Abstract
Natural CRISPR-Cas9 systems provides diverse properties for genome editing, yet finding compact variants remains a priority. In this study, we screened a panel of 11 CjCas9 orthologous using a GFP activation assay and identified seven active nucleases. Among these, Cj4Cas9 stood out as particularly noteworthy due to its compact genome size (985 amino acids) and unique PAM preference (5’-NNNGRY-3’). Cj4Cas9 demonstrates efficient disruption of the Tyr gene in mouse zygotes, resulting in an albino phenotype. Furthermore, when delivered via AAV8, Cj4Cas9 achieves efficient genome editing of the Pcsk9 gene in mouse liver, leading to reduced serum cholesterol and LDL-C levels. Seeking to further expand its utility, we engineered Cj4Cas9 for higher activity by introducing L58Y/D900K mutations, resulting in a variant termed enCj4Cas9. This variant exhibits a two-fold increase in nuclease activity compared to the wild-type Cj4Cas9 and recognizes a simplified N3GG PAM, considerably expanding its targeting scope. These findings establish Cj4Cas9 and its engineered variants for fundamental research and therapeutic applications.
Similar content being viewed by others
Introduction
CRISPR-Cas9 is a widely used genome-editing tool that introduces precise modifications across diverse organisms. This system comprises a Cas9 nuclease and a single-guide RNA (sgRNA), which together form an RNA-protein complex (RNP)1. Within this complex, the sgRNA directs the Cas9 nuclease to a specific DNA target (protospacer), where it introduces double-strand breaks (DSBs). These breaks are subsequently repaired by cellular repair mechanisms, leading to desired genetic changes2.
Among the various Cas9 nucleases, Streptococcus pyogenes Cas9 (1368 amino acids) is the most widely used for genome editing2,3. However, SpCas9 has limitations, including inefficiency at certain genomic sites and a strict requirement for an NGG protospacer-adjacent motif (PAM) at the 3’ end of the target sequence2,4. This PAM constraint restricts the applicability of SpCas9 in precision genome editing techniques such as base editing5, prime editing6, and site-specific DNA integration7. In addition, its large protein size poses a significant challenge for delivery using adeno-associated virus (AAV) vectors, which have a packaging limit of ~4.7 kb. To overcome these challenges, researchers have turned to the diverse Cas9 orthologs in public databases, many of which recognize alternative PAMs8,9,10,11,12.
One such ortholog is Campylobacter jejuni Cas9 (CjCas9), which belongs to the Class 2, Type II-C CRISPR-Cas system and is one of the most compact Cas9 nucleases identified to date, consisting of only 984 amino acids13. However, CjCas9 exhibits relatively low activity and requires a long NNNNRYAC PAM sequence, which limits its utility. To address these limitations, researchers have pursued two main strategies. One strategy involves engineering CjCas9 to improve its activity. For example, Nakagawa et al. introduced L58Y/D900K mutations into CjCas9, resulting in a high-activity variant named enCjCas914; Ruta et al. employed directed evolution to develop another high-activity variant, UltraCjCas9, which incorporates five mutations (L58K, E189G, F214I, S492V, D900K, and K913S)15. Alternatively, researchers have explored CRISPR-Cas9 tools derived from CjCas9 orthologs to recognize non-NNNNRYAC PAM sequences. For instance, Chen et al. identified Cj2Cas9 and Cj3Cas9 from CjCas9 orthologs, which recognize an N4CYA PAM16; our previous work led to the development of Hsp1Cas9, Hsp2Cas9, and CcuCas9, which recognize N4RAA, N4CC, and N4CNA PAMs, respectively17. These advancements have expanded the versatility and utility of compact Cas9 nucleases for genome editing applications.
In this study, we screened a total of 11 CjCas9 orthologs and identified Cj4Cas9, a nuclease that recognizes an N3GRY PAM and demonstrates high activity at multiple genomic sites. Notably, we show that Cj4Cas9 can effectively disrupt the mouse Tyrosinase (Tyr) gene ex vivo in zygotes and the proprotein convertase subtilisin/kexin type 9 (Pcsk9) gene in vivo in adult mice. We further engineered Cj4Cas9 to improve the activity. These findings highlight the potential of Cj4Cas9 and its high-activity variants as a powerful tool for genome editing in both basic research and therapeutic applications.
Results
Investigation of CjCas9 ortholog activity
To explore the functional diversity of CjCas9-family nucleases, we searched the NCBI Gene database using the canonical CjCas9 sequence as a query and identified orthologs across various Campylobacter and Helicobacter strains. Based on the initial search results, we selected 11 orthologs with a broad range of amino acid sequence identities to CjCas9 (44.08–92.90%) for further characterization (Table 1). We selected orthologs based on two criteria: broad phylogenetic diversity to explore PAM variation, and presence of full-length cas9 genes with conserved domains. Two orthologs (Cj4Cas9 and Cj5Cas9) clustered with canonical CjCas9, while the remaining nine formed distinct clades (Supplementary Fig. S1). Alignment of the PAM‑interacting (PI) domains revealed that, across all screened CjCas9 orthologs, there are varied at 2–3 of the key residues (Arg866, Thr913, Ser915, and Ser951) compared with the reference CjCas9 (Supplementary Fig. S2). These residues are located within the PI domain and are known from the crystal structure of CjCas9 to be critical for PAM recognition18. This suggests that these orthologs may recognize different PAM sequences, which motivated us to experimentally assess their genome editing properties.
