosphorylation as presented in the Protein Data Bank (PDB)47,62. The 2D HSQC spectra were acquired for both pHP1α and HP1α. Comparison of the two spectra (Fig. S3) revealed excellent peak overlap for the majority of residues, except for a few peaks that belong to residues of or near the phosphorylated sites. As HSQC is highly sensitive to tertiary structures, these results indicate that the phosphorylation does not change the overall structure of HP1α. Next, using those NMR peak assignments, we delineated the interaction of pHP1α self-association and the association with chromatin fibers and DNA in a site-specific manner in LLPS environments.
The interactions between the NTE and the hinge region are the primary driving factors for pHP1α self-condensation
Our previous studies have shown that pHP1α exhibits a pronounced LLPS behavior at various protein and salt concentrations63. We first tracked the phase separation behavior of pHP1α in the presence of the monovalent KCl salt. Optical microscopy showed that pHP1α spontaneously displayed liquid droplets, which dissipated concomitantly as the KCl concentration increased (Fig. 2a). Fluorescence recovery after photobleaching (FRAP) studies (Fig. 2b, c) verified the dynamics of droplets formed by pHP1α. At 150 mM KCl, where droplets are disrupted, the protein is expected to be in a relatively native state, allowing the resolution of site-specific NMR peaks. We investigated NMR signals and chemical shifts of 15N labeled pHP1α under LLPS and non-LLPS conditions (Fig. 2f–h). The 2D 1H − 15N HSQC spectrum exhibited a significant uniform reduction in intensities for all residues, indicative of the gel-like state characterized by transient oligomerization or clustering of pHP1α in the absence of KCl. In particular, the phosphorylated serines (pSer) at the NTE, and residues in the CD and hinge regions showed a decrease in intensity, suggesting their involvement in intermolecular interactions within the condensed phase. As expected, the intensity of residues at 75 mM KCl is higher than in the KCl-free state but lower than at 150 mM, implying that the domains are partially restricted by interactions in this intermediate ionic strength regime. This agrees with that, although LLPS phenomena were observed at KCl concentration less than 150 mM KCl, pHP1α molecules exhibit distinct molecular motions at different ion strengths as revealed by our solid-state NMR study of the protein condensates63.
a Representative optical microscopy images depict the LLPS of pHP1α (150 μM) at varying KCl concentrations (0, 75, 150, 300 mM). Statistical analysis of droplet area fraction is shown in Fig. S4. The scale bar is 10 μm. b Fluorescence intensity recovery of pHP1α after photobleaching. Scale bar: 2 μm. c Quantification of FRAP data for images in (b). Bleaching event occurs at t = 0 s. Data plotted as means ± SD. d MD simulations snapshot at 80 ns under 30 mM KCl. Two different representations are shown with the particle representation on the left and cartoon representation on the right. A, B, C are monomers of pHP1α. e The involvement of pSer residues and basic residues at the intermonomer interfaces obtained from contact map analyses of simulations with 30 mM KCl. f Overlay of 2D HSQC spectra of 1H-15N-pHP1α at 150 mM KCl concentration (grey) and at 75 mM (light blue), 0 mM (dark blue) KCl concentration. g The relative peak intensity ratios in pHP1α from NMR 1H-15N HSQC experiments are shown under different KCl concentrations (0, 75 mM, 300 mM, and 500 mM). Red asterisks represent disappearing peaks. The solid line represents the average of the intensity ratios, while the dashed lines represent one standard deviation above. h The averaged CSPs of resonances in 2D HSQC spectra of uniformly 15N-labeled pHP1α are presented under the influence of KCl concentrations. The solid line represents the average of the CSPs, while the dashed lines represent one standard deviation above.
Notably, as KCl concentration increases to 300 mM, we observed a progressive increase in peak intensities. This implies that the coulombic interactions between pHP1α and K+/Cl− ions become dominant, disrupting the intricate electrostatic network that stabilizes the condensed phase. Although spectral overlap prevented full assignment of some residues, the resolved residues in 2D 1H−15 N HSQC spectra show a progressive increase in chemical shift perturbations (CSPs) with increasing KCl concentration. Upon scrutinizing the residue profiles, we found that a number of charged and aromatic amino acids (such as Arg, pSer, Lys, Asp, Trp, Val, Ile, and Tyr) in the NTE, CD, and hinge regions either disappeared or underwent intensity and CSP changes during phase separation. Among those, significant CSPs were observed for residues including K6, R7, pS11-14, and D16 in the NTE, K69 sitting near the beginning of the hinge region, and K72, R90, D99, and K102 in the hinge regions. In addition, aliphatic and aromatic residues, Y20, V21, H48, W51, and Y70, showed less pronounced CSP changes, potentially indicating their role in forming hydrophobic cores and contributing to LLPS through π-π stacking and cation-π interactions64,65. Remarkably, CSPs of pSer residues in the NTE were nearly four times higher than those in the CD, accompanied by substantial intensity changes. This suggests that negatively charged pSer residues in the NTE are restricted by surrounding positively charged amino acids (Arg and Lys) in the hinge region, forming electrostatically stabilized complexes that are disrupted at high ionic strength. At high pHP1α concentrations, LLPS can readily occur at low salt concentrations, as previously reported44, and pHP1α at lower concentrations still exhibits LLPS propensity, with pSer residues and the positively charged amino acids in the hinge region, along with sparsely distributed hydrophobic residues, acting as adhesive elements that promote phase separation. These interactions are disrupted by high KCl concentrations, confirming the electrostatic nature of the phase separation process. Taken together, the NMR data suggest that full-length pHP1α undergoes conformational changes during phase transition, which depends on phosphorylated NTE and hinge region. The pSer patch increases multivalency and promotes LLPS through diverse intermolecular interactions.
As some residues in the NMR spectra cannot be fully resolved due to peak overlapping, we further explored the intermolecular contacts of pHP1α in the condensed state by performing MD simulations as a complementary approach, corroborating the low salt LLPS to investigate the self-association process among multiple pHP1α monomers, and further conducted contact analyses. The 3D structure of pHP1α monomer was predicted by AlphaFold and then further calculated (Fig. S5a). Based on the 100-ns MD simulations, oligomers containing three pHP1α monomers are obtained after about 30 ns (see time series and simulation snapshots in Fig. S5), with phosphorylation of the hydroxyl amino acid serine shown in the right panel in Fig. 2d at the 80 ns snapshot. At a low KCl concentration (30 mM), electrostatic attraction occurs between E17, pS13 around the NTE of the pHP1α monomer A (red) and K84, K104, and K105 in the hinge region of the pHP1α monomer B (grey) (Fig. 2e and contact maps in Fig. S6). Additionally, the negatively charged phosphate groups of pS12-13 in the NTE of the pHP1α monomer C (orange) establish electrostatic interactions with R82 and R115 in or near the hinge region of the pHP1α monomer A, respectively (Fig. 2e). The observed interaction of pSer with positively charged residues at the inter-molecular interfaces from the MD data agrees with the NMR data based on CSPs of pSer residues at different KCl concentrations, suggesting that pSer in NTE and residues in the hinge region are involved in intermolecular interactions within the condensed phase.
