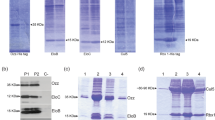
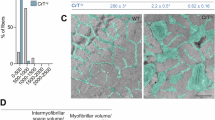
Abstract
High-energy-demanding tissues, such as skeletal muscle, rely on mitochondrial proteostasis for proper function. Two key quality-control mechanisms -the ubiquitin proteasome system (UPS) and the release of mitochondria-derived vesicles- help maintain mitochondrial proteostasis, but whether these processes interact remains unclear. Here, we show that CRL5Ozz and its substrate, Alix, localize to mitochondria and together regulate the levels and distribution of the mitochondrial solute carrier Slc25A4, which is essential for ATP production. In Ozz−/− or Alix−/− mice, skeletal muscle mitochondria exhibit similar morphological abnormalities, including swelling and dysmorphism, along with partially overlapping metabolomic alterations. We demonstrate that CRL5Ozz ubiquitinates Slc25A4, targeting it for proteasomal degradation, while Alix facilitates Slc25A4 loading into exosomes for lysosomal degradation. Loss of Ozz or Alix in vivo disrupts the steady-state levels of Slc25A4, impairing mitochondrial metabolism and triggering a switch in muscle fiber composition from oxidative, mitochondria-rich slow to glycolytic fast fibers.

Similar content being viewed by others
Data availability
All data supporting the findings of this study are available in the article and its supplementary information. Additional information can be obtained from the corresponding authors upon request. Metabolomics data have been uploaded to the Metabolomics Workbench; the accession number is: ST004190. All numerical source data underlying Figs. 3–6 and Supplementary Figs. S3, S4, S6, and S7, together with the complete metabolomic dataset, are supplied in the Supplementary Data. Uncropped blot and gel images can be found in the Supplementary Information.
References
Mukund, K. & Subramaniam, S. Skeletal muscle: A review of molecular structure and function, in health and disease. Wiley Interdiscip. Rev. Syst. Biol. Med. 12, e1462 (2020).
Schiaffino, S. & Reggiani, C. Fiber types in mammalian skeletal muscles. Physiol. Rev. 91, 1447–1531 (2011).
Ortenblad, N. et al. The muscle fiber profiles, mitochondrial content, and enzyme activities of the exceptionally well-trained arm and leg muscles of elite cross-country skiers. Front. Physiol. 9, 1031 (2018).
A. Damirchi, P. Babaei, M. Gholamali, K. Ranjbar mitochondrial biogenesis in skeletal muscle: exercise and aging, in Skeletal Muscle - From Myogenesis to Clinical Relations (IntechOpen, 2012).
Hood, D. A., Memme, J. M., Oliveira, A. N. & Triolo, M. Maintenance of skeletal muscle mitochondria in health, exercise, and aging. Annu. Rev. Physiol. 81, 19–41 (2019).
Mishra, P., Varuzhanyan, G., Pham, A. H. & Chan, D. C. Mitochondrial dynamics is a distinguishing feature of skeletal muscle fiber types and regulates Organellar compartmentalization. Cell Metab. 22, 1033–1044 (2015).
Krylova, S.V. & Feng, D. The machinery of exosomes: biogenesis, release, and uptake. Int. J. Mol. Sci. 24 (2023).
Picca, A. et al. Mitochondrial-derived vesicles in skeletal muscle remodeling and adaptation. Semin. Cell Dev. Biol. 143, 37–45 (2023).
Romanello, V. The interplay between mitochondrial morphology and Myomitokines in aging Sarcopenia. Int. J. Mol. Sci. 22 (2020).
Ahmed, S. T., Craven, L., Russell, O. M., Turnbull, D. M. & Vincent, A. E. Diagnosis and treatment of mitochondrial myopathies. Neurotherapeutics 15, 943–953 (2018).
Brunel-Guitton, C., Levtova, A. & Sasarman, F. Mitochondrial diseases and Cardiomyopathies. Can. J. Cardiol. 31, 1360–1376 (2015).
Gehrig, S. M. et al. Altered skeletal muscle (mitochondrial) properties in patients with mitochondrial DNA single deletion myopathy. Orphanet J. Rare Dis. 11, 105 (2016).
Bragoszewski, P., Turek, M. & Chacinska, A. Control of mitochondrial biogenesis and function by the ubiquitin-proteasome system. Open Biol. 7 (2017).
