Abstract
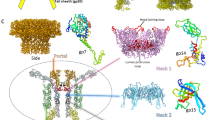
The virion of Staphylococcus phage 812 is formed by a capsid and a contractile tail joined together by neck proteins. The neck proteins are crucial for virion assembly, DNA packaging, and the regulation of genome release, but their functions are not completely understood. Here, we show that the neck of phage 812 consists of portal, adaptor, stopper, tail terminator, and two types of decoration proteins. A dodecameric DNA-binding site at the surface of the portal complex anchors the phage genome inside the capsid. The adaptor complex induces a local B-to-A form transition of the DNA in the neck channel that could slow or pause genome translocation during ejection. The central channel of a stopper complex that is not attached to the tail terminator complex is closed by gating loops. In contrast, in the phage 812 virion, the gating loops are in an open conformation, and the DNA extends into the tail. The structure of neck proteins is not affected by tail sheath contraction. Therefore, the expulsion of tail tape measure proteins triggers the genome release.

Similar content being viewed by others
Data availability
The GenBank/ENA/DDBJ accession number of the phage 812 strain K1/420 genome is KJ206563.2. The whole-genome sequencing reads (NCBI BioSample ID SAMN53543416) and residual phage DNA sequencing reads (NCBI BioSample ID SAMN48784668) were deposited in the NCBI under the BioProject ID PRJNA1269267. Cryo-EM density maps are available in the Electron Microscopy Data Bank under accession IDs EMD-18048, EMD-18065, EMD-18213, EMD-18369, EMD-18372, EMD-18385, EMD-18395, EMD-18445, EMD-18462, EMD-18489, EMD-18516, EMD-18912, and EMD-18919. Atomic coordinates and X-ray structure factors are available in the Protein Data Bank under accession IDs 8Q01, 8Q1I, 8Q7D, 8QEM, 8QEK, 8QGR, 8QJE, 8QKH, 8R5G, and 8R69. Mass spectrometry data were deposited to the ProteomeXchange Consortium via PRIDE97 partner repository under dataset identifier PXD071586. Unedited image of SDS-PAGE of phage 812 is included in Supplementary Information (Fig. S27). Numeric source data for plots are supplied as Supplementary Data 1, and processed mass spectrometry data of phage 812 as Supplementary Data 2. All other data are available from the corresponding author on reasonable request.
References
McCallin, S., Sarker, S. A., Sultana, S., Oechslin, F. & Brüssow, H. Metagenome analysis of Russian and Georgian Pyophage cocktails and a placebo-controlled safety trial of single phage versus phage cocktail in healthy Staphylococcus aureus carriers. Environ. Microbiol. 20, 3278–3293 (2018).
Botka, T. et al. Lytic and genomic properties of spontaneous host-range Kayvirus mutants prove their suitability for upgrading phage therapeutics against staphylococci. Sci. Rep. 9, 5475 (2019).
Arroyo-Moreno, S. et al. Insights into gene transcriptional regulation of kayvirus bacteriophages obtained from therapeutic mixtures. Viruses 14, 626 (2022).
Nováček, J. et al. Structure and genome release of Twort-like Myoviridae phage with a double-layered baseplate. Proc. Natl. Acad. Sci. USA 113, 9351–9356 (2016).
Tavares, P., Zinn-Justin, S. & Orlova, E. V. Genome gating in tailed bacteriophage capsids. In Viral Molecular Machines (eds Rossmann, M. G. & Rao, V. B.) 585–600 (Springer US, 2012).
Chaban, Y. et al. Structural rearrangements in the phage head-to-tail interface during assembly and infection. Proc. Natl. Acad. Sci. USA 112, 7009–7014 (2015).
Bárdy, P. et al. Structure and mechanism of DNA delivery of a gene transfer agent. Nat. Commun. 11, 3034 (2020).
Orlov, I. et al. CryoEM structure and assembly mechanism of a bacterial virus genome gatekeeper. Nat. Commun. 13, 7283 (2022).
Yang, F. et al. Fine structure and assembly pattern of a minimal myophage Pam3. Proc. Natl. Acad. Sci. USA 120, e2213727120 (2023).
Li, F. et al. High-resolution cryo-EM structure of the Pseudomonas bacteriophage E217. Nat. Commun. 14, 4052 (2023).
Hsiao, C. L. & Black, L. W. Head morphogenesis of bacteriophage T4 II. The role of gene 40 in initiating prehead assembly. Virology 91, 15–25 (1978).
Prevelige, P. E. Jr, Thomas, D. & King, J. Nucleation and growth phases in the polymerization of coat and scaffolding subunits into icosahedral procapsid shells. Biophys. J. 64, 824–835 (1993).