The present 100-ns all-atom MD simulation likely only captured limited contacts for this self-association, as longer simulation times are required to ensure adequate conformation sampling, which hinges on future advances in hardware and methods66,67. Coarse-grained MD simulations have been developed recently to study the phase behavior of proteins, including the HP1 family. The combination of all-atom and coarse-grained MD simulations suggested that the KSKKKR (aa. 102-107), KRK (aa. 89-91) and KKYKK (aa. 68-72) in the hinge region can interact with many negatively charged patches such as those in the NTE (EDEEEE, aa. 15-19) and CTE regions; however, when the protein is phosphorylated, pS11-14 are the more dominant residues interacting with those positively charged patches in the hinge region17. Residues identified by our NMR and MD studies as critical for LLPS largely coincide with these patches. Therefore, LLPS of pHP1α is resulted from a network of dynamic cooperative contacts between several patches, dominated by those in the NTE and hinge region, which is also suggested by another theoretical study68. Furthermore, it has been recently recognized that several systems, including pHP1α, of which protein molecules in the LLPS droplets form clusters exhibiting complex and heterogeneous dynamical states63,69. The question remains about the contributions of different multivalent contacts in such systems to the formation and phase properties of the LLPS droplets. This can be addressed by future studies with assessment of the detailed dynamical states and the involved cooperative interactions in the LLPS processes using a combination of approaches such as NMR, proper mutagenesis, and super-resolution imaging methods.
Overall, the above experiments suggest that the NTE and hinge region form dominant interactions leading to self-association of pHP1α. We next set to elucidate the interactions between pHP1α with nucleosome arrays of different compaction states, as well as with DNA in LLPS environments.
Condensation of pHP1α and chromatin fibers is driven by self-association of pHP1α mediated by the NTE and hinge regions, and the specific interaction of pHP1α CD with H3K9me3 nucleosome array
Previous studies have demonstrated that HP1α can potentially undergo phase separation with nucleosomes41. Solid-state NMR also demonstrated that pHP1α interacting with nucleosomes does not cause major rearrangements to the nucleosome core at the LLPS state45, but there is a lack of experimental analysis on the binding manner of pHP1α to nucleosomes. Here, to elucidate the molecular features that contribute to this process, we set to explore the molecular interactions between pHP1α and nucleosome arrays in a LLPS environment. We first employ confocal microscopy to examine the phase separation of pHP1α with nucleosome arrays containing telomeric DNA and H3K9me3. The successful installation of H3K9me3 modification was confirmed by mass spectrometry (Fig. S7). Compared to the H3K9C precursor (Fig. S7a), the alkylated product exhibited a characteristic mass shift (Fig. S7b), corresponding to the expected increase of mass upon installation of the trimethyl-lysine analog, confirming that trimethylation was installed on the majority of H3K9C residues in the nucleosome arrays. As shown in Fig. 3a, confocal microscopy analysis revealed that adding pHP1α to the 10×157 bp telomeric nucleosome array (Fig. S8) solution induces the formation of liquid droplets. The sizes of droplets escalated commensurately with increasing pHP1α: telomeric nucleosome array ratio, reaching maximal extent at a pHP1α: telomeric nucleosome array stoichiometric ratio of 10. The dynamic mobility of these condensates was also observed by the two-color mixing assay using confocal microscopy. The nucleosome arrays with either Alexa Fluor 488 or 647 dye were incubated to form droplets. Subsequently, the two samples were combined and analyzed by confocal microscopy. The complete fusion of the red and green droplets was observed, suggesting the fluidity of the formed droplets.
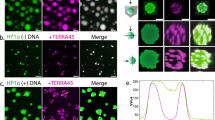
a Dual-color fluorescence images of pHP1α-mediated telomeric nucleosome array condensation. Telomeric nucleosome arrays were labeled with Alexa Fluor 488 (green, top) and Alexa Fluor 647 (red, bottom). Images show mixed labeled arrays at increasing pHP1α:nucleosome ratios (0-10) in 100 mM KCl. Scale bar: 20 μm. b Overlay of 2D HSQC spectra of 1H-15N-pHP1α in absence (grey) and in presence of telomeric tetra-nucleosome array (pHP1α:nucleosome ratios (37.5:1) is green and pHP1α:nucleosome ratios (10:1) is yellow). Telomeric tetra-nucleosome array induced relative peak intensity change (c) in pHP1α at 75 mM KCl. Red asterisks represent disappearing peaks. The solid line represents the average of the intensity ratios, while the dashed lines represent one standard deviation above. Averaged 1H,15N CSP of resonances in 2D 1H-15N HSQC spectra of uniformly 15N-labeled pHP1α (d), at increasing pHP1α: telomeric tetra-nucleosome array molar ratios (250:1, 125:1, 37.5:1, 10:1). The solid line represents the average of the CSPs, while the dashed lines represent one standard deviation above.
We then further investigated the interactions between telomeric nucleosome arrays with pHP1α using NMR (Fig. 3b–d). While titrating with the telomeric nucleosome array, the pHP1α NMR HSQC signals were attenuated significantly at a ratio of 1:37.5, likely attributed to the specific binding of the CD domain with H3K9me3. Concomitantly, significant CSPs in the CD region were observed, further confirming the specific recognition of H3K9me3 in nucleosomes by the CD domain of pHP1α. The very variable regions are mainly oriented around Y20, W41, K42, W51, which in accordance with the notion that HP1α interacts with chromatin mainly through the cation-π interaction of the aromatic “cage” with H3K9me370,71. Additionally, phosphorylation of the serine residues in the NTE of HP1α decreases the dynamics of the NTE, causing a more extended string-like conformation47. In the NMR HSQC spectrum, the relative change in intensity and CSPs of the NTE and hinge region may indicate a weakly dynamic interaction between the N-terminus in a more extended conformation and the hinge region of neighboring pHP1α. This electrostatic interaction may participate in the compaction of a tightly stacked telomeric nucleosome array under the phase separation environment.
A previous cryo-EM study revealed that HP1 forms a symmetric dimer linking two nucleosomes containing H3K9me3 without directly interacting with the linker DNA between the nucleosomes72. Therefore, the packing between different nucleosome core particle (NCP) units in the arrays may potentially impact the interactions with pHP1α. As revealed by cryo-EM studies, unlike canonical nucleosome arrays adopting a conventional zig-zag arrangement of the NCP units, telomeric nucleosomes form a columnar structure, giving its different packing between nucleosome units and distinct conformational dynamics properties60,73. Thus, in a solution, in comparison with conventional nucleosome arrays, telomeric nucleosomes potentially are more tightly stacked in the columnar arrangement. Therefore, we investigated whether the interactions between phosphorylated HP1α and telomeric nucleosome arrays extend to conventional nucleosome arrays exhibiting varying degrees of compaction. The addition of pHP1α to 12 × 177 bp Widom 601 nucleosome arrays (Fig. S9) induced condensate formation, as demonstrated by two-color mixing assays (Fig. S10). While titrating isotopically labeled pHP1α with 12 × 177 bp Widom 601 nucleosome arrays harboring H3K9me3, the NMR signals of pHP1α indicate a stabilized interaction with 12 × 177 bp Widom 601 nucleosome arrays (Fig. S11), which is generally consistent with the previous observations for the telomeric arrays. Interestingly, the signal intensity of the pHP1α CD domain starts decreasing even at the remarkably low ratio of nucleosome arrays, 250:1. As expected, we observed a gradual decrease in all peak intensities with the added nucleosome arrays, indicating the formation of large pHP1α-NCP array complexes. The uniform reduction in the intensity of residues 20-70 confirms that pHP1α interacts with the H3K9me3 through its CD domain. Furthermore, we discovered that some peaks in the CD domain such as V21, E23, L26, K32, L39, W41, K42, G43, E52, D58, M67, and Y70, vanished when the pHP1α and the nucleosome array were in a ratio of 37.5:1. Most of the assigned peaks in the CSD region also disappeared upon binding the nucleosome array. The NMR data prove that pHP1α can exert binding effects on nucleosomes through critical segments in the CSD domain and mainly depends on the CD region. In contrast to titration with telomeric nucleosome arrays, the addition of Widom 601 nucleosome arrays to pHP1α elicits relatively modest changes in intensity and CSPs within the NTE and hinge region. As a putative explanation that is not in conflict with this observation, we propose that the self-interactions within pHP1α, mediated by the NTE and hinge regions, are relatively weaker during the compaction of the more relaxed Widom 601 nucleosome arrays compared to the more tightly stacked telomeric nucleosome arrays.