Lehmann, G., Udasin, R. G. & Ciechanover, A. On the linkage between the ubiquitin-proteasome system and the mitochondria. Biochem. Biophys. Res. Commun. 473, 80–86 (2016).
Jeon, H. B. et al. A proteomics approach to identify the ubiquitinated proteins in mouse heart. Biochem. Biophys. Res. Commun. 357, 731–736 (2007).
Lavie, J. et al. Ubiquitin-dependent degradation of mitochondrial proteins regulates energy metabolism. Cell Rep. 23, 2852–2863 (2018).
Alsayyah, C., Ozturk, O., Cavellini, L., Belgareh-Touze, N. & Cohen, M. M. The regulation of mitochondrial homeostasis by the ubiquitin proteasome system. Biochim. Biophys. Acta Bioenerg. 1861, 148302 (2020).
Dimmer, K. S. & Scorrano, L. (De)constructing mitochondria: what for?. Physiology 21, 233–241 (2006).
Soubannier, V. et al. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr. Biol. 22, 135–141 (2012).
Boudreau, L. H. et al. Platelets release mitochondria serving as substrate for bactericidal group IIA-secreted phospholipase A2 to promote inflammation. Blood 124, 2173–2183 (2014).
Mathivanan, S., Fahner, C. J., Reid, G. E. & Simpson, R. J. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 40, D1241–D1244 (2012).
Ng, M. Y. W., Wai, T. & Simonsen, A. Quality control of the mitochondrion. Dev. Cell 56, 881–905 (2021).
Puhm, F. et al. Mitochondria are a subset of extracellular vesicles released by activated monocytes and Induce Type I IFN and TNF responses in endothelial cells. Circ. Res. 125, 43–52 (2019).
Ashrafi, G. & Schwarz, T. L. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 20, 31–42 (2013).
Chen, C. C. W., Erlich, A. T., Crilly, M. J. & Hood, D. A. Parkin is required for exercise-induced mitophagy in muscle: impact of aging. Am. J. Physiol. Endocrinol. Metab. 315, E404–E415 (2018).
Ma, K. et al. Mitophagy, mitochondrial homeostasis, and cell fate. Front. Cell Dev. Biol. 8, 467 (2020).
Pickles, S., Vigie, P. & Youle, R. J. Mitophagy and quality control mechanisms in mitochondrial. Maint. Curr. Biol. 28, R170–R185 (2018).
Romanello, V. & Sandri, M. Mitochondrial quality control and muscle mass maintenance. Front. Physiol. 6, 422 (2015).
Song, J., Herrmann, J. M. & Becker, T. Quality control of the mitochondrial proteome. Nat. Rev. Mol. Cell Biol. 22, 54–70 (2021).
Campos, Y. et al. Biophysical and functional study of CRL5(Ozz), a muscle-specific ubiquitin ligase complex. Sci. Rep. 12, 7820 (2022).
Nastasi, T. et al. Ozz-E3, a muscle-specific ubiquitin ligase, regulates beta-catenin degradation during myogenesis. Dev. Cell 6, 269–282 (2004).
Lee, J. et al. Neur1 and Neur2 are required for hippocampus-dependent spatial memory and synaptic plasticity. Hippocampus 30, 1158–1166 (2020).
Liu, S. & Boulianne, G. L. The NHR domains of Neuralized and related proteins: Beyond Notch signalling. Cell Signal 29, 62–68 (2017).
Campos, Y. et al. Ozz-E3 ubiquitin ligase targets sarcomeric embryonic myosin heavy chain during muscle development. PloS One 5, e9866 (2010).
Bongiovanni, A. et al. Alix protein is substrate of Ozz-E3 ligase and modulates actin remodeling in skeletal muscle. J. Biol. Chem. 287, 12159–12171 (2012).
Cabezas, A., Bache, K. G., Brech, A. & Stenmark, H. Alix regulates cortical actin and the spatial distribution of endosomes. J. Cell Sci. 118, 2625–2635 (2005).
Campos, Y. et al. Alix-mediated assembly of the actomyosin-tight junction polarity complex preserves epithelial polarity and epithelial barrier. Nat. Commun. 7, 11876 (2016).
Carlton, J. G. & Martin-Serrano, J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 316, 1908–1912 (2007).
Katzmann, D. J., Odorizzi, G. & Emr, S. D. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3, 893–905 (2002).