We further analyzed the genomic architecture of these CRISPR-Cas9 systems. Among the orthologs, ten exhibited a conserved arrangement, with the Cas9 ortholog followed by Cas1, Cas2, and a CRISPR array (Supplementary Fig. S3). The number of spacers within these arrays varied significantly, ranging from 4 to 40. Interestingly, Cco2Cas9 lacked the Cas1 and Cas2 genes, which may be lost or moved to other genomic loci during evolution. Alignment of repeat and tracrRNA sequences revealed a moderate level of conservation across the orthologs (Supplementary Fig. S4).
To assess the activity of these orthologs, we employed a previously developed GFP-activation assay8. In this system, a protospacer with a 7 bp random downstream sequence is inserted between the ATG start codon and the GFP coding sequence, disrupting GFP expression. The reporter construct is integrated into HEK293T cells via lentiviral transduction. If a Cas9 ortholog is functional, it will induce insertions or deletions (indels) at the target site, restoring GFP expression in a subset of cells (Fig. 1A, B). We synthesized each CjCas9 ortholog with human codon optimization, cloned them into mammalian expression plasmids, and co-transfected them with the corresponding sgRNA constructs into HEK293T cells. SpCas9 was included as a positive control. Five days post-transfection, GFP-positive cells were observed for seven out of the 11 CjCas9 orthologs, demonstrating their genome-editing activity (Fig. 1C). These results underscore the potential utility of CjCas9 orthologs for genome-editing applications.
A Schematic representation of the GFP-activation assay. A 7-bp random sequence followed by a 25-bp protospacer (target sequence) was inserted between the ATG start codon and the GFP-coding sequence, disrupting GFP expression. The reporter construct was packaged into a lentivirus vector and transfected into cells. sgRNA/Cas9-expressing plasmids were subsequently transfected into the reporter cells. Successful editing of the target sequence restored GFP expression. GFP-positive cells were sorted and analyzed via PCR amplification and deep sequencing to assess the activity of CjCas9 orthologs. B Schematic of the GFP-activation assay designed to assess the activity of CjCas9 orthologs. The Cas9 expression plasmid and the sgRNA expression plasmid are co-transfected into reporter cells for genome editing. C Representative images demonstrating GFP expression induced by CjCas9 orthologs. Cells transfected with SpCas9 served as a positive control for GFP activation. The percentage of GFP-positive cells is shown in the figure. BF bright field, GFP green fluorescent protein.
Analysis of CjCas9 ortholog PAM preferences
To determine the PAM preferences of the CjCas9 orthologs, GFP-positive cells were sorted using flow cytometry. The target sequences from these cells were then PCR-amplified and analyzed by deep sequencing. Using the sequencing data, we constructed PAM logos and PAM wheels to visualize the PAM preferences of each ortholog (Fig. 2A, B). As anticipated, SpCas9 exhibited a strong preference for the canonical NGG PAM, consistent with prior studies1,3, thereby validating the reliability of our experimental system. Comparative analysis of the PAM profiles across the CjCas9 orthologs revealed significant differences in their sequence preferences, reflecting evolutionary adaptations unique to each variant. Among the orthologs, Cj4Cas9 stood out due to its recognition of a unique NNNGRY PAM and its ability to induce a higher proportion of GFP-positive cells compared to the others. We selected Cj4Cas9 for further investigation in subsequent studies.
Optimization of Cj4Cas9 for genome editing
To evaluate whether Cj4Cas9 can perform genome editing using its own single guide RNA (sgRNA) scaffold, we designed a custom sgRNA scaffold for Cj4Cas9. This scaffold was created by fusing the 3′ end of a direct repeat with the 5′ end of the tracrRNA through a 4-nucleotide (nt) linker, following the design reported in a previous study of CjCas913. Sequence alignment revealed that the CjCas9 sgRNA scaffold and the Cj4Cas9 sgRNA scaffold differed by only a single mismatch (Supplementary Fig. S5A). Additionally, both scaffolds formed similar secondary RNA structures (Supplementary Fig. S5B), suggesting functional compatibility. We then transfected HEK293T cells with Cj4Cas9 and its sgRNA scaffold, using the CjCas9 sgRNA scaffold as a control, to edit the AAVS1-TS1 locus. The results demonstrated that the Cj4Cas9 sgRNA scaffold exhibited activity comparable to that of the CjCas9 sgRNA scaffold (Supplementary Fig. S5C). Based on these findings, we proceeded with the CjCas9 sgRNA scaffold for subsequent experiments.
