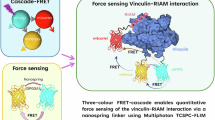
Abstract
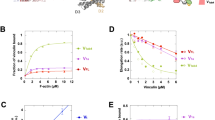
Focal adhesions (FAs) are mechanosensitive structures that mediate force transmission between cells and the extracellular matrix. While Traction Force Microscopy (TFM) quantifies cellular tractions exerted on deformable substrates, Förster Resonance Energy Transfer (FRET)-based tension probes, such as vinculin tension sensors, measure molecular-scale forces within FA proteins. Despite their potential synergy, these methods have rarely been combined to explore the interplay between molecular tension and cellular tractions. Here, we introduce a framework integrating TFM and FRET-based vinculin tension sensors to investigate FA mechanics across scales. At cell level, tractions and vinculin tension increased with substrate stiffness. At FA level, vinculin tension correlated solely with vinculin density, while tractions scaled with FA area, orientation, total vinculin content and vinculin density. Direct comparison of tractions to vinculin tension revealed a complex, heterogenous relationship between these forces, possibly linked to diverse cell and FA maturation states. Sub-FA analysis revealed conserved spatial patterns, with both tension and traction increasing towards the cell periphery. This multiscale approach provides an integrated workflow for studying focal adhesion forces, helping to bridge the gap between vinculin tension and cellular tractions.
Similar content being viewed by others
Data availability
Source Data files including the data points for the generation of figures and graphs, and the corresponding statistical analysis, are accessible at Zenodo (https://doi.org/10.5281/zenodo.14692589)76. Raw microscopy files are available from the corresponding authors upon request.
Code availability
The TFM code to analyze 2D traction microscopy data, as well as the custom-written FRET code to calculate FRET efficiencies from FLIM-Phasor data, and the custom-written TFM-FRET code for the simultaneous analysis of molecular tension and cell traction is open source and can be accessed through Zenodo (https://doi.org/10.5281/zenodo.14692589)76.
References
Uhler, C. & Shivashankar, G. V. Nuclear mechanopathology and cancer diagnosis. Trends Cancer 4, 320–331 (2018).
Maurer, M. & Lammerding, J. The driving force: nuclear mechanotransduction in cellular function, fate, and disease. Annu. Rev. Biomed. Eng. 21, 443–468 (2019).
Dwivedi, A., Kiely, P. A. & Hoey, D. A. Mechanically stimulated osteocytes promote the proliferation and migration of breast cancer cells via a potential CXCL1/2 mechanism. Biochem. Biophys. Res. Commun. 534, 14–20 (2021).
Nelson, C. M. Mechanical control of cell differentiation: insights from the early embryo. Annu. Rev. Biomed. Eng. 24, 307–322 (2022).
Nho, R. S., Ballinger, M. N., Rojas, M. M., Ghadiali, S. N. & Horowitz, J. C. Biomechanical force and cellular stiffness in lung fibrosis. Am. J. Pathol. 192, 750–761 (2022).
Case, L. B. et al. Molecular mechanism of vinculin activation and nanoscale spatial organization in focal adhesions. Nat. Cell Biol. 17, 880–892 (2015).
Saraswathibhatla, A., Indana, D. & Chaudhuri, O. Cell–extracellular matrix mechanotransduction in 3D. Nat. Rev. Mol. Cell Biol. 24, 495–516 (2023).
Möhl, C. et al. Becoming stable and strong: the interplay between vinculin exchange dynamics and adhesion strength during adhesion site maturation. Cell Motil. 66, 350–364 (2009).
Tao, A. et al. Identifying constitutive and context-specific molecular-tension-sensitive protein recruitment within focal adhesions. Dev. Cell 58, 522–534.e7 (2023).
Chen, H., Cohen, D. M., Choudhury, D. M., Kioka, N. & Craig, S. W. Spatial distribution and functional significance of activated vinculin in living cells. J. Cell Biol. 169, 459–470 (2005).
Chen, H., Choudhury, D. M. & Craig, S. W. Coincidence of actin filaments and talin is required to activate vinculin*. J. Biol. Chem. 281, 40389–40398 (2006).
