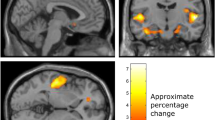
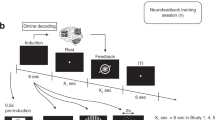
Abstract
The development of algorithms to accurately decode neural information has long been a research focus in the field of neuroscience. Brain decoding typically involves training machine learning models to map neural data onto a preestablished vector representation of stimulus features. These vectors are usually derived from image- and/or text-based feature spaces. Nonetheless, the intrinsic characteristics of these vectors might fundamentally differ from those that are encoded by the brain, limiting the ability of decoders to accurately learn this mapping. To address this issue, we propose a framework, called brain-aligning of semantic vectors, that fine-tunes pretrained feature vectors to better align with the structure of neural representations of visual stimuli in the brain. We trained this model with functional magnetic resonance imaging (fMRI) and then performed zero-shot brain decoding on fMRI, magnetoencephalography (MEG), and electrocorticography (ECoG) data. fMRI-based brain-aligned vectors improved decoding performance across all three neuroimaging datasets when accuracy was determined by calculating the correlation coefficients between true and predicted vectors. Additionally, when decoding accuracy was determined via stimulus identification, this accuracy increased in specific category types; improvements varied depending on the original vector space that was used for brain-alignment, and consistent improvements were observed across all neuroimaging modalities.
Similar content being viewed by others
Data availability
The datasets supporting the findings of this study include fMRI, MEG, and ECoG data. The fMRI dataset used in this study is available at15. Source data underlying the figures are available in Figshare with the identifier https://doi.org/10.6084/m9.figshare.30845336.
Code availability
The code used for data analysis in this study is available on our repository (https://github.com/yanagisawa-lab). For any inquiries, please contact the corresponding author.
References
Stavisky, S. D. & Wairagkar, M. Listening in to perceived speech with contrastive learning. Nat. Mach. Intell. https://doi.org/10.1038/s42256-023-00742-1 (2023).
Lebedev, M. A. & Nicolelis, M. A. L. Brain–machine interfaces: past, present and future. Trends Neurosci. 29, 536–546 (2006).
Willett, F. R. et al. A high-performance speech neuroprosthesis. Nature 620, 1031–1036 (2023).
Willsey, M. S. et al. Real-time brain-machine interface in non-human primates achieves high-velocity prosthetic finger movements using a shallow feedforward neural network decoder. Nat. Commun. 13, 6899 (2022).
Haynes, J.-D. & Rees, G. Decoding mental states from brain activity in humans. Nat. Rev. Neurosci. 7, 523–534 (2006).
Naselaris, T., Kay, K. N., Nishimoto, S. & Gallant, J. L. Encoding and decoding in fMRI. Neuroimage 56, 400–410 (2011).
Haxby, J. V. et al. Distributed and overlapping representations of faces and objects in ventral temporal cortex. Science 293, 2425–2430 (2001).
Yamins, D. L. K. et al. Performance-optimized hierarchical models predict neural responses in higher visual cortex. Proc. Natl. Acad. Sci. USA 111, 8619–8624 (2014).
Kellis, S. et al. Decoding spoken words using local field potentials recorded from the cortical surface. J. Neural Eng. 7, 056007 (2010).
Brouwer, G. J. & Heeger, D. J. Decoding and reconstructing color from responses in human visual cortex. J. Neurosci. 29, 13992–14003 (2009).
Sitaram, R. et al. Closed-loop brain training: the science of neurofeedback. Nat. Rev. Neurosci. 18, 86–100 (2017).
Fukuma, R. et al. Voluntary control of semantic neural representations by imagery with conflicting visual stimulation. Commun. Biol. 5, 214 (2022).
Chaudhary, U. et al. Spelling interface using intracortical signals in a completely locked-in patient enabled via auditory neurofeedback training. Nat. Commun. 13, 1236 (2022).
Cortese, A., Amano, K., Koizumi, A., Kawato, M. & Lau, H. Multivoxel neurofeedback selectively modulates confidence without changing perceptual performance. Nat. Commun. 7, 13669 (2016).
Horikawa, T. & Kamitani, Y. Generic decoding of seen and imagined objects using hierarchical visual features. Nat. Commun. 8, 15037 (2017).
Haynes, J.-D. & Rees, G. Predicting the orientation of invisible stimuli from activity in human primary visual cortex. Nat. Neurosci. 8, 686–691 (2005).
Kamitani, Y. & Tong, F. Decoding the visual and subjective contents of the human brain. Nat. Neurosci. 8, 679–685 (2005).
