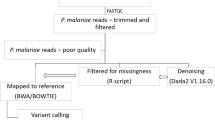
Abstract
Understanding evolution of human pathogens requires looking beyond the effects of recent interventions. To study malaria parasites prior to widespread drug selection, Plasmodium falciparum genomes were sequenced from the oldest population-based set of archived research samples yet identified, placental blood collected in the Gambia between 1966 and 1971. High-quality data were obtained from 54 infected samples, showing that genomic complexity within infections was high, most infections were genetically unrelated, and no drug resistance alleles were detected. Strong signatures of positive selection are clearly seen at multiple loci throughout the genome, most of which encode surface proteins that bind erythrocytes and are targets of acquired antibody responses. Comparison of population samples obtained over a following period of almost 50 years revealed major directional allele frequency changes at several loci apart from drug resistance genes. Exceptional changes over this time are seen at gdv1 that regulates the rate of parasite sexual conversion required for transmission, and at the unlinked Pfsa1 and Pfsa3 loci previously associated with infection of individuals with sickle-cell trait. Other affected loci encode surface and transporter proteins warranting targeted functional analyses. This identification of key long-term adaptations is important for understanding and managing future evolution of malaria parasites.
Similar content being viewed by others
Data availability
All original data reported here are fully available as outlined in the tables and data spreadsheets in the Supplementary Data 1–9. This includes the parasite genome sequences from each of the individual samples, for which the European Nucleotide Archive (ENA) accession numbers are listed in Supplementary Data 1 and 6.
References
Amambua-Ngwa, A. et al. Chloroquine resistance evolution in Plasmodium falciparum is mediated by the putative amino acid transporter AAT1. Nat. Microbiol 8, 1213–1226 (2023).
Pelleau, S. et al. Adaptive evolution of malaria parasites in French Guiana: Reversal of chloroquine resistance by acquisition of a mutation in pfcrt. Proc. Natl. Acad. Sci. USA 112, 11672–11677 (2015).
Carrasquilla, M. et al. Resolving drug selection and migration in an inbred South American Plasmodium falciparum population with identity-by-descent analysis. PLoS Pathog. 18, e1010993 (2022).
Florimond, C. et al. Impact of piperaquine resistance on malaria treatment effectiveness in The Guianas: a descriptive epidemiological study. Lancet Infect. Dis. 24, 161–171 (2024).
MalariaGen et al. Pf7: an open dataset of Plasmodium falciparum genome variation in 20,000 worldwide samples. Wellcome Open Res. 8, 22 (2023).
Fola, A. A. et al. Plasmodium falciparum resistant to artemisinin and diagnostics have emerged in Ethiopia. Nat. Microbiol 8, 1911–1919 (2023).
Conrad, M. D. et al. Evolution of partial resistance to Artemisinins in malaria parasites in Uganda. N. Engl. J. Med. 389, 722–732 (2023).
Band, G. et al. Malaria protection due to sickle haemoglobin depends on parasite genotype. Nature 602, 106–111 (2021).
Michel, M. et al. Ancient genomes shed light on the history of human malaria. Nature 631, 125–133 (2024).
Enabulele, E. E. et al. Prospecting for zoonotic pathogens by using targeted DNA enrichment. Emerg. Infect. Dis. 29, 1566–1579 (2023).
Nelder, M. P., Schats, R., Poinar, H. N., Cooke, A. & Brickley, M. B. Pathogen prospecting of museums: Reconstructing malaria epidemiology. Proc. Natl. Acad. Sci. USA 121, e2310859121 (2024).
Amambua-Ngwa, A. et al. Consistent signatures of selection from genomic analysis of pairs of temporal and spatial Plasmodium falciparum populations from The Gambia. Sci. Rep. 8, 9687 (2018).
Amambua-Ngwa, A. et al. SNP genotyping identifies new signatures of selection in a deep sample of West African Plasmodium falciparum malaria parasites. Mol. Biol. Evol. 29, 3249–3253 (2012).