Subsequently, we investigated the optimal spacer length for Cj4Cas9-mediated genome editing. We designed ten spacers ranging from 18 to 25-nt targeting the AAVS1-TS1 site. All sgRNAs were transcribed using the U6 promoter, and a 5′ guanine (G) was added when necessary to ensure efficient transcription. This additional G was not counted as part of the spacer length. Targeted deep sequencing revealed that spacers of 22- to 25-nt achieved higher editing efficiency compared to shorter spacers (Supplementary Fig. S6A). Consequently, we selected the 22-nt spacer for further studies. HEK293T cells were then transfected with Cj4Cas9 and its corresponding sgRNA, and samples were collected at 3-, 5-, and 7-days post-transfection. Targeted deep sequencing results indicated that Cj4Cas9-mediated gene editing became detectable by day 5 and remained stable thereafter (Supplementary Fig. S6B). In summary, these optimization experiments demonstrate that Cj4Cas9 is a functional genome-editing tool.
Test of Cj4Cas9 specificity
To test the mismatch tolerance of Cj4Cas9, we designed a panel of 11 sgRNAs with dinucleotide mismatches to target the AAVS1-TS1 locus. Five days after co-transfection of Cj4Cas9 with individual sgRNAs, targeted deep sequencing was performed to detect indel rates. The results showed that Cj4Cas9 exhibited moderate activity with mismatched sgRNAs MS2 and MS4, but minimal activity with others (Supplementary Fig. S7A, left panel). This low tolerance for mismatches ensures high specificity for precise genome-editing applications.
We then evaluated the genome-wide off-target effects of Cj4Cas9 at the EMX1-TS10 locus using GUIDE-seq technology19. After transfecting cells with the Cj4Cas9 plasmid, sgRNA, and GUIDE-seq oligonucleotides, we prepared sequencing libraries for deep sequencing. Analysis of the sequencing data demonstrated efficient on-target cleavage by Cj4Cas9, as evidenced by the high GUIDE-seq read counts at the target site (Supplementary Fig. S7B). Notably, only one off-target site was detected with this specific sgRNA. These results highlight the high specificity of Cj4Cas9.
Cj4Cas9 enables genome editing at endogenous sites
To further test the genome editing capability of Cj4Cas9, we selected a panel of 10 endogenous sites with NNNGRY PAMs targeting the AAVS1 locus in HEK293T cells. Five days after the co-transfection of CjCas9 and sgRNA plasmids (Fig. 3A, B), cells were harvested, and genomic DNA was extracted for target deep sequencing. Cj4Cas9 achieved robust editing efficiencies, with indel rates up to 32.81% (Fig. 3C). In parallel, we selected a panel of seven endogenous sites with NNNGRY PAMs targeting the Rosa26 locus in mouse neuroblastoma (N2a) cells. Cj4Cas9 achieved indel rates up to 29.18% (Fig. 3D). These results underscore Cj4Cas9 as a promising tool for genome editing in diverse cellular contexts.
A Schematic representation of the experimental design to assess Cj4Cas9 editing efficiency. B Illustration of the workflow for editing efficiency and mismatch tolerance assays. C Genome-editing efficiency of Cj4Cas9 in HEK293T cells, showing indel rates at target sites. D Editing efficiency of Cj4Cas9 in N2a cells, demonstrating comparable activity across mammalian cell types.
Cj4Cas9 enables genome editing in zygotes
Previous studies have shown that bi-allelic inactivation of Tyr disrupts melanin production, resulting in albino pups11,20. To achieve this, we designed five Tyr-targeting sgRNAs for Cj4Cas9 and tested their activity in N2a cells (Fig. 4A). Among them, Tyr-TS3 demonstrated the highest editing efficiency and was selected to evaluate the potential of Cj4Cas9 for genome editing in mouse zygotes (Fig. 4B). Cj4Cas9 mRNA, sgRNA targeting the Tyr gene, and GFP mRNA were in vitro transcribed and co-microinjected into pronuclear-stage embryos. GFP expression was observed 24 h after injection, indicating successful mRNA delivery (Supplementary Fig. S8A).
A Schematic diagram of the mouse Tyrosinase (Tyr) gene. The sgRNA target loci are shown. Red lines indicate PAMs. B Indel frequencies of Cj4Cas9 at 5 Tyr sites in N2a cell line (n = 3). Data are presented as the mean ± SD. C Workflow for genome editing in mouse zygotes. D Photograph of mice at one week after birth. Mice with mutations in Tyr presented the phenotype of albino. E Deep sequencing results showed indels in edited mouse pups.
Following this, we optimized the concentrations of Cj4Cas9 mRNA and sgRNA for microinjection by testing three combinations: 25/10, 50/25, and 100/50 ng/µL. The 25/10 ng/µL group showed the highest blastocyst development rate, with four out of ten injected zygotes developing into blastocysts (Supplementary Fig. S8B, C). In contrast, the 100/50 ng/µL group showed no blastocyst development. Both the 25/10 ng/µL and 50/25 ng/µL groups exhibited similar editing efficiencies in blastocysts (Supplementary Fig. S8D).
Based on the combination of highest embryo viability and efficient editing, we selected the 25/10 ng/µL condition for subsequent zygote injections. In the embryo transfer experiment, a total of 40 pronuclear-stage embryos were injected, of which 33 developed to the 2-cell stage. These embryos were transferred unilaterally into the oviducts of pseudo pregnant females (11 embryos per side). Three surrogate mothers gave birth to six pups in total. Analysis of the resulting pups’ coat color confirmed successful Tyr gene editing, as evidenced by the absence of black pigmentation in the edited mice (Fig. 4D). Targeted deep sequencing revealed the corresponding indel frequencies for each pup, with the highest observed in an albino pup at 98.83% (Fig. 4E). These results demonstrate that Cj4Cas9 is an effective tool for genome editing in mouse zygotes.