Martin-Serrano, J., Yarovoy, A., Perez-Caballero, D. & Bieniasz, P. D. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. USA 100, 12414–12419 (2003).
Morita, E. et al. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26, 4215–4227 (2007).
Pan, S. et al. Involvement of the conserved adaptor protein Alix in actin cytoskeleton assembly. J. Biol. Chem. 281, 34640–34650 (2006).
Raiborg, C., Rusten, T. E. & Stenmark, H. Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 15, 446–455 (2003).
Schmidt, M. H. H., Chen, B., Randazzo, L. M. & Bogler, O. SETA/CIN85/Ruk and its binding partner AIP1 associate with diverse cytoskeletal elements, including FAKs, and modulate cell adhesion. J. Cell Sci. 116, 2845–2855 (2003).
Strack, B., Calistri, A., Craig, S., Popova, E. & Gottlinger, H. G. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114, 689–699 (2003).
Sun, S. et al. ALG-2 activates the MVB sorting function of ALIX through relieving its intramolecular interaction. Cell Discov. 1, 15018 (2015).
Romancino, D. P. et al. Identification and characterization of the nano-sized vesicles released by muscle cells. FEBS Lett. 587, 1379–1384 (2013).
Drake, J. C. & Yan, Z. Mitophagy in maintaining skeletal muscle mitochondrial proteostasis and metabolic health with ageing. J. Physiol. 595, 6391–6399 (2017).
Guan, Y., Drake, J. C. & Yan, Z. Exercise-induced mitophagy in skeletal muscle and heart. Exerc. Sport Sci. Rev. 47, 151–156 (2019).
Leduc-Gaudet, J.P., Hussain, S.N.A., Barreiro, E. & Gouspillou, G. Mitochondrial dynamics and mitophagy in skeletal muscle health and aging. Int. J. Mol. Sci. 22 (2021).
McWilliams, T. G. et al. mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J. Cell Biol. 214, 333–345 (2016).
Gutierrez-Aguilar, M. & Baines, C. P. Physiological and pathological roles of mitochondrial SLC25 carriers. Biochem J. 454, 371–386 (2013).
Kunji, E. R. S., King, M. S., Ruprecht, J. J. & Thangaratnarajah, C. The SLC25 carrier family: important transport proteins in mitochondrial physiology and pathology. Physiology 35, 302–327 (2020).
Palmieri, F. Mitochondrial transporters of the SLC25 family and associated diseases: a review. J. Inherit. Metab. Dis. 37, 565–575 (2014).
Ruprecht, J. J. & Kunji, E. R. S. The SLC25 Mitochondrial Carrier Family: Structure and Mechanism. Trends Biochem Sci. 45, 244–258 (2020).
Chandel, N.S. Glycolysis. Cold Spring Harb. Perspect. Biol. 13 (2021).
Moreno-Justicia, R. et al. Human skeletal muscle fiber heterogeneity beyond myosin heavy chains. Nat. Commun. 16, 1764 (2025).
de Boer, E. et al. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc. Natl. Acad. Sci. USA 100, 7480–7485 (2003).
Driegen, S. et al. A generic tool for biotinylation of tagged proteins in transgenic mice. Transgen. Res 14, 477–482 (2005).
van de Vlekkert, D. et al. Excessive exosome release is the pathogenic pathway linking a lysosomal deficiency to generalized fibrosis. Sci. Adv. 5, eaav3270 (2019).
Jezek, P., Jaburek, M., Holendova, B., Engstova, H. & Dlaskova, A. Mitochondrial Cristae morphology reflecting metabolism, superoxide formation, redox homeostasis, and pathology. Antioxid. Redox Signal 39, 635–683 (2023).
Joubert, F. & Puff, N. Mitochondrial Cristae Architecture and Functions: Lessons from Minimal Model Systems. Membranes 11 (2021).
Patten, D. A. et al. OPA1-dependent cristae modulation is essential for cellular adaptation to metabolic demand. EMBO J. 33, 2676–2691 (2014).
Nederveen, J. P. et al. Variability in skeletal muscle fibre characteristics during repeated muscle biopsy sampling in human vastus lateralis. Appl Physiol. Nutr. Metab. 45, 368–375 (2020).
Van de Casteele, F. et al. Does one biopsy cut it? Revisiting human muscle fiber type composition variability using repeated biopsies in the vastus lateralis and gastrocnemius medialis. J. Appl Physiol. (1985) 137, 1341–1353 (2024).