Histone-free DNA mediates pHP1α condensation mainly through competing with the phosphorylated NTE to interact with the hinge region
When HP1 proteins bridge nucleosomes in chromatin fiber, the histone-free DNA is available for interacting with additional HP1 molecules, which is potentially important for forming an intermolecular network in LLPS72. HP1α has also been reported to also exhibit LLPS phenomena in the presence of short DNA at low stoichiometric ratios, which is driven by electrostatic interactions between the hinge region and DNA74. However, introducing local negative charges through a protein phosphorylation of NTE can modulate DNA binding affinity, positively or negatively75,76. Previous studies have also found that the wide type (WT) HP1α-DNA interaction mainly depends on the electrostatic attraction between the positively charged side chains in the NTE of HP1α and the negatively charged backbone phosphates of DNA71. We therefore wondered how DNA interactions, modulated by phosphorylation, affect pHP1α dynamics in this system.
Optical microscopy observation showed that the addition of DNA perturbs the LLPS propensity of pHP1α (Fig. 4a). To further delineate the interactions that mediate this LLPS attenuation, we then analyzed the changes in HSQC peaks of pHP1α in the presence of different ratios of 177 bp Widom 601 DNA (Fig. 4b–d). Unlike DNA-induced overall stronger signal attenuation in WTHP1α71, the affected regions in pHP1α mainly include positively charged amino acids K6, R7, K42, K69, K71, K80, the negatively charged residues pSer, E15, D16, E23, and the aromatic residues Y20 and W51. The DNA-induced large CSPs and slight signal intensity attenuation observed for NTE suggest that the NTE is mostly affected in this DNA-mediated protein LLPS. The introduction of four consecutive phosphorylations on Ser residues results in a tighter binding of the NTE region with the positively charged hinge regions to drive the protein condensation. This intermolecular interaction can be attenuated by additional DNA through competitively binding to the hinge region, thus leading to the disruption of the LLPS formation. The pSer and negatively charged residues in the NTE directly block the electrostatic attraction or π–cation interactions between DNA and side chains of Arg/Lys residues in the pHP1α hinge region77. In addition, the pronounced CSPs and intensity changes observed for the positively charged amino acids in the NTE are likely due to that this region is involved in electrostatic interactions with the DNA.
a Light microscopy image of pHP1α droplets formed with different DNA concentrations. Scale bar is 20 μm. b Overlay of 2D HSQC spectra of 1H-15N-pHP1α in absence (grey) and in presence of Widom 601 177 bp DNA (blue and yellow). c DNA induced relative peak intensity change in pHP1α. Red asterisks represent disappearing peaks. The solid line represents the average of the intensity ratios, while the dashed lines represent one standard deviation above. d Averaged 1H,15N CSP of resonances in 2D 1H-15N HSQC spectra of uniformly 15N-labeled pHP1α, at increasing pHP1α:DNA molar ratios (125:1, 10:1, 6:1, 2:1). The solid line represents the average of the intensity ratios, while the dashed lines represent one standard deviation above.
Non-specific interactions between pHP1α and DNA are considerably weaker than protein-protein interactions; therefore, the observed intensity changes are relatively modest compared to the pure pHP1α aggregation state. Overall, the pHP1α condensation is suppressed with the presence of DNA. The histone-free DNA can compete to form electrostatic attractions with positively charged amino acids in the NTE and hinge regions, as well as aromatic residues (Y, W) in the CD region, resulting in the disruption of pHP1α self-association.
Discussion
Recent studies have extensively investigated the interactions between HP1 family proteins and chromatin, revealing their ability to form dynamic condensates both in vitro and in vivo. The canonical mechanism for these interactions involves HP1 dimerization through its CSD domain, coupled with binding to H3K9me3 histone tails via its CD domain, establishing a multivalent binding network that promotes phase separation78,79,80,81,82,83. Importantly, heterochromatin components such as nucleosomes and DNA preferentially partition into HP1α droplets, while transcriptional machinery proteins like TFIIB are excluded, suggesting selective molecular partitioning that reinforces heterochromatin function79. But beyond this, emerging research has identified additional proteins that participate in HP1α-mediated heterochromatin phase separation. In-vivo studies have revealed that 53BP1 forms nuclear punctate structures with HP1 family proteins that partially colocalize with heterochromatin markers84. Notably, 53BP1 and HP1α exhibit a mutually dependent relationship in liquid droplet formation. This cooperative behavior suggests a novel regulatory mechanism for heterochromatin organization. Similarly, the transcription factor Prospero (Pros) undergoes LLPS and is retained at pericentromeric heterochromatin regions in dividing neural precursors, where it specifically interacts with the N-terminal portion of HP1α. This interaction enables Pros to remodel H3K9me3-positive heterochromatin by recruiting HP1α, thereby driving terminal neuronal differentiation85. The N-terminal region of HP1α has emerged as particularly significant for phase separation behavior. This region contains key phosphorylation sites that modulate HP1α‘s capacity for LLPS and its interactions with other proteins. The post-translational modification status of this domain may therefore represent a critical regulatory mechanism for heterochromatin organization and function. Collectively, these findings highlight the complex interplay between HP1 proteins, chromatin, and various nuclear factors in establishing phase-separated heterochromatin, revealing how biophysical properties of proteins influence chromatin organization and cell fate decisions.
In this study, we have elucidated key molecular determinants governing the phase separation behavior and interactions of pHP1α and telomeric as well as canonical (represented by Widom 601 nucleosome arrays) chromatin in an LLPS environment. The phosphorylated NTE and the hinge region play critical roles in KCl-mediated reversible LLPS of pHP1α through electrostatic interactions. Notably, the CD domain facilitates specific recognition and binding to H3K9me3-containing nucleosomes, while the NTE and hinge region regulate the compaction of distinct chromatin configurations. Interestingly, we observed that introducing phosphorylated serine residues in the NTE inhibits non-specific interactions with DNA. This finding implies that phosphorylation of the NTE may modulate the affinity of pHP1α for different chromatin, potentially influencing its function. Based on our findings, we proposed the interaction modes between pHP1α with nucleosomes (for both Widom 601 nucleosomes and telomeric nucleosomes) in the context of LLPS, as illustrated in Fig. 5. We also found that the unique columnar structure of telomeric chromatin facilitates a stronger interaction with pHP1α compared to the Widom 601 DNA nucleosome arrays under LLPS conditions. This might be attributed to the orientation of H3 tails in the columnar stacks that are close to each other60. Such close proximity of the trimethylated H3 tails would bring the two pHP1α bound to the individual tails in close proximity to each other and facilitate the observed stronger interaction. We further postulate that this stronger interaction might contribute to the maintenance of the telomere cap. This study provided valuable insights into the molecular basis underlying the diverse functions of the HP1α in DNA interaction. The elucidation of the distinct LLPS behaviors of pHP1α and their interactions with chromatin and histone-free DNA contributes to a deeper understanding of the nuanced mechanisms governing higher-order chromatin structure and epigenetic regulation. Future studies await the exploration of the detailed molecular information for HP1β and HP1γ interacting with nucleosomes to further understand their distinct biological activities.
Created in BioRender. Shi, X. (2025) https://BioRender.com/n588571.