Cj4Cas9 enables in vivo genome editing
To investigate the therapeutic potential, we evaluated the efficacy of Cj4Cas9 for in vivo genome editing. The compact size of Cj4Cas9 enabled the co-packaging of both a cytomegalovirus (CMV)-driven Cj4Cas9 expression cassette and a U6-driven sgRNA into a single AAV vector (Fig. 5A). To assess Cj4Cas9 activity in vivo, we targeted Pcsk9, a therapeutically relevant gene involved in cholesterol homeostasis21. Pcsk9 encodes a protein that regulates the degradation of LDL receptors, thereby influencing the levels of low-density lipoprotein cholesterol (LDL-C), a major carrier of cholesterol (CHO) in the blood. Elevated LDL-C levels are associated with an increased risk of cardiovascular diseases, whereas reduced PCSK9 activity has been shown to lower LDL-C and total cholesterol levels, offering protective benefits against cardiovascular conditions22,23.
A Schematic diagram of the AAV8-Cj4Cas9 vectors. CMV, cytomegalovirus. U6, U6 promoter. ITR, inverted terminal repeats. B The procedure of in vivo genome editing in mice. Viruses are administered intravenously via tail vein injection in 4–5 weeks C57BL/6J mice. C, D Time course of total serum cholesterol and LDL-C in animals treated with AAV8-Cj4Cas9 respectively (n = 3). E, F Liver function ALT and AST level in the AAV-Cj4Cas9 and PBS group (n = 3). G Indel efficiencies of Pcsk9 in mouse liver. The PBS-injected group serves as a control (n = 3). Band intensities were quantified by ImageJ and normalized to GAPDH, with values shown relative to the PBS group (Lane 2). H Analysis of PCSK9 expression in mouse liver by Western blot. Data represent means ± SD. NS no significant, ∗p < 0.05.
We designed five Pcsk9-targeting sgRNAs and confirmed their activity in N2a cells (Supplementary Fig. S9A). All five sgRNAs exhibited comparable editing efficiencies (Supplementary Fig. S9B). Based on previous studies indicating that using multiple sgRNAs can enhance editing efficiency24,25, we packaged each sgRNA individually with Cj4Cas9 into a hepatotropic high-expression AAV serotype, AAV826. The AAV-Cj4Cas9 constructs targeting Pcsk9 were mixed at equal titers and administered via tail vein injection into mice at a dose of 2 × 1011 genome copies (GCs). A control group received PBS. Serum samples were collected at 0-, 14-, 28-, and 42-days post-injection for analysis, and all mice were euthanized at 42 days (Fig. 5B).
The results revealed a 42.68% reduction in serum LDL-C levels and a 34.04% reduction in serum cholesterol in the AAV-Cj4Cas9-treated group, whereas PBS-treated mice maintained normal LDL-C and cholesterol levels at 42 days post-injection (Fig. 5C, D). To preliminarily evaluate in vivo safety, serum aspartate aminotransferase (AST) and alanine aminotransferase (ALT) levels were measured at the endpoint, showing no significant differences between the AAV-Cj4Cas9 and PBS groups (Fig. 5E, F). Targeted deep sequencing demonstrated an average editing efficiency of 24.43% (Fig. 5G). Consistent with these findings, western blot analysis confirmed a reduction in PCSK9 protein levels in Cj4Cas9-treated mice compared to PBS-treated controls (Fig. 5H, Supplementary Fig. S12). These results collectively demonstrate the efficacy of Cj4Cas9 for in vivo genome editing and its potential therapeutic application in modulating cholesterol metabolism.
Engineering of Cj4Cas9 for enhanced activity
To improve the editing efficiency, we engineered Cj4Cas9 based on previously developed high-activity enCjCas914 and ultraCjCas915. They contain mutations that enhance interactions between CjCas9 and the nucleic acids and thereby improve the DNA cleavage activity14,15,27. We aligned CjCas9 to enCjCas9 and ultraCjCas9 and identified corresponding mutations (Supplementary Fig. S10). First, we introduced L58Y/D900K mutations into Cj4Cas9 to generate enCj4Cas9. Subsequently, we incorporated E189G/F214I/S492V/K913S mutations identified from ultraCjCas9 into enCj4Cas9 to generate ultraCj4Cas9 (Fig. 6A).