Nwakanma, D. C. et al. Changes in malaria parasite drug resistance in an endemic population over a 25-year period with resulting genomic evidence of selection. J. Infect. Dis. 209, 1126–1135 (2014).
Ceesay, S. J. et al. Changes in malaria indices between 1999 and 2007 in The Gambia: a retrospective analysis. Lancet 372, 1545–1554 (2008).
Ceesay, S. J. et al. Continued decline of malaria in The Gambia with implications for elimination. PLoS One 5, e12242 (2010).
Greenwood, B. M. et al. Mortality and morbidity from malaria among children in a rural area of The Gambia, West Africa. T R. Soc. Trop. Med H. 81, 478–485 (1987).
McGregor, I. A., Gilles, H. M., Walters, J. H., Davies, A. H. & Pearson, F. A. Effects of heavy and repeated malarial infections on Gambian infants and children; effects of erythrocytic parasitization. Br. Med J. 2, 686–692 (1956).
Mwesigwa, J. et al. On-going malaria transmission in The Gambia despite high coverage of control interventions: a nationwide cross-sectional survey. Malar. J. 14, 314 (2015).
Amambua-Ngwa, A. et al. Population genomic scan for candidate signatures of balancing selection to guide antigen characterization in malaria parasites. PLoS Genet 8, e1002992 (2012).
Duffy, C. W. et al. Population genetic structure and adaptation of malaria parasites on the edge of endemic distribution. Mol. Ecol. 26, 2880–2894 (2017).
Mobegi, V. A. et al. Genome-wide analysis of selection on the malaria parasite Plasmodium falciparum in West African populations of differing infection endemicity. Mol. Biol. Evol. 31, 1490–1499 (2014).
Carter, R. & McGregor, I. A. Enzyme variation in Plasmodium falciparum in The Gambia. T R. Soc. Trop. Med H. 67, 830–837 (1973).
Wilson, R. J., McGregor, I. A., Hall, P., Williams, K. & Bartholomew, R. Antigens associated with Plasmodium falciparum infections in man. Lancet 2, 201–205 (1969).
Wilson, R. J. M. Antigens and antibodies associated with Plasmodium falciparum infections in West Africa. T R. Soc. Trop. Med H. 64, 547 (1970).
Michel-Todo, L. et al. Patterns of heterochromatin transitions linked to changes in the expression of Plasmodium falciparum clonally variant genes. Microbiol Spectr. 11, e0304922 (2023).
Naung, M. T. et al. Global diversity and balancing selection of 23 leading Plasmodium falciparum candidate vaccine antigens. PLoS Comput Biol. 18, e1009801 (2022).
Weedall, G. D. & Conway, D. J. Detecting signatures of balancing selection to identify targets of anti-parasite immunity. Trends Parasitol. 26, 363–369 (2010).
Amambua-Ngwa, A. et al. Major subpopulations of Plasmodium falciparum in sub-Saharan Africa. Science 365, 813–816 (2019).
Zhu, S. J. et al. The origins and relatedness structure of mixed infections vary with local prevalence of P. falciparum malaria. Elife 8, e40845 (2019).
Eksi, S. et al. Plasmodium falciparum gametocyte development 1 (Pfgdv1) and gametocytogenesis early gene identification and commitment to sexual development. PLoS Pathog. 8, e1002964 (2012).
Filarsky, M. et al. GDV1 induces sexual commitment of malaria parasites by antagonizing HP1-dependent gene silencing. Science 359, 1259–1263 (2018).
Freville, A. et al. Expression of the MSPDBL2 antigen in a discrete subset of Plasmodium falciparum schizonts is regulated by GDV1 but may not be linked to sexual commitment. mBio, 15, e0314023 (2024).
Lamarque, M. et al. Food vacuole proteome of the malarial parasite Plasmodium falciparum. Proteom. Clin. Appl 2, 1361–1374 (2008).