Methods
Preparation of histone proteins
Human histone proteins (H3, H4, H2A, and H2B) were expressed in E. coli BL21 (DE3) cells transformed with plasmids encoding the respective genes. The transformed cells were grown in LB medium supplemented with appropriate antibiotics at 37 °C until the culture reached an optical density of 0.6–0.8 (OD 600). Protein expression was induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at 18 °C for 16 hours. The cells were harvested by centrifugation at 5000 × g for 10 minutes at 4 °C. The harvested cell pellets were resuspended in lysis buffer containing 50 mM Tris-HCl (pH 7.5), 500 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 5 mM Beta-mercaptoethanol (BME). Cells were lysed by sonication on ice (10 seconds on, 10 seconds off, 5 minutes). The lysate was clarified by centrifugation at 18,000 × g for 30 minutes at 4 °C. The resulting pellet containing inclusion bodies was washed twice with washing buffer (50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 1% Triton X-100, and 1 mM EDTA). After washing, the pellet was resuspended in DMSO and solubilization buffer containing 7 M urea, 20 mM Tris-HCl (pH 7.5), 10 mM BME, 200 mM NaCl and 1 mM EDTA (SAUDE-200 buffer). The mixture was incubated with gentle stirring at room temperature for 1 hour to solubilize the histone proteins. Aggregates were removed by centrifugation at 20,000 × g for 10 minutes. The histones were purified using fast protein liquid chromatography (FPLC) on a HiLoad Superdex 75 size-exclusion column (SEC).
Histone H3 C9 trimethylation
Histone H3 was dissolved in 4.9 mL alkylation buffer (6M Guanidinium Hydrochloride (Gdn·HCl), 1 M HEPES, pH 7.8, 10 mM D/L Methionine) to a final concentration of 8 mg/mL. Disulfide bonds were reduced by introducing 100 μL of 1 M dithiothreitol (DTT) to the solution. Subsequently, 300 mg of (2-bromoethyl) trimethylammonium bromide was added, and the reaction mixture underwent incubation at 50 °C with periodic mixing. 100 mg of (2-bromoethyl) trimethylammonium bromide and 50 μL of 1 M DTT were supplemented, and the reaction was allowed to proceed for 3 more hours. The reaction was then quenched by adding 50 μL of neat BME and incubating for 30 minutes. For purification of the trimethylated histone, the reaction mixture was dialyzed overnight against distilled water containing 0.1% trifluoroacetic acid (TFA) at 4 °C.
Preparation of 4×157 bp and 10×157 bp telomeric DNA and 12 × 177 Widom 601 DNA
Competent SURE2 colonies containing the puC57 plasmid harboring telomeric 10x or 4 × 157 bp DNA was grown in 15 L of TB media and incubated overnight at 30 °C in an orbital shaker at 220 rpm. Cultured cells were harvested at 12,000 g for 10 minutes at 20 °C using a Beckman-Coulter Allegra J-25 Centrifuge and 9.1-JLA rotor (Beckman-Coulter, USA). The plasmid was isolated using the alkaline-lysis and phenol-chloroform isoamyl alcohol (CIA) extraction method86. A 37 °C overnight test digestion was performed with 75 U/mg of EcoRV-HF, Dpn1, and HaeIII on the extracted DNA (2000 ng/µl). Appropriate polyethylene glycol (PEG) was used for large-scale precipitation of the DNA to separate DNA templates from the vector backbone. Centrifugation was done at 10,000 × g for 10 minutes. The pellet obtained was dissolved in TE buffer. Ion exchange chromatography (IEX) was performed using ÄKTA pure and MonoQ 5/10 GL Column (Cytiva, Sweden). The plasmid containing 12 × 177 bp Widom 601 DNA was digested with EcoRV to release the DNA fragment, followed by purification using PEG precipitation. The purified DNA was resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) and quantified by UV absorbance at 260 nm. For nucleosome reconstitution, the DNA was ethanol-precipitated and resuspended in TE buffer to the desired concentration.
Histone octamer refolding
Histone octamers were assembled by mixing histones H3, H4, H2A, and H2B in equimolar amounts. The histones were first dissolved in unfolding buffer (10 mM Tris-HCl, 7 M guanidinium hydrochloride, 10 mM DTT, pH 7.5) and then were extensively dialyzed at 4 °C against refolding buffer containing 20 mM Tris-HCl (pH 7.5), 2 M NaCl, 1 mM EDTA, and 5 mM BME. Aggregates were removed by centrifugation at 20,000 × g for 10 minutes, and the supernatant containing properly folded histone octamers was used for further applications. The refolded histone octamers were purified using SEC on a HiLoad Superdex 200 column equilibrated with 20 mM Tris-HCl (pH 7.5), 2 M NaCl, 1 mM EDTA, and 1 mM DTT.
Reconstitution of telomeric 10x, 4×157 bp nucleosome arrays and 12 × 177 bp Widom 601 nucleosome array
The telomeric 10x, 4 × 157 bp DNA comprising repeats of the TTAGGG sequence or 12 × 177 bp 601 DNA was mixed respectively with histone octamers in a molar ratio optimized for efficient nucleosome saturation (Figs. S7, S8). Reconstitution was carried out by stepwise salt gradient dialysis starting from 2 M KCl to 10 mM KCl (LiCl was used for telomeric nucleosome array) over 48 hours at 4 °C. The reconstituted nucleosome arrays were purified by centrifugation to remove aggregates and were analyzed by agarose gel electrophoresis.
Preparation of pHP1α
The pNIC28-Bsa4 vector harboring the HP1α construct, and the pET duet-1 vector containing the CK2 kinase construct were co-transformed into the E. coli BL21 (DE3) expression strain. Transformant selection was facilitated by plating the cells on LB agar plates supplemented with kanamycin (100 μg/ml) and ampicillin (100 μg/ml) to maintain the respective plasmids. Isolated colonies were inoculated into a starter culture comprising 20 ml LB broth supplemented with the appropriate antibiotics and incubated overnight at 37 °C with vigorous shaking at 220 rpm. The pre-cultured E. coli BL21 cells were subsequently transferred to 1 L of LB medium containing 1 ml ampicillin and 1 ml kanamycin for large-scale protein expression. Cultivation proceeded at 37 °C with agitation at 220 rpm until the OD600 reached approximately 0.7, indicating mid-logarithmic growth phase. At this stage, IPTG was added to a final concentration of 0.2 mM to induce the expression of the HP1 protein. The induced culture was further incubated for 4 hours at 37 °C with continuous shaking at 220 rpm to facilitate protein production.
Following the expression period, the cell culture was harvested by centrifugation at 7000 rpm for 10 minutes. The supernatant was discarded, and the cell pellets were resuspended in 30 ml lysis buffer (50 mM Tris-HCl, 500 mM NaCl, 1 mM PMSF, 2 mM BME, 10% glycerol, pH 7.5). Cell lysis was achieved through sonication at low amplitude (300 W, with a 5 s on/off cycle, 4 °C) for 1 hour until the solution became homogeneous, transparent, and exhibited reduced viscosity, indicating effective cell disruption. To prevent protein denaturation, the sample containing pHP1α was maintained in an ice-water mixture throughout the purification process. The lysate was subsequently clarified by centrifugation at 17,000 rpm and 4 °C for 25 minutes. A 5 ml Ni-NTA column (1 ml of beads for 30 mg of protein, Qiagen, Hilden, Germany) was equilibrated with 5 column volumes of lysis buffer, and the clarified supernatant was incubated with the Ni-NTA resin for 1 hour at 4 °C to facilitate binding of the polyhistidine-tagged HP1. The mixture was then loaded onto the column, and the unbound fraction was collected. After draining the unbound fraction, the resin was washed with 5 ml of lysis buffer to remove residual contaminants. Subsequently, the column was washed with a buffer (50 mM Tris-HCl, 500 mM NaCl, 2 mM BME, 10% glycerol, 30 mM imidazole, pH 7.5) using at least 5 column volumes to eliminate non-specifically bound proteins. To achieve a higher degree of purity, the column was further washed with a stepwise gradient of imidazole concentrations (100 mM, 200 mM, 300 mM, 400 mM, and 500 mM) in wash buffer (50 mM Tris-HCl, 500 mM NaCl, 2 mM BME, 10% glycerol, pH 7.5), with each wash fraction collected separately. The protein composition and purity of the eluted fractions were assessed using nanodrop spectrophotometry and 15% SDS-PAGE analysis.