Thirion, B. et al. Inverse retinotopy: inferring the visual content of images from brain activation patterns. Neuroimage 33, 1104–1116 (2006).
Cox, D. D. & Savoy, R. L. Functional magnetic resonance imaging (fMRI) “brain reading”: detecting and classifying distributed patterns of fMRI activity in human visual cortex. NeuroImage 19, 261–270 (2003).
Nakai, T., Koide-Majima, N. & Nishimoto, S. Correspondence of categorical and feature-based representations of music in the human brain. Brain Behav. 11, e01936 (2021).
Koide-Majima, N., Nishimoto, S. & Majima, K. Mental image reconstruction from human brain activity: Neural decoding of mental imagery via deep neural network-based Bayesian estimation. Neural Networks 170, 349–363 (2024).
Miyawaki, Y. et al. Visual image reconstruction from human brain activity using a combination of multiscale local image decoders. Neuron 60, 915–929 (2008).
Shen, G., Dwivedi, K., Majima, K., Horikawa, T. & Kamitani, Y. End-to-end deep image reconstruction from human brain activity. Front. Comput. Neurosci. 13, 21 (2019).
Shen, G., Horikawa, T., Majima, K. & Kamitani, Y. Deep image reconstruction from human brain activity. PLoS Comput. Biol. 15, e1006633 (2019).
Liu, Y., Ma, Y., Zhou, W., Zhu, G. & Zheng, N. BrainCLIP: bridging brain and visual-linguistic representation Via CLIP for generic natural visual stimulus decoding. Preprint at https://doi.org/10.48550/arXiv.2302.1297 (2023).
Radford, A. et al. Learning transferable visual models from natural language supervision. in 8748–8763 (PMLR, 2021).
Pereira, F. et al. Toward a universal decoder of linguistic meaning from brain activation. Nat. Commun. 9, 963 (2018).
Mikolov, T., Chen, K., Corrado, G. & Dean, J. Efficient estimation of word representations in vector space. arXiv preprint arXiv:1301.3781 (2013).
Pennington, J., Socher, R. & Manning, C. GloVe: Global Vectors for Word Representation. in Proc. 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP) 1532–1543 (Association for Computational Linguistics, 2014).
Shirakawa, K. et al. Spurious reconstruction from brain activity. Neural Netw. 190, 107515 (2025).
Federer, C., Xu, H., Fyshe, A. & Zylberberg, J. Improved object recognition using neural networks trained to mimic the brain’s statistical properties. Neural Netw. 131, 103–114 (2020).
Muttenthaler, L. et al. Improving neural network representations using human similarity judgments. Advances in neural information processing systems 36, 50978–51007 (2023).
Schneider, S., Lee, J. H. & Mathis, M. W. Learnable latent embeddings for joint behavioural and neural analysis. Nature 617, 360–368 (2023).
Kay, K. N., Naselaris, T., Prenger, R. J. & Gallant, J. L. Identifying natural images from human brain activity. Nature 452, 352–355 (2008).
Ogawa, S., Lee, T.-M., Kay, A. R. & Tank, D. W. Brain magnetic resonance imaging with contrast dependent on blood oxygenation. Proc. Natl. Acad. Sci. USA 87, 9868–9872 (1990).
Penfield, W. & Jasper, H. Epilepsy and the functional anatomy of the human brain. (Little, Brown & Co., Boston, 1954).
Cohen, D. Magnetoencephalography: evidence of magnetic fields produced by alpha-rhythm currents. Science 161, 784–786 (1968).
Deng, J. et al. ImageNet: a large-scale hierarchical image database. in 2009 IEEE Conference on Computer Vision and Pattern Recognition 248–255 https://doi.org/10.1109/CVPR.2009.5206848 (2009).
Kriegeskorte, N., Mur, M. & Bandettini, P. Representational similarity analysis—connecting the branches of systems neuroscience. Front. Syst. Neurosci. 2, 4 (2008).
Kourtzi, Z. & Kanwisher, N. Cortical regions involved in perceiving object shape. J. Neurosci. 20, 3310–3318 (2000).
Kanwisher, N., McDermott, J. & Chun, M. M. The fusiform face area: a module in human extrastriate cortex specialized for face perception. J. Neurosci. 17, 4302–4311 (1997).
Epstein, R. & Kanwisher, N. A cortical representation of the local visual environment. Nature 392, 598–601 (1998).
Gifford, A. T., Jastrzębowska, M. A., Singer, J. J. D. & Cichy, R. M. In silico discovery of representational relationships across visual cortex. Nat. Hum. Behav. https://doi.org/10.1038/s41562-025-02252-z (2025).