Duffy, C. W. et al. Comparison of genomic signatures of selection on Plasmodium falciparum between different regions of a country with high malaria endemicity. BMC Genomics 16, 527 (2015).
Weedall, G. D., Preston, B. M., Thomas, A. W., Sutherland, C. J. & Conway, D. J. Differential evidence of natural selection on two leading sporozoite stage malaria vaccine candidate antigens. Int J. Parasitol. 37, 77–85 (2007).
Cheeseman, I. H. et al. Population structure shapes copy number variation in malaria parasites. Mol. Biol. Evol. 33, 603–620 (2016).
Martin, R. E., Ginsburg, H. & Kirk, K. Membrane transport proteins of the malaria parasite. Mol. Microbiol 74, 519–528 (2009).
Kenthirapalan, S., Waters, A. P., Matuschewski, K. & Kooij, T. W. Functional profiles of orphan membrane transporters in the life cycle of the malaria parasite. Nat. Commun. 7, 10519 (2016).
Stewart, L. B. et al. Multiplication rate variation of malaria parasites from hospital cases and community infections. Sci. Rep. 15, 666 (2025).
Kwiatkowski, D. P. How malaria has affected the human genome and what human genetics can teach us about malaria. Am. J. Hum. Genet 77, 171–192 (2005).
Kariuki, S. N. & Williams, T. N. Human genetics and malaria resistance. Hum. Genet 139, 801–811 (2020).
McGregor, I. A. Studies in the acquisition of immunity to Plasmodium falciparum infections in Africa. T R. Soc. Trop. Med H. 58, 80–92 (1964).
Massamba, J. E., Djontu, J. C., Vouvoungui, C. J., Kobawila, C. & Ntoumi, F. Plasmodium falciparum multiplicity of infection and pregnancy outcomes in Congolese women from southern Brazzaville, Republic of Congo. Malar. J. 21, 114 (2022).
Mayengue, P. I. et al. Submicroscopic Plasmodium falciparum infections and multiplicity of infection in matched peripheral, placental and umbilical cord blood samples from Gabonese women. Trop. Med Int Health 9, 949–958 (2004).
Duffy, C. W. et al. Multi-population genomic analysis of malaria parasites indicates local selection and differentiation at the gdv1 locus regulating sexual development. Sci. Rep. 8, 15763 (2018).
Usui, M. et al. Plasmodium falciparum sexual differentiation in malaria patients is associated with host factors and GDV1-dependent genes. Nat. Commun. 10, 2140 (2019).
Chourasia, B. K. et al. Plasmodium falciparum Clag9-associated PfRhopH complex is involved in merozoite binding to human erythrocytes. Infect. Immun. 88, e00504-19 (2020).
Goel, S. et al. Dual stage synthesis and crucial role of cytoadherence-linked asexual gene 9 in the surface expression of malaria parasite var proteins. Proc. Natl. Acad. Sci. USA 107, 16643–16648 (2010).
McGregor, I. A., Williams, K., Billewicz, W. Z. & Thomson, A. M. Haemoglobin concentration and anaemia in young West African (Gambian) children. Trans. R. Soc. Trop. Med Hyg. 60, 650–667 (1966).
Stevens, G. A. et al. Global, regional, and national trends in haemoglobin concentration and prevalence of total and severe anaemia in children and pregnant and non-pregnant women for 1995-2011: a systematic analysis of population-representative data. Lancet Glob. Health 1, e16–e25 (2013).
Pasricha, S. R., Armitage, A. E., Prentice, A. M. & Drakesmith, H. Reducing anaemia in low income countries: control of infection is essential. BMJ 362, k3165 (2018).
Goheen, M. M. et al. Anemia offers stronger protection than Sickle Cell Trait against the erythrocytic stage of falciparum malaria and this protection Is reversed by iron supplementation. EBioMedicine 14, 123–130 (2016).
Spyrou, M. A., Bos, K. I., Herbig, A. & Krause, J. Ancient pathogen genomics as an emerging tool for infectious disease research. Nat. Rev. Genet 20, 323–340 (2019).