Cohen, D. M., Kutscher, B., Chen, H., Murphy, D. B. & Craig, S. W. A conformational switch in vinculin drives formation and dynamics of a talin-vinculin complex at focal adhesions*. J. Biol. Chem. 281, 16006–16015 (2006).
Dumbauld, D. W. et al. How vinculin regulates force transmission. Proc. Natl. Acad. Sci. 110, 9788–9793 (2013).
Humphries, J. D. et al. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J. Cell Biol. 179, 1043–1057 (2007).
Dumbauld, D. W., Michael, K. E., Hanks, S. K. & García, A. J. Focal adhesion kinase-dependent regulation of adhesive forces involves vinculin recruitment to focal adhesions. Biol. Cell 102, 203–213 (2010).
Pasapera, A. M., Schneider, I. C., Rericha, E., Schlaepfer, D. D. & Waterman, C. M. Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J. Cell Biol. 188, 877–890 (2010).
Thievessen, I. et al. Vinculin–actin interaction couples actin retrograde flow to focal adhesions, but is dispensable for focal adhesion growth. J. Cell Biol. 202, 163–177 (2013).
Mierke, C. T. The role of vinculin in the regulation of the mechanical properties of cells. Cell Biochem. Biophys. 53, 115–126 (2009).
Goldmann, W. H. Role of vinculin in cellular mechanotransduction. Cell Biol. Int. 40, 241–256 (2016).
Butler, J. P., Tolić-Nørrelykke, I. M., Fabry, B. & Fredberg, J. J. Traction fields, moments, and strain energy that cells exert on their surroundings. Am. J. Physiol. Cell Physiol. 282, C595–C605 (2002).
Sabass, B., Gardel, M. L., Waterman, C. M. & Schwarz, U. S. High resolution traction force microscopy based on experimental and computational advances. Biophys. J. 94, 207–220 (2008).
Denisin, A. K., Kim, H., Riedel-Kruse, I. H. & Pruitt, B. L. Field guide to traction force microscopy. Cell Mol. Bioeng. 17, 87–106 (2024).
van Hoorn, H. et al. The nanoscale architecture of force-bearing focal adhesions. Nano Lett. 14, 4257–4262 (2014).
Sarangi, B. R. et al. Coordination between intra- and extracellular forces regulates focal adhesion dynamics. Nano Lett. 17, 399–406 (2017).
Roca-Cusachs, P., Iskratsch, T. & Sheetz, M. P. Finding the weakest link—exploring integrin-mediated mechanical molecular pathways. J. Cell Sci. 125, 3025–3038 (2012).
Grashoff, C. et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 466, 263–266 (2010).
Fischer, L. S., Rangarajan, S., Sadhanasatish, T. & Grashoff, C. Molecular force measurement with tension sensors. Annu. Rev. Biophys. 50, 595–616 (2021).
Chang, C.-W. & Kumar, S. Vinculin tension distributions of individual stress fibers within cell–matrix adhesions. J. Cell Sci. 126, 3021–3030 (2013).
Rothenberg, K. E., Neibart, S. S., LaCroix, A. S. & Hoffman, B. D. Controlling cell geometry affects the spatial distribution of load across vinculin. Cel. Mol. Bioeng. 8, 364–382 (2015).
LaCroix, A. S., Lynch, A. D., Berginski, M. E. & Hoffman, B. D. Tunable molecular tension sensors reveal extension-based control of vinculin loading. eLife 7, e33927 (2018).
Rothenberg, K. E., Scott, D. W., Christoforou, N. & Hoffman, B. D. Vinculin force-sensitive dynamics at focal adhesions enable effective directed cell migration. Biophys. J. 114, 1680–1694 (2018).
Kanoldt, V. et al. Metavinculin modulates force transduction in cell adhesion sites. Nat. Commun. 11, 6403 (2020).
Shoyer, T. C. et al. Coupling during collective cell migration is controlled by a vinculin mechanochemical switch. Proc. Natl. Acad. Sci. USA 120, e2316456120 (2023).
Chirasani, V. R. et al. Molecular basis and cellular functions of vinculin-actin directional catch bonding. Nat. Commun. 14, 8300 (2023).