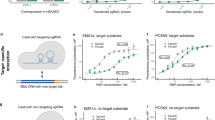
A Schematic representation of the mutation sites introduced in enCj4Cas9 and ultraCj4Cas9, highlighting the key amino acid changes engineered to improve activity. B Indel efficiencies across 16 target sites, demonstrating the enhanced performance of enCj4Cas9 and ultraCj4Cas9 compared to wild-type Cj4Cas9. Statistical significance was determined using two-sided Student’s t tests with FDR correction for multiple comparisons. C Box plots summarizing the editing efficiencies of wild-type Cj4Cas9, enCj4Cas9, and ultraCj4Cas9, illustrating the overall increase in activity of the engineered variants. D Western blot analysis confirming comparable protein expression levels of Cj4Cas9, enCj4Cas9, and ultraCj4Cas9 in transfected cells. GAPDH was used as a loading control to ensure equal protein loading. Data represent mean ± SD for n = 3 biologically independent experiments. p values were determined using a two-sided Student’s t test. NS no significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
The editing efficiencies of wild-type Cj4Cas9 and its engineered variants, enCj4Cas9 and ultraCj4Cas9, were systematically evaluated across 16 target sites. Targeted deep sequencing revealed that both enCj4Cas9 and ultraCj4Cas9 demonstrated significantly enhanced editing efficiencies compared to the wild-type enzyme (Fig. 6B). In particular, enCj4Cas9 and ultraCj4Cas9 exhibited 2.03-fold and 2.14-fold higher overall activity, respectively, relative to the wild-type (Fig. 6C). Western blot analysis confirmed that the protein expression levels of all three variants were comparable, eliminating the possibility that increased activity was due to differential expression (Fig. 6D, Supplementary Fig. S13).
Additionally, we investigated the specificity of enCj4Cas9 and ultraCj4Cas9 by designing sgRNAs with dinucleotide mismatches targeting the AAVS1-TS1 locus. The results indicated that Cj4Cas9 and enCj4Cas9 exhibited similar specificity profiles, whereas ultraCj4Cas9 exhibited reduced specificity at MS2-8 sites (Supplementary Fig. S7A). To assess genome-wide off-target effects, we performed the GUIDE-seq assay using a sgRNA targeting the EMX1-TS10 locus. Five days after co-transfection of HEK293T cells with Cj4Cas9/sgRNA-expressing plasmids and GUIDE-seq oligos, genomic DNA was extracted and subjected to deep sequencing. GUIDE-seq revealed one off-target site (OT) for wild-type Cj4Cas9, two sites for enCj4Cas9, and four sites for ultraCj4Cas9 (Supplementary Fig. S7B). A shared off target site, EMX1-TS10-OT1 was further validated by targeted amplicon sequencing (Supplementary Fig. S7C). The off target indel levels were ≤1% for enCj4Cas9 but averaged ~4% for Cj4Cas9 and ultraCj4Cas9. These data suggest that the ostensibly low number of off-targets in Cj4Cas9 may partly reflect its lower cleavage activity, whereas the engineered variants enCj4Cas9 and ultraCj4Cas9 exhibit higher on-target activity accompanied by additional off-target detection. Importantly, enCj4Cas9 maintained a balance of improved activity and minimal off-target editing, supporting its selection as the optimized variant for further application. To evaluate whether off-target effects might accumulate over time, we performed a time-course analysis of editing efficiencies at 3-, 5-, and 7-days post-transfection. The results showed that indel frequencies plateaued after day 3 across all variants, with no significant increase at later time points (Supplementary Fig. S7D), suggesting that off-target accumulation is minimal under our transient transfection conditions. These results highlight the superior performance of the engineered Cj4Cas9 variants in genome editing applications. Given these results, we focused on enCj4Cas9 for subsequent characterization.
The PAM preference of the enCj4Cas9 variant was further characterized using the GFP-activation assay. Intriguingly, the results demonstrated that enCj4Cas9 preferentially recognized a simple NNNGG PAM (Fig. 7A, B). We proceeded to compare the editing activity of WT Cj4Cas9, enCj4Cas9, SpRY, a previously engineered SpCas9 variant with relaxed PAM specificity28 and WT SpCas9. We designed a panel of 13 endogenous sites containing NGGGRY PAMs. Five days post-transfection, genomic DNA was extracted and subjected to targeted deep sequencing. The results revealed that Cj4Cas9 exhibited editing activity comparable to that of SpRY and SpCas9 across the tested sites (Fig. 7C, D). To ensure consistency, all four Cas9 orthologs were cloned into identical construct backbones, and Western blot analysis confirmed comparable protein expression levels among Cj4Cas9, enCj4Cas9, SpRY and WT SpCas9 (Fig. 7E, Supplementary Fig. S14).
A PAM logo and B PAM wheel diagrams show that enCj4Cas9 recognizes an NNNGG PAM. C Indel efficiencies of Cj4Cas9, enCj4Cas9, SpRY and SpCas9 at selected target sites. Statistical significance was assessed using two-sided Student’s t tests with FDR correction for multiple comparisons. D Box plots illustrate no significant differences in editing efficiencies between enCj4Cas9 and SpRY. E Western blot analysis showing equivalent protein expression levels of Cj4Cas9, enCj4Cas9, SpRY and SpCas9 in transfected cells. Data represent mean ± SD for n = 3 biologically independent experiments. p values were determined using a two-sided Student’s t test. NS no significant, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Enhanced Cj4Cas9 activity by fusion to non-specific DNA-binding proteins
In parallel, to explore additional strategies for enhancing activity, we constructed fusion proteins by linking non-sequence-specific DNA-binding domains (DBDs), Sso7d and HMG-D, to Cj4Cas9 (Supplementary Fig. S11A), as previously described by Li et al.23. Although these DBDs improved editing activity at certain loci, neither fusion variant showed a significant overall enhancement in editing efficiency (Supplementary Fig. S11B, C). Western blot analysis confirmed comparable protein expression levels among the three variants, ruling out expression differences as the cause of increased activity (Supplementary Figs. S11D, S15). These findings suggest that the intrinsic structural modifications in enCj4Cas9 and ultraCj4Cas9 play a more pivotal role in enhancing activity compared to fusion strategies involving DBDs.