Duchene, S., Ho, S. Y. W., Carmichael, A. G., Holmes, E. C. & Poinar, H. The recovery, interpretation and use of ancient pathogen genomes. Curr. Biol. 30, R1215–R1231 (2020).
Martin, M. D. et al. Reconstructing genome evolution in historic samples of the Irish potato famine pathogen. Nat. Commun. 4, 2172 (2013).
Campos, P. E. et al. Herbarium specimen sequencing allows precise dating of Xanthomonas citri pv. citri diversification history. Nat. Commun. 14, 4306 (2023).
Woods, W. A., Watson, M., Ranaweera, S., Tajuria, G. & Sumathipala, A. Under-representation of low and middle income countries (LMIC) in the research literature: Ethical issues arising from a survey of five leading medical journals: have the trends changed?. Glob. Public Health 18, 2229890 (2023).
Oyola, S. O. et al. Whole genome sequencing of Plasmodium falciparum from dried blood spots using selective whole genome amplification. Malar. J. 15, 597 (2016).
Lee, S. & Bahlo, M. moimix: an R package for assessing clonality in high-througput sequencing data (v0.0.1.9001). Zenodo (2016). https://doi.org/10.5281/zenodo.58257.
Chang, H. H. et al. THE REAL McCOIL: A method for the concurrent estimation of the complexity of infection and SNP allele frequency for malaria parasites. PLoS Comput Biol. 13, e1005348 (2017).
Siewert, K. M. & Voight, B. F. Detecting Long-Term Balancing Selection Using Allele Frequency Correlation. Mol. Biol. Evol. 34, 2996–3005 (2017).
Gautier, M., Klassmann, A. & Vitalis, R. REHH 2.0: a reimplementation of the R package REHH to detect positive selection from haplotype structure. Mol. Ecol. Resour. 17, 78–90 (2017).
Aurrecoechea, C. et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res 37, D539–D543 (2009).
Kadekoppala, M., Ogun, S. A., Howell, S., Gunaratne, R. S. & Holder, A. A. Systematic genetic analysis of the Plasmodium falciparum MSP7-like family reveals differences in protein expression, location, and importance in asexual growth of the blood-stage parasite. Eukaryot. Cell 9, 1064–1074 (2010).
Acknowledgements
We are grateful to current and previous members of staff at the MRC Unit in The Gambia and the London School of Hygiene and Tropical Medicine who, at different times enabled the maintenance of research sample archives. We thank Lindsay Stewart for support with DNA extraction processing, as well as Eleanor Drury, Victoria Simpson and Sonia Goncalves at the Wellcome Sanger Institute for support with processing of samples for sequencing. This research was supported by a European and Developing Countries Clinical Trials Partnership (EDCTP) Senior Fellowship Plus award (TMA2019SFP-2843-EGSAT) and a Wellcome Sanger Institute Senior International Fellowship award (S4739-IF-A.A.-N.) to A.A.-N., and an MRC Project Grant (MR/S009760/1) to D.J.C.
Author information
Authors and Affiliations
Contributions
A.A.-N.: Conceptualization, Resources, Methodology, Data curation, Investigation, Formal analysis, Validation, Funding acquisition, Writing – original draft, Writing – review and editing. M.F.D.: Investigation. C.J.D.: Resources, Writing – review and editing. U.d’A.: Resources, Writing – review and editing. D.P.K.: Methodology, Resources. D.J.C.: Conceptualization, Resources, Methodology, Data curation, Investigation, Formal analysis, Validation, Funding acquisition, Writing – original draft, Writing – review and editing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Nishith Gupta and Johannes Stortz. [A peer review file is available].
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Amambua-Ngwa, A., Diop, M.F., Drakeley, C.J. et al. Major features of parasite adaptation revealed by genomes of Plasmodium falciparum population samples archived for over 50 years. Commun Biol (2026). https://doi.org/10.1038/s42003-025-09460-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-025-09460-3