Kobatake, E. & Tanaka, K. Neuronal selectivities to complex object features in the ventral visual pathway of the macaque cerebral cortex. J. Neurophysiol. 71, 856–867 (1994).
Binder, J. R. et al. Toward a brain-based componential semantic representation. Cogn. Neuropsychol. 33, 130–174 (2016).
Chersoni, E., Santus, E., Huang, C.-R. & Lenci, A. Decoding word embeddings with brain-based semantic features. Comput. Linguist. 47, 663–698 (2021).
Li, Y., Yang, H. & Gu, S. Enhancing neural encoding models for naturalistic perception with a multi-level integration of deep neural networks and cortical networks. Sci. Bull. https://doi.org/10.1016/j.scib.2024.02.035 (2024).
Haxby, J. V. et al. A common, high-dimensional model of the representational space in human ventral temporal cortex. Neuron 72, 404–416 (2011).
Guntupalli, J. S. et al. A model of representational spaces in human cortex. Cereb. cortex 26, 2919–2934 (2016).
Cichy, R. M. & Pantazis, D. Multivariate pattern analysis of MEG and EEG: A comparison of representational structure in time and space. NeuroImage 158, 441–454 (2017).
Salmela, V., Salo, E., Salmi, J. & Alho, K. Spatiotemporal dynamics of attention networks revealed by representational similarity analysis of EEG and fMRI. Cereb. Cortex 28, 549–560 (2018).
Sereno, M. I. et al. Borders of multiple visual areas in humans revealed by functional magnetic resonance imaging. Science 268, 889–893 (1995).
Tadel, F., Baillet, S., Mosher, J. C., Pantazis, D. & Leahy, R. M. Brainstorm: a user-friendly application for MEG/EEG analysis. Comput. Intell. Neurosci. 2011, 879716 (2011).
Yoshioka, T. et al. Evaluation of hierarchical Bayesian method through retinotopic brain activities reconstruction from fMRI and MEG signals. NeuroImage 42, 1397–1413 (2008).
Glasser, M. F. et al. A multi-modal parcellation of human cerebral cortex. Nature 536, 171–178 (2016).
Dale, A. M., Fischl, B. & Sereno, M. I. Cortical surface-based analysis: I. segmentation and surface reconstruction. NeuroImage 9, 179–194 (1999).
Papademetris, X. et al. BioImage suite: an integrated medical image analysis suite: an update. Insight J. 2006, 209 (2006).
Groppe, D. M. et al. iELVis: an open source MATLAB toolbox for localizing and visualizing human intracranial electrode data. J. Neurosci. Methods 281, 40–48 (2017).
Fukuma, R. et al. Image retrieval based on closed-loop visual–semantic neural decoding. Preprint at https://doi.org/10.1101/2024.08.05.606113 (2024).
Acknowledgements
We acknowledge the use of open-source code from the Kamitani Lab. Specifically, we used the Brain Decoding Toolbox (BDPy; https://github.com/KamitaniLab/bdpy) for neuroimaging data processing and analysis, and adapted decoding algorithms from the Generic Object Decoding repository (https://github.com/KamitaniLab/GenericObjectDecoding; Horikawa & Kamitani, 2017). We thank the Kamitani Lab for making these resources publicly available. We also thank all the subjects for their participation. This research was supported by the Japan Science and Technology Agency (JST) Moonshot R&D (JPMJMS2012), the JST Core Research for Evolutional Science and Technology (CREST) (JPMJCR18A5), the JST AIP Acceleration Research (JPMJCR24U2), K Program (JPMJKP25Y7), and the Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research (KAKENHI) (JP26560467 and JP20H05705).
Author information
Authors and Affiliations
Contributions
S.V. and T.Y. conceptualized the project. S.V. was responsible for the theory. S.V., R.F., and T.Y. were responsible for the methodology. S.V. undertook the analysis and investigation. R.F. and H.Y. were responsible for the MEG and ECoG experiments. S.V. was responsible for data preprocessing and curation. S.V. wrote the original draft and created the figures. S.V. and T.Y. edited the final version of the article. S.O., N.T., H.M.K., H.S., Y.I., H.S., M.N., H.K., and K.T. performed the neurosurgery of ECoG experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Peer review
Peer review information
Communications Biology thanks Marijn van Vliet and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Shenbing Kuang and Jasmine Pan. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Vafaei, S., Fukuma, R., Yanagisawa, T. et al. Brain-aligning of semantic vectors improves neural decoding of visual stimuli. Commun Biol (2026). https://doi.org/10.1038/s42003-025-09482-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-025-09482-x