Discussion
Among the numerous CRISPR-Cas9 tools developed to date, CjCas9 stands out due to its exceptionally compact size (984 amino acids), which is smaller than other Cas9 orthologs. To expand its targeting scope, we and others have previously identified five closely related CjCas9 orthologs that exhibit activity in human cells. For instance, Cj2Cas9 and Cj3Cas9 recognize an N4CYA PAM16, while Hsp1Cas9, Hsp2Cas9, and CcuCas9 recognize N4RAA, N4CC, and N4CNA PAMs, respectively17. In this study, we screened a panel of 11 CjCas9 orthologs and identified seven active nucleases, which displayed highly diverse PAM preferences, consistent with our earlier findings29. These orthologs represent a valuable resource for future genetic engineering efforts aimed at developing novel genome-editing tools with expanded capabilities.
Among these orthologs, Cj4Cas9 is particularly noteworthy due to its compact size (985 amino acids) and unique PAM preference (NNNGRY). Its small genome size makes it highly suitable for AAV delivery, a critical advantage for therapeutic applications. Moreover, Cj4Cas9 demonstrated robust editing efficiency in both in vitro and in vivo settings, including the successful disruption of the Tyr gene in mouse zygotes and the Pcsk9 gene in adult mice. Although our in vivo experiments were performed with a modest sample size, as is typical in early-stage AAV-mediated CRISPR studies, no overt signs of toxicity or behavioral abnormalities were observed. In addition, preliminary serum biomarker analysis (ALT and AST) showed no significant liver injury, and targeted deep sequencing of potential off-target sites did not reveal detectable editing events. However, more comprehensive safety evaluations, including expanded toxicity profiling, immunogenicity assessment (e.g., cytokine release, anti-Cas9 antibody formation), and unbiased genome-wide off-target analysis, will be important priorities for future studies.
To further enhance its utility, we engineered a high-activity variant, enCj4Cas9, by introducing L58Y/D900K mutations, which resulted in a two-fold increase in nuclease activity compared to the wild-type enzyme. Notably, enCj4Cas9 exhibited a simplified PAM preference (NNNGG), significantly broadening its targeting scope. Collectively, these advancements highlight the potential of Cj4Cas9 and its engineered variants for more efficient genome-editing applications in both research and therapy.
Conclusion
In summary, we developed a novel small Cj4Cas9 that can efficiently induce genome editing in vivo. We further generated enCj4Cas9 with improved activity and a larger targeting scope. Cj4Cas9 and enCj4Cas9 are promising tools for basic research and therapeutic applications.
Materials and methods
Animals
Age-matched 4–8 weeks C57BL/6 and ICR mice (Institute of Laboratory Animal Sciences, JSJ, China) were used as controls. All experimental mice were maintained in the animal facility of School of Life Sciences, Fudan University. Mice were housed in a 12-h light/dark cycle, with enough water and food. The mouse procedures were approved by the Institutional Animal Care and Use Committee of Fudan University (No.: 2024JS075). We have complied with all relevant ethical regulations for animal use. All procedures were performed in accordance with the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research.
Cas9 expression plasmid construction
The plasmid Cas9-AAV was amplified by the primers Cas9-F/Cas9-R to obtain the Cas9-AAV backbone. The human codon optimized Cas9 gene (Supplementary Data 1) was synthesized by HuaGene (Shanghai, China) and cloned into the Cas9-AAV backbone by the NEBuilder assembly tool (NEB) according to the manufacturer’s instructions. Sequences of Cas9 were confirmed by Sanger sequencing (Azenta, Suzhou, China).
sgRNA expression plasmid construction
The sgRNA expression plasmids were constructed by ligating sgRNA into the Bbs I-digested U6-Cj_scaffold plasmid, which is the same as CjCas9-scaffold. The primer sequences and target sequences are listed in Supplementary Data 2 and Supplementary Data 3, respectively.
Cell culture and transfection
The cell culture reagents were purchased from Gibco unless otherwise indicated. Human embryonic kidney 293T (HEK293T) and mouse neuroblastoma (N2a) cell lines were obtained from American Type Culture Collection (ATCC). Both cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM). All cell cultures were supplemented with 10% fetal bovine serum (FBS) (Gibco) that was inactivated at 56 °C for 30 min and 1% penicillin-streptomycin (Gibco). All cells were cultured in a humidified incubator at 37 °C and 5% CO2. All cell line identities were validated by STR profiling (ATCC) and repeatedly tested for mycoplasma by PCR.
HEK293T and N2a cells were transfected with Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. For transient transfection, a total of 500 ng Cas9-expressing plasmid and 300 ng sgRNA plasmid were co-transfected into a 24-well plate. For Cas9 PAM sequence screening, 1.2 × 107 HEK293T cells were transfected with 10 μg of Cas9 plasmid and 5 μg of sgRNA plasmid in 10 cm dishes.
