Abstract
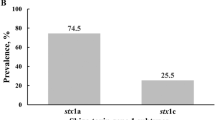
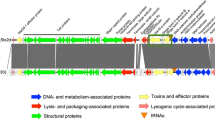
Shiga toxin-producing Escherichia coli (STEC) is detected in healthy individuals, who face work restrictions to prevent secondary transmission. To assess the virulence potential, we sequence 495 STEC isolates from healthy food handlers and social welfare workers in 2021 and compare them with 250 isolates from symptomatic patients. Nineteen serotypes (e.g., O156:H25, O174:H21, O105:H7) are significantly associated with asymptomatic carriers (SAAC), while five serotypes (O157:H7, O26:H11, O111:H8, O121:H19, O145:H28) are significantly associated with symptomatic patients (SAPA). SAPA strains frequently carry major virulence factors, including the LEE-encoded type III secretion system (100%) and Shiga toxin 2a (63.2%), which are less common in SAAC strains (38.3% and 3.7%). Among the 495 carrier isolates, 35 (7.1%) are high-risk, 178 (36.0%) moderate-risk, and 282 (57.0%) low-risk based on serotype and virulence markers. These findings suggest many strains in asymptomatic carriers have limited virulence, underscoring the need for risk-based strategies that avoid unnecessary restrictions.
Similar content being viewed by others
Data availability
All sequence data generated in this study have been deposited in the NCBI BioProject database under accession number PRJDB18641. The source data underlying all figures and analyses in the manuscript are provided in Supplementary Data 1. Any additional data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Terajima, J., Izumiya, H., Hara-Kudo, Y. & Ohnishi, M. Shiga Toxin (Verotoxin)-producing Escherichia coli and Foodborne Disease: A Review. Food Saf (Tokyo) 5, 35–53 (2017).
Freedman, S. B., van de Kar, N. & Tarr, P. I. Shiga Toxin-Producing Escherichia coli and the Hemolytic-Uremic Syndrome. N Engl J Med 389, 1402–1414 (2023).
Bai, X., Scheutz, F., Dahlgren, H. M., Hedenstrom, I. & Jernberg, C. Characterization of Clinical Escherichia coli Strains Producing a Novel Shiga Toxin 2 Subtype in Sweden and Denmark. Microorganisms 9, 2374 (2021).
Gill, A. et al. Characterization of Atypical Shiga Toxin Gene Sequences and Description of Stx2j, a New Subtype. J Clin Microbiol 60, e0222921 (2022).
Hughes, A. C. et al. Structural and Functional Characterization of Stx2k, a New Subtype of Shiga Toxin 2. Microorganisms 8, 4 (2019).
Scheutz, F. et al. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J Clin Microbiol 50, 2951–2963 (2012).
Yang, X. et al. Genomic Characterization of Escherichia coli O8 Strains Producing Shiga Toxin 2l Subtype. Microorganisms 10, 1245 (2022).
Schmidt, H. Shiga-toxin-converting bacteriophages. Res Microbiol 152, 687–695 (2001).
De Rauw, K., Buyl, R., Jacquinet, S. & Pierard, D. Risk determinants for the development of typical haemolytic uremic syndrome in Belgium and proposition of a new virulence typing algorithm for Shiga toxin-producing Escherichia coli. Epidemiol Infect 147, e6 (2018).
Tack, D. M. et al. Shiga Toxin-Producing Escherichia coli Outbreaks in the United States, 2010–2017. Microorganisms 9, 1529 (2021).
Buchholz, U. et al. German outbreak of Escherichia coli O104:H4 associated with sprouts. N Engl J Med. 365, 1763–1770 (2011).
Brooks, J. T. et al. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983-2002. J Infect Dis. 192, 1422–1429 (2005).
Schmidt, M. A. LEEways: tales of EPEC, ATEC and EHEC. Cell Microbiol 12, 1544–1552 (2010).
Tobe, T. et al. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc Natl Acad Sci USA 103, 14941–14946 (2006).
Hayashi, T. et al. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8, 11–22 (2001).
Armstrong, G. L., Hollingsworth, J. & Morris, J. G. Jr Emerging foodborne pathogens: Escherichia coli O157:H7 as a model of entry of a new pathogen into the food supply of the developed world. Epidemiol Rev. 18, 29–51 (1996).
Karmali, M. A. Infection by verocytotoxin-producing Escherichia coli. Clin Microbiol Rev 2, 15–38 (1989).
Kintz, E., Brainard, J., Hooper, L. & Hunter, P. Transmission pathways for sporadic Shiga-toxin producing E. coli infections: A systematic review and meta-analysis. Int J Hyg Environ Health 220, 57–67 (2017).
Sayk, F., Hauswaldt, S., Knobloch, J. K., Rupp, J. & Nitschke, M. Do asymptomatic STEC-long-term carriers need to be isolated or decolonized? New evidence from a community case study and concepts in favor of an individualized strategy. Front Public Health 12, 1364664 (2024).
Terajima, J., Iyoda, S., Ohnishi, M. & Watanabe, H. Shiga Toxin (Verotoxin)-Producing Escherichia coli in Japan. Microbiol Spectr 2, (2014).
Harada, T. et al. Laboratory investigation of an Escherichia coli O157:H7 strain possessing a vtx2c gene with an IS1203 variant insertion sequence isolated from an asymptomatic food handler in Japan. Diagn Microbiol Infect Dis. 77, 176–178 (2013).