Sun, S. et al. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell 135, 1251–1262 (2008).
Daudén, M. I. et al. Large terminase conformational change induced by connector binding in bacteriophage T7. J. Biol. Chem. 288, 16998–17007 (2013).
Woodson, M. et al. A viral genome packaging motor transitions between cyclic and helical symmetry to translocate dsDNA. Sci. Adv. 7, eabc1955 (2021).
Ray, K., Ma, J., Oram, M., Lakowicz, J. R. & Black, L. W. Single-molecule and FRET fluorescence correlation spectroscopy analyses of phage DNA packaging: colocalization of packaged phage T4 DNA ends within the capsid. J. Mol. Biol. 395, 1102–1113 (2010).
Orlova, E. V. et al. Structure of the 13-fold symmetric portal protein of bacteriophage SPP1. Nat. Struct. Biol. 6, 842–846 (1999).
Lokareddy, R. K. et al. Portal protein functions akin to a DNA-sensor that couples genome-packaging to icosahedral capsid maturation. Nat. Commun. 8, 14310 (2017).
Casjens, S. R. Bacteriophage lambda FII gene protein: Role in head assembly. J. Mol. Biol. 90, 1–20 (1974).
Lhuillier, S. et al. Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating. Proc. Natl. Acad. Sci. UAE 106, 8507–8512 (2009).
Kizziah, J. L., Mukherjee, A., Parker, L. K. & Dokland, T. Structure of the Staphylococcus aureus bacteriophage 80α neck shows details of the DNA, tail completion protein, and tape measure protein. Structure 33, 1063–1073.e2 (2025).
Gu, Z., Wu, K. & Wang, J. Structural morphing in the viral portal vertex of bacteriophage lambda. J. Virol. 98, e00068–24 (2024).
Liu, Y.-T., Jih, J., Dai, X., Bi, G.-Q. & Zhou, Z. H. Cryo-EM structures of herpes simplex virus type 1 portal vertex and packaged genome. Nature 570, 257–261 (2019).
Ayala, R. et al. Nearly complete structure of bacteriophage DT57C reveals architecture of head-to-tail interface and lateral tail fibers. Nat. Commun. 14, 8205 (2023).
Iglesias, S. M. et al. Cryo-EM analysis of Pseudomonas phage Pa193 structural components. Commun. Biol. 7, 1275 (2024).
Li, X. et al. Cryo-EM Reveals structural diversity in prolate-headed mycobacteriophage Mycofy1. J. Mol. Biol. 437, 169126 (2025).
Leiman, P. G. & Shneider, M. M. Contractile tail machines of bacteriophages. In Viral Molecular Machines (eds Rossmann, M. G. & Rao, V. B.) 93–114 (Springer US, Boston, MA, 2012).
Sinden, R. R., Pearson, C. E., Potaman, V. N. & Ussery, D. W. DNA: structure and function. In Advances in Genome Biology (eds. Verma, R. S.) 1–141 (Elsevier, 1998).
Łobocka, M. et al. Chapter 5 - Genomics of staphylococcal twort-like phages - potential therapeutics of the post-antibiotic era. In Advances in Virus Research (eds Łobocka, M. & Szybalski, W.) 143–216 (Academic Press, 2012).
Arsuaga, J., Vázquez, M., Trigueros, S., Sumners, D. W. & Roca, J. Knotting probability of DNA molecules confined in restricted volumes: DNA knotting in phage capsids. Proc. Natl. Acad. Sci. USA 99, 5373–5377 (2002).
Guasch, A. et al. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage φ29 connector particle. J. Mol. Biol. 315, 663–676 (2002).
Sun, L. et al. Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution. Nat. Commun. 6, 7548 (2015).
Kulkarni, M. & Mukherjee, A. Sequence dependent free energy profiles of localized B-to A-form transition of DNA in water. J. Chem. Phys. 139, 10B617_1 (2013).
Tolstorukov, M. Y., Jernigan, R. L. & Zhurkin, V. B. Protein–DNA Hydrophobic Recognition in the Minor Groove is Facilitated by Sugar Switching. J. Mol. Biol. 337, 65–76 (2004).
Mirzabekov, A. D. & Rich, A. Asymmetric lateral distribution of unshielded phosphate groups in nucleosomal DNA and its role in DNA bending. Proc. Natl. Acad. Sci. USA 76, 1118–1121 (1979).
Shui, X., McFail-Isom, L., Hu, G. G. & Williams, L. D. The B-DNA Dodecamer at high resolution reveals a spine of water on sodium. Biochemistry 37, 8341–8355 (1998).