Stanton, A. E., Tong, X. & Yang, F. Varying solvent type modulates collagen coating and stem cell mechanotransduction on hydrogel substrates. APL Bioeng. 3, 036105 (2019).
Alvarez, L. Application note: SP8 FALCON: a novel concept in fluorescence lifetime imaging enabling video-rate confocal FLIM. Nature https://www.nature.com/articles/d42473-019-00261-x (2019).
Reissaus, C. A. et al. PIE-FLIM measurements of two different fret-based biosensor activities in the same living cells. Biophys. J. 118, 1820–1829 (2020).
Datta, R., Heaster, T. M., Sharick, J. T., Gillette, A. A. & Skala, M. C. Fluorescence lifetime imaging microscopy: fundamentals and advances in instrumentation, analysis, and applications. JBO 25, 071203 (2020).
Torrado, B., Pannunzio, B., Malacrida, L. & Digman, M. A. Fluorescence lifetime imaging microscopy. Nat. Rev. Methods Prim. 4, 1–23 (2024).
Izquierdo-Álvarez, A. et al. Spatiotemporal analyses of cellular tractions describe subcellular effect of substrate stiffness and coating. Ann. Biomed. Eng. 47, 624–637 (2019).
Jorge-Peñas, A. et al. Free form deformation–based image registration improves accuracy of traction force microscopy. PLoS One 10, e0144184 (2015).
Digman, M. A., Caiolfa, V. R., Zamai, M. & Gratton, E. The phasor approach to fluorescence lifetime imaging analysis. Biophys. J. 94, l14–l16 (2008).
Hinde, E., Digman, M. A., Welch, C., Hahn, K. M. & Gratton, E. Biosensor Förster resonance energy transfer detection by the phasor approach to fluorescence lifetime imaging microscopy. Microsc. Res. Tech. 75, 271–281 (2012).
Ranjit, S., Malacrida, L., Jameson, D. M. & Gratton, E. Fit-free analysis of fluorescence lifetime imaging data using the phasor approach. Nat. Protoc. 13, 1979–2004 (2018).
Coucke, Q. et al. Particle-based phasor-FLIM-FRET resolves protein-protein interactions inside single viral particles. Biophys. Rep. 3, 100122 (2023).
Li, D., Liu, X., Dong, F. & Li, W. Advancements in phasor-based FLIM: multi-component analysis and lifetime probes in biological imaging. J. Mater. Chem. B 13, 472–484 (2025).
Berg, S. et al. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232 (2019).
Martin, K. J. et al. Accepting from the best donor; analysis of long-lifetime donor fluorescent protein pairings to optimise dynamic FLIM-based FRET experiments. PLOS ONE 13, e0183585 (2018).
Gates, E. M., LaCroix, A. S., Rothenberg, K. E. & Hoffman, B. D. Improving quality, reproducibility, and usability of FRET-based tension sensors. Cytom. Part A 95, 201–213 (2019).
McCullock, T. W., MacLean, D. M. & Kammermeier, P. J. Comparing the performance of mScarlet-I, mRuby3, and mCherry as FRET acceptors for mNeonGreen. PLOS ONE 15, e0219886 (2020).
Esposito, A. How many photons are needed for FRET imaging? Biomed. Opt. Express 11, 1186–1202 (2020).
Oakes, P. W., Beckham, Y., Stricker, J. & Gardel, M. L. Tension is required but not sufficient for focal adhesion maturation without a stress fiber template. J. Cell Biol. 196, 363–374 (2012).
Weng, S., Shao, Y., Chen, W. & Fu, J. Mechanosensitive subcellular rheostasis drives emergent single-cell mechanical homeostasis. Nat. Mater. 15, 961–967 (2016).
Baumann, H. et al. Biphasic reinforcement of nascent adhesions by vinculin. J. Mol. Recognit. 36, e3012 (2023).
Beningo, K. A., Dembo, M., Kaverina, I., Small, J. V. & Wang, Y. Nascent focal adhesions are responsible for the generation of strong propulsive forces in migrating fibroblasts. J. Cell Biol. 153, 881–888 (2001).