The pooled protein sample was subsequently dialyzed against 2 L of buffer containing 25 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, pH 7.5, to remove imidazole and exchange the buffer conditions. The dialyzed sample was then incubated with Tobacco Etch Virus (TEV) protease (Sangong Biotech Co. Ltd., Shanghai, China) to cleave the polyhistidine tag. Further purification was achieved through FPLC using a HiLoad 16/600 Superdex 75 pg SEC (Cytiva, Massachusetts, USA). The column was equilibrated with a running buffer composed of 20 mM HEPES-HCl, 150 mM KCl, 10 mM DTT, pH 7.5, and the purification was performed at a flow rate of 1 ml/min. The purity of the eluted protein fractions was assessed by 15% SDS-PAGE.
Expression and purification of 13C, 15N-pHP1α, 15N-pHP1α
Similar to the expression of non-labeled HP1, cells were first cultivated in 1 L rich LB medium at 37 °C (220 rpm) supplied with ampicillin and kanamycin. When the OD reached about 0.8, the cultivation medium was centrifuged for 10 min at 5000 rpm and 20 °C. The sedimented cells were gently resuspended in 1 L M9 minimum medium (For 13C, 15N-pHP1α, supplemented with 2 g 13C-Glucose, 1 g 15N- NH4Cl, 0.25 x trace metal, 2.5 mL BME vitamins stock solution (Sigma-Aldrich), 2 mM MgSO4 and 0.2 mM CaCl2, for 15N-pHP1α, supplemented with 4 g Glucose, 1 g 15N- NH4Cl, 0.25 x trace metal, 2.5 mL BME vitamins stock solution (Sigma-Aldrich), 2 mM MgSO4 and 0.2 mM CaCl2.). The cells were further cultivated at 37 °C (220 rpm) for 2 h and IPTG was added at a final concentration of 0.2 mM. The cells were cultivated at 37 °C for an additional 4 hours after induction at 220 rpm before harvest. The rest of the cell processing and protein purification was the same as described above.
Sample preparation for solution-state NMR spectroscopy
NMR spectra for comparison of full-length pHP1α and full-length HP1α were acquired at 298 K on 600 MHz Brucker spectrometer. Samples contained 0.25 mM protein in buffer consisting of 20 mM HEPES (pH 7.5), 150 mM KCl, 10 mM DTT, and 10% D2O. NMR spectra for backbone resonance assignment of full-length pHP1α were acquired at 298 K on 800 MHz Brucker spectrometers equipped with 5 mm cryogenic probes. To obtain the chemical shift assignments of pHP1α, several NMR samples were prepared with protein concentrations of about 0.5–0.7 μM in 20 mM HEPES (pH 7.5), 150 mM KCl, 10 mM DTT, 0.04% NaN3 and 10% D2O. The backbone resonances were assigned using the following NMR experiments: 2D 1H-15N HSQC, 3D HNCO, 3D HNCA, 3D HNCOCA, 3D HNCACO, 3D HNCACB. The recorded spectra were analyzed by CCPNmr Analysis version 3.187. Spectra were processed with TopSpin 4.1 (Bruker) and analyzed using Sparky88.
For the KCl titration experiments, samples were prepared in the following way. Freshly prepared pHP1α samples were concentrated to a final volume of 450 μL 150 μM in 20 mM HEPES, (0, 75 mM, 150 mM, 300 mM, 500 mM KCl), 10 mM DTT, pH 7.5. 10% D2O and 0.04% NaN3 were added to the tube and 1H-15N HSQC experiments were obtained. 2D 1H-15N HSQC for 15N-pHP1α were recorded for the proteins (150 μM) with increasing molar ratios of DNA in 20 mM HEPES, pH 7.5, 75 mM KCl, 10 mM DTT and 10% D2O. NMR titrations of 15N-pHP1α in 20 mM HEPES (pH 7.5), 75 mM KCl, 10 mM DTT, and 10% D2O with nucleosome arrays were acquired at 298 K on a Bruker 800 and 600 MHz spectrometers.
NMR data analysis
The NMR spectral data for pHP1α were processed using TopSpin 4.1 and analyzed with CCPNmr 3.1 and NMRFAM-SPARKY software. The quantitative analysis of peak intensities was performed by extracting the peak height for each cross-peak. To establish a reference for comparing the effects of different conditions, the intensity of each cross-peak in the HSQC spectrum was measured for the pHP1α in the absence of any interaction partners (KCl, DNA, and nucleosome arrays). In addition, to meet the requirement of nucleosome array quantity for NMR experiments, we chose to use 4 × 157 bp telomeric nucleosome array harboring H3K9me3 in the NMR titration experiments, as this construct results in superior yield (compared to the 10 × 157 bp telomeric array). For each set of experiments, we measured the intensity of each peak in the control sample containing HP1 protein (I0). Subsequently, titration experiments were conducted by incrementally adding KCl, DNA, 4 × 157 bp telomeric nucleosomes or 12 × 177 bp Widom 601 nucleosomes, and the intensities of the cross-peaks were measured again in the bound state (Ibound). The normalized peak intensity was analyzed by dividing the Ibound values by the corresponding I0 values for each residue, allowing for the identification of residues exhibiting significant intensity changes upon molecular binding.
The chemical shift perturbations (CSPs) of individual residues, which serve as indicators of structural and dynamic changes, were quantified according to the following equation:
where δN and δH are the changes in 15N and 1H dimensions in units of ppm.
The mean and standard deviation of all measured CSPs were calculated to establish significance thresholds. Residues exhibiting CSPs greater than the mean plus one standard deviation were considered significantly perturbed. For intensity changes, the ratio of peak intensities (I/I₀) between bound and unbound states was determined for each residue, and the mean and standard deviation were calculated across all monitored signals. Residues showing intensity changes beyond the mean plus one standard deviation threshold were identified as significantly affected by the binding event, providing complementary information about interaction dynamics and potential conformational changes.
Fluorescence recovery after photobleaching (FRAP) analysis
Microscopy experiments were conducted using a ZEISS Axio Observer 7 + LSM 800 inverted light microscope equipped with 63X oil immersion objectives, enabling high-resolution imaging of the phase-separated condensates. The 384-Well Microscopy Plates (Cellvis, #P384-1.5H-N) were washed and then pre-treated with 1 M NaOH and 5 K mPEG-silane as described in one of our recent studies36. In preparation for imaging, the substrates were repeatedly rinsed with a buffer containing 20 mM HEPES, 5 mM tris (2-carboxyethyl) phosphine (TCEP), pH 7.5, and the buffer was left inside until the samples were ready for addition. Before observing the phase-separated condensates with a microscope, all the buffer in the well was carefully removed, and the sample was quickly transferred to minimize potential evaporation or environmental alterations. After adding the microscope samples to the treated plates, they were sealed with transparent tape to prevent volatilization and maintain the desired sample conditions during the imaging process.