Hancock, S. P., Hiller, D. A., Perona, J. J. & Jen-Jacobson, L. The energetic contribution of induced electrostatic asymmetry to dna bending by a site-specific protein. J. Mol. Biol. 406, 285–312 (2011).
Minchenkova, L. E., Schyolkina, A. K., Chernov, B. K. & Ivanov, V. I. CC/GG contacts facilitate the B to A transition of DMA in solution. J. Biomol. Struct. Dyn. 4, 463–476 (1986).
Tang, J. et al. DNA poised for release in bacteriophage ø29. Structure 16, 935–943 (2008).
Xu, J., Wang, D., Gui, M. & Xiang, Y. Structural assembly of the tailed bacteriophage ϕ29. Nat. Commun. 10, 2366 (2019).
Lanni, Y. T. First-step-transfer deoxyribonucleic acid of bacteriophage T5. Bacteriol. Rev. 32, 227–242 (1968).
de Frutos, M., Letellier, L. & Raspaud, E. DNA Ejection from bacteriophage T5: analysis of the kinetics and energetics. Biophys. J. 88, 1364–1370 (2005).
Molineux, I. J. & Panja, D. Popping the cork: mechanisms of phage genome ejection. Nat. Rev. Microbiol. 11, 194–204 (2013).
Chen, Y.-J. et al. Two-stage dynamics of in vivo bacteriophage genome ejection. Phys. Rev. X 8, 021029 (2018).
Katsura, I. & Hendrix, R. W. Length determination in bacteriophage lambda tails. Cell 39, 691–698 (1984).
Hendrix, R. W. & Casjens, S. R. Protein cleavage in bacteriophage λ tail assembly. Virology 61, 156–159 (1974).
Xu, J., Hendrix, R. W. & Duda, R. L. Chaperone–protein interactions that mediate assembly of the bacteriophage lambda tail to the correct length. J. Mol. Biol. 426, 1004–1018 (2014).
Seul, A. et al. Biogenesis of a bacteriophage long non-contractile tail. J. Mol. Biol. 433, 167112 (2021).
Kizziah, J. L., Manning, K. A., Dearborn, A. D. & Dokland, T. Structure of the host cell recognition and penetration machinery of a Staphylococcus aureus bacteriophage. PLoS Pathog. 16, e1008314 (2020).
Wingett, S. W. & Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 7, 1338 (2018).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595 (2017).
Brettin, T. et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5, 8365 (2015).
NCBI Resource Coordinators Database resources of the national center for biotechnology information. Nucleic Acids Res. 41, D8–D20 (2013).
Mitchell, A. L. et al. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47, D351–D360 (2019).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Tang, G. et al. EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Zhang, K. Gautomatch. MRC Laboratory of Molecular Biology. https://www2.mrc-lmb.cam.ac.uk/ (2020).
Scheres, S. H. W. A bayesian view on cryo-EM structure determination. J. Mol. Biol. 415, 406–418 (2012).
de la Rosa-Trevín, J. M. et al. Xmipp 3.0: An improved software suite for image processing in electron microscopy. J. Struct. Biol. 184, 321–328 (2013).
R Core Team. R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, Vienna, Austria, 2022).
Ilca, S. L. et al. Localized reconstruction of subunits from electron cryomicroscopy images of macromolecular complexes. Nat. Commun. 6, 8843 (2015).
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. D. Features and development of Coot. Acta Crystallogr. Sect. D 66, 486–501 (2010).
Wang, S., Sun, S., Li, Z., Zhang, R. & Xu, J. Accurate de novo prediction of protein contact map by ultra-deep learning model. PLoS Comput. Biol. 13, e1005324 (2017).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Croll, T. I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. Sect. D 74, 519–530 (2018).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. Sect. D 75, 861–877 (2019).
Williams, C. J. et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Černý, J. et al. Structural alphabets for conformational analysis of nucleic acids available at dnatco.datmos.org. Acta Crystallogr. Sect. D 76, 805–813 (2020).
Li, S., Olson, W. K. & Lu, X.-J. Web 3DNA 2.0 for the analysis, visualization, and modeling of 3D nucleic acid structures. Nucleic Acids Res. 47, W26–W34 (2019).
Sambrook, J. E., Fritsch, E. F. & Maniatis, T. Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1989).
Studier, F. W. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. Sect. D 69, 1204–1214 (2013).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D 67, 235–242 (2011).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D 67, 355–367 (2011).