Liu, J. et al. Coupling of mitochondrial function and skeletal muscle fiber type by a miR-499/Fnip1/AMPK circuit. EMBO Mol. Med. 8, 1212–1228 (2016).
Coyne, L. P. & Chen, X. J. Consequences of inner mitochondrial membrane protein misfolding. Mitochondrion 49, 46–55 (2019).
Gariballa, N. & Ali, B. R. Endoplasmic Reticulum Associated Protein Degradation (ERAD) in the Pathology of Diseases Related to TGFbeta Signaling Pathway: Future Therapeutic Perspectives. Front. Mol. Biosci. 7, 575608 (2020).
Olzmann, J.A., Kopito, R.R. & Christianson, J.C. The mammalian endoplasmic reticulum-associated degradation system. Cold Spring Harb. Perspect. Biol. 5 (2013).
Azzu, V. & Brand, M. D. Degradation of an intramitochondrial protein by the cytosolic proteasome. J. Cell Sci. 123, 578–585 (2010).
Azzu, V., Mookerjee, S. A. & Brand, M. D. Rapid turnover of mitochondrial uncoupling protein 3. Biochem. J. 426, 13–17 (2010).
Mookerjee, S. A. & Brand, M. D. Characteristics of the turnover of uncoupling protein 3 by the ubiquitin proteasome system in isolated mitochondria. Biochim. Biophys. Acta 1807, 1474–1481 (2011).
Klingenberg, M. The ADP and ATP transport in mitochondria and its carrier. Biochim. Biophys. Acta 1778, 1978–2021 (2008).
Hers, H. G. & Van Schaftingen, E. Fructose 2,6-bisphosphate 2 years after its discovery. Biochem. J. 206, 1–12 (1982).
Maldonado, E. N. & Lemasters, J. J. ATP/ADP ratio, the missed connection between mitochondria and the Warburg effect. Mitochondrion 19 Pt A, 78–84 (2014).
Mor, I., Cheung, E. C. & Vousden, K. H. Control of glycolysis through regulation of PFK1: old friends and recent additions. Cold Spring Harb. Symp. Quant. Biol. 76, 211–216 (2011).
Clemencon, B., Babot, M. & Trezeguet, V. The mitochondrial ADP/ATP carrier (SLC25 family): pathological implications of its dysfunction. Mol. Asp. Med. 34, 485–493 (2013).
Graham, B. H. et al. A mouse model for mitochondrial myopathy and cardiomyopathy resulting from a deficiency in the heart/muscle isoform of the adenine nucleotide translocator. Nat. Genet. 16, 226–234 (1997).
Wang, X. & Chen, X. J. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature 524, 481–484 (2015).
Liu, Y. et al. Mitochondrial carrier protein overloading and misfolding induce aggresomes and proteostatic adaptations in the cytosol. Mol. Biol. Cell 30, 1272–1284 (2019).
Thompson, K. et al. Recurrent De Novo Dominant Mutations in SLC25A4 Cause Severe Early-Onset Mitochondrial Disease and Loss of Mitochondrial DNA Copy Number. Am. J. Hum. Genet. 99, 860–876 (2016).
Park, K. P. et al. SLC25A4 and C10ORF2 mutations in autosomal dominant progressive external Ophthalmoplegia. J. Clin. Neurol. 7, 25–30 (2011).
Hua, Y., Laserstein, P. & Helmstaedter, M. Large-volume en-bloc staining for electron microscopy-based connectomics. Nat. Commun. 6, 7923 (2015).
Pachitariu, M. & Stringer, C. Cellpose 2.0: how to train your own model. Nat. Methods 19, 1634–1641 (2022).
Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021).
Chiu, C.-L. C.N. Napari: a Python Multi-Dimensional Image Viewer Platform for the Research Community. Microsc. Microanal. 28, 1576–1577 (2022).
Lewiner, T., Lopes, H., Vieira, A. W. & Tavares, G. Efficient implementation of Marching Cubes’ cases with topological guarantees. J. Graph. Tools 8, 1–15 (2003).
Wadell, H. Volume, shape, and roundness of quartz particles. J. Geol. 43, 250–280 (1935).
Wieckowski, M. R., Giorgi, C., Lebiedzinska, M., Duszynski, J. & Pinton, P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat. Protoc. 4, 1582–1590 (2009).