To observe the phase-separated condensates of the pHP1α, cy3-labeled pHP1α protein solutions (0.15 mM) were mixed with varying concentrations of KCl (0, 30 mM, 75 mM, 150 mM, and 300 mM) in a phase separation buffer consisting of 20 mM HEPES at pH 7.5. The samples were thoroughly mixed to ensure homogeneity, and 50 μL were carefully loaded onto a microscope plate well for image collection. All the images were obtained at resolution of 512 × 512 pixels. Quantification of LLPS droplet area fraction for 0.15 mM pHP1α at 75 mM KCl, as shown in Fig. S4. For FRAP experiments, pHP1α condensates were formed with 0.15 mM pHP1α labeling with Cy3 at 75 mM KCl concentration. Samples were photobleached using the 561 nm-excitation laser of the LSM 800 confocal microscope (100% intensity, 50 iterations), and then fluorescence recovery was recorded for 300 seconds with 2-second intervals at 0.2% of the total laser power at room temperature. For quantification, background intensity was subtracted, and each image was normalized to correct for any photobleaching caused by the image acquisition itself. The data represent 3 replicates of the FRAP experiment. The recovery curves were plotted with GRAPHPAD PRISM 10 software.
Liquid–Liquid Phase Separation of pHP1α and nucleosome array
Confocal microscopy (63x magnification) was conducted with Zeiss Laser Scanning Microscope 800 (ZEISS, Germany) by Wayne Shum (research associate). Varying molar ratio of pHP1α to telomeric 10 × 157 bp or 12 × 177 bp Widom 601 nucleosome array were used with two sets of reaction mixtures corresponding to the two types of arrays (AF488-green and AF647-red arrays). To fluorescently label the H2B_T116C histone variant, the cysteine residue was first reduced with TCEP, followed by maleimide-based conjugation with Alexa Fluor 488 or Alexa Fluor 647 fluorophores36. Unreacted dye was removed, and the labeled histones were purified by desalting chromatography and dialysis to ensure optimal preparation for downstream applications. Green LLPS reaction mixtures were transferred to the red LLPS reaction mixtures in the confocal plate for dual-color imaging. 15-minute incubation was done before imaging.
MD simulation
MD simulations of phosphorylated HP1α monomers and the self-association process of three monomers were performed following the same protocol in our previous study63. Briefly, the unphosphorylated HP1α monomer structure was first predicted using AlphaFold structure prediction server and simulated in water under NPT ensemble (303 K and 1 bar) using the atomistic CHARMM27 force-field using GROMACS version 2018.2 MD software for 200 ns89,90,91. The equilibrated structure was then phosphorylated using CHARMM-GUI webserver, and then three copies of the phosphorylated HP1α were placed in a simulation box with water molecules and KCl at 30 mM concentration92,93. Simulation was then carried out under similar NPT conditions for 100 ns. Molecular figures of the oligomer were generated using Visual Molecular Dynamics software94.
Inter-monomer interactions at the residue level were determined from pair-wise contact map analyses using the MAPIYA Contact Map Server95. Contact maps and residues identified to be at the inter-monomer interfaces were presented for the oligomer at 80 ns as inter-monomer electrostatics interaction between monomers converged by 80 ns. Molecular figures were then generated from selected contact map regions within the MAPIYA web interface to show residue sidechains involved in the identified inter-monomer interactions.
Statistical analysis and reproducibility
More than 15 optical microsopy images on average were recorded for each sample. For quantifying pHP1α LLPS droplet area fraction at 75 mM KCl, individual data points represent the area fraction occupied by liquid droplets in each image field (n = 15). For FRAP quantification, background intensity was subtracted, and each image was normalized to correct for any photobleaching caused by the image acquisition itself. The data represent three independent replicates of the FRAP experiments. All statistical analyses were performed with GRAPHPAD PRISM 10 software or Python. The presented mean values and SD were explained in each figure legend.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The chemical shift assignments of pHP1α obtained in this study are available in the BMRB database under entry number 52976. Other data and information can be obtained from the corresponding author on reasonable request.
References
Ruthenburg, A. J., Li, H., Patel, D. J. & Allis, C. D. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8, 983–994 (2007).
Bannister, A. J. & Kouzarides, T. Regulation of Chromatin by histone modifications. Cell Res. 21, 381–395 (2011).
Tang, X. et al. The PTM Profiling of CTCF reveals the regulation of 3D Chromatin structure by O-GlcNAcylation. Nat. Commun. 15, 2813 (2024).
Reinberg, D. & Vales, L. D. Chromatin domains rich in inheritance. Science 361, 33–34 (2018).
Berger, S. L. The complex language of chromatin regulation during transcription. Nature 447, 407–412 (2007).
Kilic, S., Bachmann, A. L., Bryan, L. C. & Fierz, B. Multivalency governs HP1α association dynamics with the silent chromatin state. Nat. Commun. 6, 7313 (2015).
Maison, C. & Almouzni, G. HP1 and the dynamics of Heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 5, 296–304 (2004).
Eissenberg, J. C. & Elgin, S. C. R. HP1a: A structural chromosomal protein regulating transcription. Trends Genet 30, 103–110 (2014).
Ma, J., Hwang, K. K., Worman, H. J., Courvalin, J. C. & Eissenberg, J. C. Expression and functional analysis of Three Isoforms of human Heterochromatin-associated Protein HP1 in Drosophila. Chromosoma 109, 536–544 (2001).
Lee, Y.-H., Kuo, C.-Y., Stark, J. M., Shih, H.-M. & Ann, D. K. HP1 Promotes tumor suppressor BRCA1 functions during the DNA damage response. Nucleic Acids Res. 41, 5784–5798 (2013).
Fraschka, S. A. et al. Comparative Heterochromatin Profiling Reveals Conserved and Unique Epigenome Signatures Linked to Adaptation and Development of Malaria Parasites. Cell Host Microbe 23, 407–420.e8 (2018).
Watanabe, S., Mishima, Y., Shimizu, M., Suetake, I. & Takada, S. Interactions of HP1 Bound to H3K9me3 Dinucleosome by Molecular Simulations and Biochemical Assays. Biophys. J. 114, 2336–2351 (2018).
Utani, K. & Shimizu, N. How transcription proceeds in a large artificial heterochromatin in human cells. Nucleic Acids Res. 37, 393–404 (2009).
Verschure, P. J. et al. In Vivo HP1 targeting causes large-scale chromatin condensation and enhanced histone lysine methylation. Mol. Cell Biol. 25, 4552–4564 (2005).
Richart, A. N., Brunner, C. I. W., Stott, K., Murzina, N. V. & Thomas, J. O. Characterization of Chromoshadow domain-mediated binding of heterochromatin protein 1α (HP1α) to Histone H3. J. Biol. Chem. 287, 18730–18737 (2012).
Mendez, D. L., Mandt, R. E. & Elgin, S. C. R. Heterochromatin Protein 1a (HP1a) partner specificity is determined by critical amino acids in the chromo shadow domain and C-terminal extension. J. Biol. Chem. 288, 22315–22323 (2013).
Her, C. et al. Molecular interactions underlying the phase separation of HP1α: role of phosphorylation, ligand and nucleic acid binding. Nucleic Acids Res. 50, 12702–12722 (2022).
Brasher, S. V. et al. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 19, 1587–1597 (2000).
Hiragami-Hamada, K. et al. Dynamic and flexible H3K9me3 bridging via HP1β Dimerization establishes a plastic state of condensed chromatin. Nat. Commun. 7, 11310 (2016).