Zwart, P., Grosse-Kunstleve, R. W. & Adams, P. Xtriage and Fest: Automatic Assessment Of X-ray Data And Substructure Structure Factor Estimation. www:https://cci.lbl.gov (2005).
Krissinel, E. & Henrick, K. Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol. 372, 774–797 (2007).
Zheng, H. et al. CheckMyMetal: a macromolecular metal-binding validation tool. Acta Crystallogr. Sect. D 73, 223–233 (2017).
Holm, L. Dali server: structural unification of protein families. Nucleic Acids Res. 50, W210–W215 (2022).
Hildebrand, A., Remmert, M., Biegert, A. & Söding, J. Fast and accurate automatic structure prediction with HHpred. Proteins Struct. Funct. Bioinforma. 77, 128–132 (2009).
Papadopoulos, J. S. & Agarwala, R. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–1079 (2007).
Crooks, G., Hon, G., Chandonia, J.-M. & Brenner, S. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Katoh, K., Rozewicki, J. & Yamada, K. D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166 (2019).
Robinson, O., Dylus, D. & Dessimoz, C. Phylo.io: Interactive viewing and comparison of large phylogenetic trees on the web. Mol. Biol. Evol. 33, 2163–2166 (2016).
Wang, S., Li, W., Liu, S. & Xu, J. RaptorX-Property: a web server for protein structure property prediction. Nucleic Acids Res. 44, W430–W435 (2016).
Pettersen, E. F. et al. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Barlow, D. J. & Thornton, J. M. Ion-pairs in proteins. J. Mol. Biol. 168, 867–885 (1983).
Laughton, C. & Luisi, B. The mechanics of minor groove width variation in DNA, and its implications for the accommodation of ligands. J. Mol. Biol. 288, 953–963 (1999).
Blanchet, C., Pasi, M., Zakrzewska, K. & Lavery, R. CURVES+ web server for analyzing and visualizing the helical, backbone and groove parameters of nucleic acid structures. Nucleic Acids Res. 39, W68–W73 (2011).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
ACD/ChemSketch. ACD/Labs. https://www.acdlabs.com/resource/acd-labs-releases-v2022-software-update/ (2022).
Perez-Riverol, Y. et al. The PRIDE database at 20 years: 2025 update. Nucleic Acids Res. 53, D543–D553 (2025).
Acknowledgements
We gratefully acknowledge the Cryo-electron Microscopy and Tomography Core Facility and Proteomics Core Facility of the CEITEC MU of CIISB, Instruct-CZ Centre, supported by the Ministry of Education, Youth and Sports of the Czech Republic (MEYS CR) infrastructure project LM2023042, and the European Regional Development Fund - Projects “UP CIISB” (No. CZ.02.1.01/0.0/0.0/18_046/0015974) and “Innovation of Czech Infrastructure for Integrative Structural Biology” (No. CZ.02.01.01/00/23_015/0008175). Computational resources were provided by the e-INFRA CZ project (ID: 90254), supported by MEYS CR. This work was supported by the project National Institute of Virology and Bacteriology (Programme EXCELES, ID Project No. LX22NPO5103) - Funded by the European Union - Next Generation EU, and the project New Technologies for Translational Research in Pharmaceutical Sciences / NETPHARM, project ID OP JAC CZ.02.01.01/00/22_008/0004607, which is co-funded by the European Union. This work also received funding from ERC Consolidator Grant No. 101043452 to P.P., and from the Ministry of Health of the Czech Republic in cooperation with the Czech Health Research Council under project No. NU21J-05–00035 to T.B.
Author information
Authors and Affiliations
Contributions
M.B., M.S., Z.C. and P.B. purified the phage samples. Z.C., J.N. and M.S. performed cryo-EM data collection and processing, and Z.C. and T.F. performed cryo-EM data analysis. B.P. cloned, purified, and crystallized the stopper protein. B.P. and P.P. collected and processed crystallographic data. Z.C. carried out X-ray data analysis, X-ray and cryo-EM structural determination, deposition of atomic coordinates and maps, bioinformatic analysis and figure preparation. T.B. performed the sequencing and assembly of the phage 812 genome, and sequencing and analysis of residual capsid DNA after genome ejection. T.B. and R.P. performed the genome annotation. P.P. and R.P. designed and supervised the research. Z.C. and P.P. wrote the manuscript. All authors participated in critical reading of the manuscript and approved the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Robert Kingsley and Mengtan Xing. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cieniková, Z., Nováček, J., Šiborová, M. et al. Genome anchoring, retention, and release by neck proteins of Staphylococcus phage 812. Commun Biol (2026). https://doi.org/10.1038/s42003-025-09477-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-025-09477-8