PAM discovery assay
The PAM discovery assays were performed essentially as previously described, using a library of 7 N. Transfected library cells with a certain percentage of GFP-positive cells were collected by centrifugation at 1000 rpm for 5 min and resuspended in PBS. Then, GFP-positive cells were collected by flow cytometry and cultured in six-well plates. Five days after culture, we extracted the genome and built deep sequencing library.
Genome editing and deep sequencing analysis of indels for endogenous sites
Cells were seeded into 24-well plates one day prior to transfection and transfected at 70–80% confluency using Lipofectamine 2000 (Life Technologies) following the manufacturer’s recommended protocol. For genome editing, 106 cells were transfected with a total of 500 ng of Cas9 plasmid and 300 ng of sgRNA plasmid in 28-well plates. Six days after transfection, the cells were harvested, and genomic DNA was extracted in QuickExtract DNA Extraction Solution (Epicenter). To measure indel frequencies, the target sites were amplified by two rounds of nested PCR to add the Illumina adaptor sequence. The PCR products (200–300 bp in length) were gel-extracted by a Gel Extraction Kit (TIANGEN) for deep sequencing.
mRNA and gRNA preparation
The Cj4Cas9 and GFP were PCR amplified with primers containing T7 promoter and 75 nt poly-A tail with KOD FX (TOYOBO) and transcribed in vitro using the RNA transcription kit (novoprotein). Cj4Cas9 gRNA was PCR amplified with primers containing T7 promoter. These in vitro transcribed mRNA and gRNA were stored at −80 °C after purification with Monarch RNA cleanup kit (NEB) according to the manufacturer’s protocol. Aliquoting was necessary for convenience and longer storage.
Microinjection of mouse zygotes and genotyping
Groups of 30 pronuclear-stage zygotes were injected with Cj4Cas9 mRNA (25 ng/µL) and sgRNA (12.5 ng/µL). The zygotes were cultured in KSOM at 37 °C with 5% CO2. All zygotes were collected individually with QuickExtract DNA Extraction Solution (Epicenter) when developed into blastocyst stage to analyze gene editing efficiencies. Embryo lysis was subjected to PCR amplification. Amplicons containing target sequences were analyzed by deep sequencing.
GUIDE-seq off-target assay
We performed a GUIDE-seq experiment with some modifications to the original protocol, as described19. On the day of the experiment, 2 × 105 HEK293T cells per target site were harvested and washed in PBS and transfected with 500 ng of Cas9 plasmid, 500 ng of sgRNA plasmid and 100 pmol annealed GUIDE-seq oligonucleotides through the Neon Transfection System. The electroporation voltage, width, and number of pulses were 1150 V, 20 ms, and 2 pulses, respectively. Genomic DNA was extracted with the DNeasy Blood and Tissue kit (QIAGEN) for 6–10 days according to cell proliferation after electroporation according to the manufacturer’s protocol. The genome library was prepared using the NEBNext Ultra II kit (NEB) and sequenced on an Illumina NovaSeq S4 platform (2 × 150 bp), generating ~20 million paired end reads per sample. Two biological replicates were included per Cas9 variant.
Western blot
Cells were collected and resuspended in lysis RIPA (Beyotime) supplemented with 1 mmol/L phenylmethanesulfonyl fluoride (PMSF) (Beyotime). Cell lysates were centrifuged at 10,000 × g for 20 min at 4 °C, and the supernatants were collected. Equal amounts of protein were separated by SDS-PAGE gel and transferred onto PVDF membranes. The membranes were blocked with 5% non-fat milk in TBS-T (0.1% Tween 20 in 1 × TBS) for 1 h at room temperature and then incubated overnight with the anti-HA antibody (1:1000, Abcam, ab236632), anti-GAPDH antibody (1:2000, CST, 5174s), or anti-PCSK9 (1:2000, ab185194) at 4 °C overnight. The membranes were washed three times in TBS-T for 5 min each time and incubated with the anti-rabbit (SA00001-2) secondary antibodies at a 1:10,000 dilution for 1 h at room temperature. The membranes were then washed with TBST buffer three times and imaged.
Histology and serum analysis
Tissues were fixed using 4% PFA at 4 °C overnight and dehydrated the next day before paraffinization. Paraffin blocks were cut into 5 µm thick sections, deparaffinized with xylene, and rehydrated. Sections were stained for DAPI and examined for transduction efficacy.
Serum levels of LDL-C were evaluated by surfactant removal method (Gcell) following the manufacturer’s instructions. Similarly, the Total Cholesterol Assay Kit (Gcell) was utilized to measure CHO levels in the serum.
Statistics
All statistical analyses were performed on data from at least n = 3 biologically independent experiments using R. For pairwise comparisons (in Figs. 6C and 7D), unpaired or paired two-tailed Student’s t tests were conducted using the dplyr package. For multi-group comparisons across multiple target sites (Figs. 6B and 7C), pairwise two-sided t-tests were performed, and p-values were adjusted using the false discovery rate (FDR) method with the rstatix package. Detailed information on samples and experimental replicates can be found in the figure legends. p values less than 0.05 were considered significant, denoted as NS. no significant, ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All NGS data have been uploaded to the NCBI Sequence Read Archive database under accession code PRJNA1307308. There are no restrictions on data availability. All raw data in this study were listed in Supplementary Data 4. Uncropped and unedited western blot images are provided in Supplementary Figs. S12–14.