Munari, F. et al. Methylation of Lysine 9 in Histone H3 directs alternative modes of highly dynamic interaction of Heterochromatin Protein hHP1β with the Nucleosome. J. Biol. Chem. 287, 33756–33765 (2012).
Festenstein, R. et al. Modulation of Heterochromatin Protein 1 dynamics in primary mammalian cells. Science 299, 719–721 (2003).
Bosch-Presegué, L. et al. Mammalian HP1 isoforms have specific roles in heterochromatin structure and organization. Cell Rep. 21, 2048–2057 (2017).
Ayoub, N., Jeyasekharan, A. D., Bernal, J. A. & Venkitaraman, A. R. HP1-β mobilization promotes chromatin changes that initiate the DNA damage response. Nature 453, 682–686 (2008).
Skourti-Stathaki, K., Kamieniarz-Gdula, K. & Proudfoot, N. J. R-Loops induce repressive chromatin marks over mammalian gene terminators. Nature 516, 436–439 (2014).
Vakoc, C. R., Mandat, S. A., Olenchock, B. A. & Blobel, G. A. Histone H3 Lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell 19, 381–391 (2005).
Lomberk, G., Bensi, D., Fernandez-Zapico, M. E. & Urrutia, R. Evidence for the existence of an HP1-mediated subcode within the histone code. Nat. Cell Biol. 8, 407–415 (2006).
Ulianov, S. V. et al. Suppression of liquid-liquid phase separation by 1,6-Hexanediol partially compromises the 3D genome organization in living cells. Nucleic Acids Res. 49, 10524–10541 (2021).
Liu, Q. et al. The Histone Methyltransferase SUVR2 promotes DSB repair via chromatin remodeling and liquid-liquid phase separation. Mol. Plant 15, 1157–1175 (2022).
Nordenskiöld, L. et al. Liquid-Liquid Phase Separation (LLPS) in DNA and Chromatin systems from the perspective of colloid physical chemistry. Adv. Colloid Interface Sci. 326, 103133 (2024).
Trojanowski, J. et al. Transcription activation is enhanced by multivalent interactions independent of phase separation. Mol. Cell 82, 1878–1893.e10 (2022).
Tong, X. et al. Liquid-liquid phase separation in tumor biology. Signal Transduct. Target Ther. 7, 221 (2022).
Gibson, B. A. et al. Organization of Chromatin by intrinsic and regulated phase separation. Cell 179, 470–484.e21 (2019).
Farr, S. E., Woods, E. J., Joseph, J. A., Garaizar, A. & Collepardo-Guevara, R. Nucleosome plasticity is a critical element of chromatin liquid-liquid phase separation and multivalent nucleosome interactions. Nat. Commun. 12, 2883 (2021).
Shakya, A., Park, S., Rana, N. & King, J. T. Liquid-liquid phase separation of histone proteins in cells: role in chromatin organization. Biophys. J. 118, 753–764 (2020).
Turner, A. L. et al. Highly disordered histone H1-DNA model complexes and their condensates. Proc. Natl Acad. Sci. USA 115, 11964–11969 (2018).
Chen, Q. et al. Chromatin Liquid-Liquid Phase Separation (LLPS) is regulated by ionic conditions and fiber length. Cells 11, 3145 (2022).
Gibson, B. A. et al. In diverse conditions, intrinsic chromatin condensates have liquid-like material properties. Proc. Natl Acad. Sci. USA. 120, e2218085120 (2023).
Strickfaden, H. et al. Condensed chromatin behaves like a solid on the mesoscale in vitro and in living cells. Cell 183, 1772–1784.e13 (2020).
Wang, L. et al. Rett syndrome-causing mutations compromise MeCP2-mediated liquid-liquid phase separation of chromatin. Cell Res. 30, 393–407 (2020).
Strom, A. R. et al. Phase separation drives heterochromatin domain formation. Nature 547, 241–245 (2017).
Sanulli, S. et al. HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature 575, 390–394 (2019).
Zhang, H. et al. MeCP2-Induced Heterochromatin organization is driven by oligomerization-based liquid-liquid phase separation and restricted by DNA methylation. Nucleus 13, 1–34 (2022).
Ackermann, B. E. & Debelouchina, G. T. Heterochromatin Protein HP1α gelation dynamics revealed by solid-state NMR Spectroscopy. Angew. Chem. Int. Ed. Engl. 58, 6300–6305 (2019).
Larson, A. G. et al. Liquid droplet formation by HP1α suggests a role for phase separation in Heterochromatin. Nature 547, 236–240 (2017).
Elathram, N., Ackermann, B. E., Clark, E. T., Dunn, S. R. & Debelouchina, G. T. Phosphorylated HP1α-Nucleosome interactions in phase separated environments. J. Am. Chem. Soc. 145, 23994–24004 (2023).
Hiragami-Hamada, K. et al. N-terminal phosphorylation of HP1{alpha} promotes its chromatin binding. Mol. Cell Biol. 31, 1186–1200 (2011).
Shimojo, H. et al. Extended string-like binding of the phosphorylated HP1α N-terminal tail to the Lysine 9-methylated Histone H3 tail. Sci. Rep. 6, 22527 (2016).
Teles, K. et al. Nucleosome binding peptide presents laudable biophysical and in vivo effects. Biomed. Pharmacother. 121, 109678 (2020).
Orsetti, A. et al. Structural dynamics in chromatin unraveling by pioneer transcription factors. Biophys. Rev. 16, 365–382 (2024).
Jaroniec, C. P. Structural and dynamic studies of chromatin by solid-state NMR Spectroscopy. Curr. Opin. Struct. Biol. 89, 102921 (2024).
Sun, W. et al. Conformational and interaction landscape of Histone H4 tails in nucleosomes probed by paramagnetic NMR Spectroscopy. J. Am. Chem. Soc. 145, 25478–25485 (2023).
Lupo, B. E., Chu, P., Harms, M. J., Morrison, E. A. & Musselman, C. A. Evolutionary conservation of structural and functional coupling between the BRM AT-Hook and Bromodomain. J. Mol. Biol. 433, 166845 (2021).
Morrison, E. A., Baweja, L., Poirier, M. G., Wereszczynski, J. & Musselman, C. A. Nucleosome composition regulates the Histone H3 tail conformational ensemble and accessibility. Nucleic Acids Res. 49, 4750–4767 (2021).
Shi, X. et al. Structure and dynamics in the nucleosome revealed by solid-state NMR. Angew. Chem. 130, 9882–9886 (2018).
Shi, X., Prasanna, C., Soman, A., Pervushin, K. & Nordenskiöld, L. Dynamic networks observed in the nucleosome core particles couple the histone globular domains with DNA. Commun. Biol. 3, 639 (2020).
Shi, X., Kannaian, B., Prasanna, C., Soman, A. & Nordenskiöld, L. Structural and dynamical investigation of Histone H2B in well-hydrated nucleosome core particles by solid-state NMR. Commun. Biol. 6, 672 (2023).
Fanti, L. & Pimpinelli, S. HP1: a functionally multifaceted protein. Curr. Opin. Genet. Dev. 18, 169–174 (2008).
Savitsky, M., Kwon, D., Georgiev, P., Kalmykova, A. & Gvozdev, V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 20, 345–354 (2006).
Blasco, M. A. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 8, 299–309 (2007).
Soman, A. et al. Columnar structure of human telomeric Chromatin. Nature 609, 1048–1055 (2022).
Shen, Y., Delaglio, F., Cornilescu, G. & Bax, A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 (2009).
Mendez, D. L. et al. The HP1a disordered C terminus and chromo shadow domain cooperate to select target peptide partners. ChemBioChem 12, 1084–1096 (2011).