Hollenbach, A. D., McPherson, C. J., Lagutina, I. & Grosveld, G. The EF-hand calcium-binding protein calmyrin inhibits the transcriptional and DNA-binding activity of Pax3. Biochim. Biophys. Acta 1574, 321–328 (2002).
Milne, S. B., Mathews, T. P., Myers, D. S., Ivanova, P. T. & Brown, H. A. Sum of the parts: mass spectrometry-based metabolomics. Biochemistry 52, 3829–3840 (2013).
Doppler, M. et al. Stable isotope-assisted plant metabolomics: combination of global and tracer-based labeling for enhanced untargeted profiling and compound annotation. Front. Plant Sci. 10, 1366 (2019).
Malik, A. N., Czajka, A. & Cunningham, P. Accurate quantification of mouse mitochondrial DNA without co-amplification of nuclear mitochondrial insertion sequences. Mitochondrion 29, 59–64 (2016).
Hallett, P. J., Collins, T. L., Standaert, D. G. & Dunah, A. W. Biochemical fractionation of brain tissue for studies of receptor distribution and trafficking. Curr. Protoc. Neurosci. Chapter 1, Unit 1 16 (2008).
Muller, L., Kutzner, C.E., Balaji, V. & Hoppe, T. In Vitro Analysis of E3 Ubiquitin Ligase Function. J. Vis. Exp. (2021).
Acknowledgements
We would like to thank Dr. George Campbell of the St. Jude Cell and Tissue Imaging Center for help with image acquisitions. We gratefully acknowledge Dr. Tommaso Nastasi for his valuable contributions during the initial phase of this project and Dr. Angela McArthur (St. Jude Department of Scientific Editing) for editing the manuscript. The C57BL/6-Gt(ROSA)26Sortm1(CAG-mCherry/GFP)Ganl/GanlH mice were obtained from the MRC Harwell Institute, which distributes this strain on behalf of the European Mouse Mutant Archive (EMMA: www.infrafrontier.eu). The repository number is EM:11343. The C57BL/6-Gt(ROSA)26Sortm1(CAG-mCherry/GFP)Ganl/GanlH mice were originally produced at The University of Dundee. A. d’Azzo holds an endowed chair in Genetics and Gene Therapy from the Jewelry Charity Fund. This study was funded in part by the National Institutes of Health grants AR049867, GM104981, and CA021764, the Assisi Foundation of Memphis, and the American Lebanese Syrian Associated Charities (ALSAC). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
Y.C. conceived the study, designed the experiments, analyzed the data, generated the mouse models, coordinated the efforts of all authors, and drafted the manuscript. G.P. designed, performed, and analyzed the soleus metabolomics and TCA data, and helped with the interpretation of the data and explanation of the methodology. E.G. maintained the animal colonies and performed surgeries. D.v.d.V. performed the isolation, purification, and analyses of exosomes from WT and mutant muscles, and helped with statistical analyses, and the editing of the manuscript and figures. K.V., J.J., and K.K. performed the 3D segmentation, rendering, and statistical analyses of muscle mitochondria from WT and mutant mice. J.W. wrote the macros for ImageJ/Fiji software used to analyze and quantify mitophagy, to determine the MyHC content in myofibers from WT and mutant muscle,s and to quantify ATPase activity in different fiber types. R. W. and C. R. obtained and processed TEM and SEM images. R.R. and S.R. helped with the mitochondrial OCR assays and the analyses of ΔΨm. X.Q. purified anti-Ozz and anti-Alix antibodies, prepared and maintained primary myoblast cultures, and performed Alix overexpression studies in C2C12 cells. J.D. performed proteomic analyses of C2C12 overexpressing tagged Ozz. J.T.O. and T.B. provided intellectual insights. G.C.G. provided intellectual insights, advised on the CRISPR/CAS9 mutagenesis, and edited the manuscript. A. d’A. conceived the study, designed the experiments, analyzed the data, oversaw all experiments, coordinated the efforts of all authors, secured the funding for this project, and wrote and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Giulia Bertolin and Dario Ummarino. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Campos, Y., Palacios, G., Gomero, E. et al. Mitochondrial proteostasis regulated by CRL5Ozz and Alix modulates skeletal muscle metabolism and fiber type. Commun Biol (2025). https://doi.org/10.1038/s42003-025-09363-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-025-09363-3