References
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Wang, D. et al. Optimized CRISPR guide RNA design for two high-fidelity Cas9 variants by deep learning. Nat. Commun. 10, 4284 (2019).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019).
Kan, Y., Ruis, B., Takasugi, T. & Hendrickson, E. A. Mechanisms of precise genome editing using oligonucleotide donors. Genome Res. 27, 1099–1111 (2017).
Hu, Z. et al. A compact Cas9 ortholog from Staphylococcus Auricularis (SauriCas9) expands the DNA targeting scope. PLoS Biol. 18, e3000686 (2020).
Hu, Z. et al. Discovery and engineering of small SlugCas9 with broad targeting range and high specificity and activity. Nucleic Acids Res. 49, 4008–4019 (2021).
Wang, S. et al. Compact SchCas9 recognizes the simple NNGR PAM. Adv. Sci. (Weinh.) 9, e2104789 (2022).
Edraki, A. et al. A compact, high-accuracy Cas9 with a dinucleotide PAM for in vivo genome editing. Mol. Cell 73, 714–726.e714 (2019).
Gasiunas, G. et al. A catalogue of biochemically diverse CRISPR-Cas9 orthologs. Nat. Commun. 11, 5512 (2020).
Kim, E. et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 8, 14500 (2017).
Nakagawa, R. et al. Engineered Campylobacter jejuni Cas9 variant with enhanced activity and broader targeting range. Commun. Biol. 5, 211 (2022).
Ruta, G. V. et al. Eukaryotic-driven directed evolution of Cas9 nucleases. Genome Biol. 25, 79 (2024).
Chen, S. et al. Compact Cje3Cas9 for efficient in vivo genome editing and adenine base editing. CRISPR J. 5, 472–486 (2022).
Gao, S. et al. Genome editing with natural and engineered CjCas9 orthologs. Mol. Ther. 31, 1177–1187 (2023).
Yamada, M. et al. Crystal structure of the minimal Cas9 from Campylobacter jejuni reveals the molecular diversity in the CRISPR-Cas9 systems. Mol. Cell 65, 1109–1121.e1103 (2017).
Tsai, S. Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015).
Wang, M. et al. Hypercompact TnpB and truncated TnpB systems enable efficient genome editing in vitro and in vivo. Cell Discov. 10, 31 (2024).
Musunuru, K. et al. In vivo CRISPR base editing of PCSK9 durably lowers cholesterol in primates. Nature 593, 429–434 (2021).
Sabatine, M. S. et al. Evolocumab and clinical outcomes in patients with cardiovascular disease. N. Engl. J. Med. 376, 1713–1722 (2017).
Yin, S. et al. Engineering of efficiency-enhanced Cas9 and base editors with improved gene therapy efficacies. Mol. Ther. 31, 744–759 (2023).
Jang, D. E. et al. Multiple sgRNAs with overlapping sequences enhance CRISPR/Cas9-mediated knock-in efficiency. Exp. Mol. Med. 50, 1–9 (2018).
Li, J., Kong, D., Ke, Y., Zeng, W. & Miki, D. Application of multiple sgRNAs boosts efficiency of CRISPR/Cas9-mediated gene targeting in Arabidopsis. BMC Biol. 22, 6 (2024).
Zincarelli, C., Soltys, S., Rengo, G. & Rabinowitz, J. E. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 16, 1073–1080 (2008).
Schmidheini, L. et al. Continuous directed evolution of a compact CjCas9 variant with broad PAM compatibility. Nat. Chem. Biol. 20, 333–343 (2024).
Walton, R. T., Christie, K. A., Whittaker, M. N. & Kleinstiver, B. P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368, 290–296 (2020).
Wei, J. et al. Closely related type II-C Cas9 orthologs recognize diverse PAMs. Elife 11, https://doi.org/10.7554/eLife.77825 (2022).
Acknowledgements
This work was supported by grants from the National Key Research and Development Program of China (2023YFC2705600, 2023YFC2705602, 2021YFA0910602); the National Natural Science Foundation of China (82370254, 82070258); and the Science and Technology Research Program of Shanghai (24HC2810100, 23ZR1426000).
Author information
Authors and Affiliations
Contributions
T.Y. Wang conceived and designed the experiments. T.Y. Wang, Y.F. Tian, R. Yin, M.R. Li, J. Luo, and Y. Yang performed the experiments. Chendong Zhang, Hongyan Chen provided resources. Yongming Wang, and Daru Lu provided experimental guidance and supervision; Yongming Wang contributed to writing and reviewing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Informed consent
Informed consents were obtained from all subjects involved in the study.
Peer review
Peer review information
Communications Biology thanks Tristan Henser-Brownhill, Cem Kuscu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Yuhong Cao and Mengtan Xing. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, T., Tian, Y., Yin, R. et al. In vivo genome editing with a novel Cj4Cas9. Commun Biol 9, 152 (2026). https://doi.org/10.1038/s42003-025-09430-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-09430-9