Baba, H. et al. Genomic analysis of Shiga toxin-producing Escherichia coli from patients and asymptomatic food handlers in Japan. PLoS One 14, e0225340 (2019).
Morita-Ishihara, T., Iyoda, S., Iguchi, A. & Ohnishi, M. Secondary Shiga Toxin-Producing Escherichia coli Infection, Japan, 2010-2012. Emerg Infect Dis. 22, 2181–2184 (2016).
Sui, X. et al. Characteristics of Shiga Toxin-Producing Escherichia coli Circulating in Asymptomatic Food Handlers. Toxins (Basel) 15, 640 (2023).
Iyoda, S. et al. Phylogenetic Clades 6 and 8 of Enterohemorrhagic Escherichia coli O157:H7 With Particular stx Subtypes are More Frequently Found in Isolates From Hemolytic Uremic Syndrome Patients Than From Asymptomatic Carriers. Open Forum Infect Dis. 1, ofu061 (2014).
Manning, S. D. et al. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc Natl Acad Sci USA 105, 4868–4873 (2008).
Bizot, E. et al. Shiga toxin-producing Escherichia coli carriage in 959 healthy French infants. Arch Dis Child 106, 1239–1240 (2021).
Harries, M., Dreesman, J., Rettenbacher-Riefler, S. & Mertens, E. Faecal carriage of extended-spectrum beta-lactamase-producing Enterobacteriaceae and Shiga toxin-producing Escherichia coli in asymptomatic nursery children in Lower Saxony (Germany), 2014. Epidemiol Infect 144, 3540–3548 (2016).
Tarr, P. I. & Freedman, S. B. Why antibiotics should not be used to treat Shiga toxin-producing Escherichia coli infections. Curr Opin Gastroenterol 38, 30–38 (2022).
Bai, X. et al. Comparative Genomics of Shiga Toxin-Producing Escherichia coli Strains Isolated from Pediatric Patients with and without Hemolytic Uremic Syndrome from 2000 to 2016 in Finland. Microbiol Spectr 10, e0066022 (2022).
Blankenship, H. M. et al. Population structure and genetic diversity of non-O157 Shiga toxin-producing Escherichia coli (STEC) clinical isolates from Michigan. Sci Rep. 11, 4461 (2021).
Kalalah, A. A., Koenig, S. S. K., Bono, J. L., Bosilevac, J. M. & Eppinger, M. Pathogenomes and virulence profiles of representative big six non-O157 serogroup Shiga toxin-producing Escherichia coli. Front Microbiol 15, 1364026 (2024).
Karmali, M. A. et al. Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J Clin Microbiol 41, 4930–4940 (2003).
Melton-Celsa, A. R. & O’Brien, A. D. Animal models for STEC-mediated disease. Methods Mol Med 73, 291–305 (2003).
Ritchie, J. M. Infant Rabbit Model for Studying Shiga Toxin-Producing Escherichia coli. Methods Mol Biol 2291, 365–379 (2021).
Thorpe, C. M., Pulsifer, A. R., Osburne, M. S., Vanaja, S. K. & Leong, J. M. Citrobacter rodentium(varphiStx2dact), a murine infection model for enterohemorrhagic Escherichia coli. Curr Opin Microbiol 65, 183–190 (2022).
Kajitani, R. et al. Platanus_B: an accurate de novo assembler for bacterial genomes using an iterative error-removal process. DNA Res 27, dsaa014 (2020).
Bessonov, K. et al. ECTyper: in silico Escherichia coli serotype and species prediction from raw and assembled whole-genome sequence data. Microb. Genom 7, 000728 (2021).
Inouye, M. et al. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 6, 90 (2014).
Page, A. J. et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693 (2015).
Page, A. J. et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb Genom 2, e000056 (2016).
Kozlov, A. M., Darriba, D., Flouri, T., Morel, B. & Stamatakis, A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455 (2019).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021).
Riordan, J. T., Viswanath, S. B., Manning, S. D. & Whittam, T. S. Genetic differentiation of Escherichia coli O157:H7 clades associated with human disease by real-time PCR. J Clin Microbiol 46, 2070–2073 (2008).
Berger, M., Aijaz, I., Berger, P., Dobrindt, U. & Koudelka, G. Transcriptional and Translational Inhibitors Block SOS Response and Shiga Toxin Expression in Enterohemorrhagic Escherichia coli. Sci Rep. 9, 18777 (2019).
Yang, X. et al. Characterization of Escherichia coli strains producing Shiga Toxin 2f subtype from domestic Pigeon. Sci Rep. 14, 24481 (2024).
Acknowledgements
We thank Ayaka Wakakuwa for providing technical assistance.
Author information
Authors and Affiliations
Contributions
Y.I., K.L., T.S., S.I., and Y.O. conceptualized the study. Y.I., M.O., Y.H., T.Y., and Y.O. curated the data. Y.I., Y.H., H.K., A.N., T.Y., H.N.I., and T.K. performed the investigation. T.S. and Y.O. administered the project. Y.I. and Y.O. wrote the original draft. All the authors were responsible for reviewing and editing the manuscript. All the authors had access to the data presented in this study and had final responsibility for the decision to submit for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Yanwen Xiong, Ying Hua and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Yu-Wei Wu and Tobias Goris. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yumi, I., Okuno, M., Hoshiko, Y. et al. Genomics of Shiga toxin-producing Escherichia coli from asymptomatic carriers in Japan highlights risk-adaptive control. Commun Biol (2025). https://doi.org/10.1038/s42003-025-09369-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-025-09369-x