Chin, S. Y. et al. Investigating different dynamic pHP1α states in their KCl-mediated liquid–liquid phase separation (LLPS) using Solid-State NMR (SSNMR) and molecular dynamic (MD) simulations. J. Phys. Chem. B 128, 10451–10459 (2024).
Hong, Y. et al. Hydrophobicity of Arginine leads to reentrant liquid-liquid phase separation behaviors of Arginine-rich proteins. Nat. Commun. 13, 7326 (2022).
Netzer, A., Baruch Leshem, A., Veretnik, S., Edelstein, I. & Lampel, A. Regulation of peptide liquid-liquid phase separation by aromatic amino acid composition. Small 20, e2401665 (2024).
Fawzi, N. L., Parekh, S. H. & Mittal, J. Biophysical studies of phase separation integrating experimental and computational methods. Curr. Opin. Struct. Biol. 70, 78–86 (2021).
Shea, J.-E., Best, R. B. & Mittal, J. Physics-based computational and theoretical approaches to intrinsically disordered proteins. Curr. Opin. Struct. Biol. 67, 219–225 (2021).
Alshareedah, I. et al. Interplay between short-range attraction and long-range repulsion controls reentrant liquid condensation of Ribonucleoprotein-RNA complexes. J. Am. Chem. Soc. 141, 14593–14602 (2019).
Gao, J., Li, H., Tan, S., Zhou, R., Lee, T.-H. Roles of Histone Chaperone Nap1 and Histone acetylation in regulating phase-separation of chromatin arrays. bioRxiv, 2025.05.09.653121. https://doi.org/10.1101/2025.05.09.653121 (2025).
Eisert, R. J., Kennedy, S. A. & Waters, M. L. Investigation of the β-sheet interactions between dHP1 Chromodomain and Histone 3. Biochemistry 54, 2314–2322 (2015).
Ukmar-Godec, T. et al. Multimodal interactions drive chromatin phase separation and compaction. Proc. Natl Acad. Sci. Usa. 120, e2308858120 (2023).
Machida, S. et al. Structural basis of heterochromatin formation by human HP1. Mol. Cell 69, 385–397.e8 (2018).
Garcia-Saez, I. et al. Structure of an H1-bound 6-Nucleosome array reveals an untwisted two-start chromatin fiber conformation. Mol. Cell 72, 902–915.e7 (2018).
Keenen, M. M. et al. HP1 proteins compact DNA into mechanically and positionally stable phase separated domains. Elife 10, e64563 (2021).
Didier, J.-P., Cozzone, A. J. & Duclos, B. Phosphorylation of the virulence regulator SarA modulates its ability to bind DNA in Staphylococcus Aureus. FEMS Microbiol. Lett. 306, 30–36 (2010).
Derouiche, A. et al. Bacillus Subtilis SalA is a phosphorylation-dependent transcription regulator that represses scoC and activates the production of the Exoprotease AprE. Mol. Microbiol. 97, 1195–1208 (2015).
Dang, M., Li, T., Zhou, S. & Song, J. Arg/Lys-containing IDRs are cryptic binding domains for ATP and nucleic acids that interplay to modulate LLPS. Commun. Biol. 5, 1315 (2022).
Tatarakis, A., Behrouzi, R. & Moazed, D. Evolving models of Heterochromatin: from Foci to liquid droplets. Mol. Cell 67, 725–727 (2017).
Wang, L. et al. Histone modifications regulate chromatin compartmentalization by contributing to a phase separation mechanism. Mol. Cell 76, 646–659.e6 (2019).
Erdel, F. et al. Mouse heterochromatin adopts digital compaction states without showing hallmarks of HP1-driven liquid-liquid phase separation. Mol. Cell 78, 236–249.e7 (2020).
Furukawa, A. et al. A dynamic structural unit of phase-separated heterochromatin Protein 1α as revealed by integrative structural analyses. Nucleic Acids Res. 53, gkaf154 (2025).
Xie, J. et al. Heterochromatin formation and remodeling by IRTKS condensates counteract cellular senescence. EMBO J. 43, 4542–4577 (2024).
Shen, S. et al. TRIM66-HP1γ remodels the chromatin through phase separation. Biophys. Rep. 11, 18–33 (2025).
Zhang, L. et al. 53BP1 regulates heterochromatin through liquid phase separation. Nat. Commun. 13, 360 (2022).
Liu, X. et al. Mitotic implantation of the transcription factor prospero via phase separation drives terminal neuronal differentiation. Dev. Cell 52, 277–293.e8 (2020).
Dyer, P. N. et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 (2004).
Vranken, W. F. et al. The CCPN data model for NMR Spectroscopy: development of a software pipeline. Proteins 59, 687–696 (2005).
Lee, W., Tonelli, M. & Markley, J. L. NMRFAM-SPARKY: enhanced software for biomolecular NMR Spectroscopy. Bioinformatics 31, 1325–1327 (2015).
Abraham, M. J. et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 (2015).
Páll, S.; Abraham, M. J.; Kutzner, C.; Hess, B.; Lindahl, E. Tackling Exascale Software Challenges in Molecular Dynamics Simulations with GROMACS. In Solving Software Challenges for Exascale; Markidis, S., Laure, E., Eds; Lecture Notes in Computer Science; Springer International Publishing: Cham, 8759, 3–27 (2015).
Pronk, S. et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29, 845–854 (2013).
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput Chem. 29, 1859–1865 (2008).
Lee, J. et al. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413 (2016).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Badaczewska-Dawid, A. E., Nithin, C., Wroblewski, K., Kurcinski, M. & Kmiecik, S. MAPIYA contact map server for identification and visualization of molecular interactions in proteins and biological complexes. Nucleic Acids Res. 50, W474–W482 (2022).
Acknowledgements
This work is supported by funds from the National Science Foundation of China (#32201006), Guangdong Province (#2021QN02Y103, #2022ZDZX2061, #2025KCXTD055), and the 2023 Stable Support Plan Program of Shenzhen Colleges and Universities Fund. Work at Nanyang Technological University was supported by the Singapore Ministry of Education (MOE) Academic Research Fund (AcRF) Tier 1 (2021-T1-002-0820), Tier 2 (2018-T2-1-112), and Tier 3 (MOE2019-T3-1-012) grants (to L.N.) The authors also acknowledge the NMR facility of the National Center for Protein Sciences at Peking University and the Nanyang Technological University (NTU) Center of High-Field NMR Spectroscopy and Imaging for the NMR usage, as well as the NTU Institute of Structural Biology (NISB). We also thank Dr. Hongwei Li for assistance with NMR experiments. A.S. acknowledges the support of an NTU Presidential Post-Doctoral Fellowship. Z.Z. and Y.C. thank the Shenzhen Municipal Government and Shenzhen MSU-BIT University for the student scholarship support.
Author information
Authors and Affiliations
Contributions
Conceptualization and study design: X.S. and A.S. Sample preparation and optical microscopy experiment conduction: Z.Z., A.S., Y.C., J.K., J.G., L.Z., D.A.P., G.S.G., and C.P. Acquisition and analysis of NMR data: Z.Z. and X.S. MD simulation: C.C., C.H., and K.X. Manuscript writing and editing: X.S., Z.Z., L.N., C.C., and C.H. All authors have given approval to the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Eitan Lerner and the other anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Kuangyu Yen and Mengtan Xing.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhai, Z., Soman, A., Kuang, J. et al. Revealing site-specific molecular features mediating contacts of heterochromatin protein 1 α (HP1α) and the interactions with chromatin and DNA in LLPS environments. Commun Biol 8, 1758 (2025). https://doi.org/10.1038/s42003-025-09236-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-09236-9