Zhou, D. W., Lee, T. T., Weng, S., Fu, J. & García, A. J. Effects of substrate stiffness and actomyosin contractility on coupling between force transmission and vinculin–paxillin recruitment at single focal adhesions. MBoC 28, 1901–1911 (2017).
Hernández-Varas, P., Berge, U., Lock, J. G. & Strömblad, S. A plastic relationship between vinculin-mediated tension and adhesion complex area defines adhesion size and lifetime. Nat. Commun. 6, 7524 (2015).
Peña Ccoa, W. J., Mukadum, F., Ramon, A., Stirnemann, G. & Hocky, G. M. A direct computational assessment of vinculin–actin unbinding kinetics reveals catch-bonding behavior. Proc. Natl. Acad. Sci. USA. 122, e2411584122 (2025).
Wang, Y. et al. Force-dependent interactions between talin and full-length vinculin. J. Am. Chem. Soc. 143, 14726–14737 (2021).
Murrell, M., Oakes, P. W., Lenz, M. & Gardel, M. L. Forcing cells into shape: the mechanics of actomyosin contractility. Nat. Rev. Mol. Cell Biol. 16, 486–498 (2015).
Wang, C., Zhang, F., Shan, B., Liu, J. & Zhu, L. Real-time measurement of cell contractile force during activation of human hepatic stellate cell line LX-2]. Sheng Wu Yi Xue Gong. Cheng Xue Za Zhi 36, 841–849 (2019).
Saraswathibhatla, A. & Notbohm, J. Tractions and stress fibers control cell shape and rearrangements in collective cell migration. Phys. Rev. X 10, 011016 (2020).
Böhringer, D. et al. Fiber alignment in 3D collagen networks as a biophysical marker for cell contractility. Matrix Biol. 124, 39–48 (2023).
Carisey, A. et al. Vinculin regulates the recruitment and release of core focal adhesion proteins in a force-dependent manner. Curr. Biol. 23, 271–281 (2013).
Ciobanasu, C., Faivre, B. & Le Clainche, C. Actomyosin-dependent formation of the mechanosensitive talin–vinculin complex reinforces actin anchoring. Nat. Commun. 5, 3095 (2014).
Austen, K. et al. Extracellular rigidity sensing by talin isoform-specific mechanical linkages. Nat. Cell Biol. 17, 1597–1606 (2015).
Ringer, P. et al. Multiplexing molecular tension sensors reveals piconewton force gradient across talin-1. Nat. Methods 14, 1090–1096 (2017).
Kim, S. H., Yasunaga, A. B., Zhang, H., Whitley, K. D. & Li, I. T. S. Quantitative super-resolution imaging of molecular tension. Adv. Sci. 12, 2408280 (2025).
Mierke, C. T. et al. Vinculin facilitates cell invasion into three-dimensional collagen matrices. J. Biol. Chem. 285, 13121–13130 (2010).
Lam, A. et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat. Methods 9, 1005–1012 (2012).
Koushik, S. V. & Vogel, S. S. Energy migration alters the fluorescence lifetime of Cerulean: implications for fluorescence lifetime imaging Forster resonance energy transfer measurements. JBO 13, 031204 (2008).
Tse, J. R. & Engler, A. J. Preparation of hydrogel substrates with tunable mechanical properties. CP Cell Biol. 47, 10.16.1–10.16.16 (2010).
Boudou, T. et al. An extended modeling of the micropipette aspiration experiment for the characterization of the Young’s modulus and Poisson’s ratio of adherent thin biological samples: Numerical and experimental studies. J. Biomech. 39, 1677–1685 (2006).
Luengo, C. DIPlib. DIPlib Quantitative Image Analysis in C + + , MATLAB and Python https://diplib.org/ (2024).
Klein, S., Staring, M., Murphy, K., Viergever, M. A. & Pluim, J. P. W. elastix: a toolbox for intensity-based medical image registration. IEEE Trans. Med. Imaging 29, 196–205 (2010).
Aytekin, S. et al. Source Data & Code - linking molecular tension and cellular tractions: a multiscale approach to focal adhesion mechanics. Zenodo https://doi.org/10.5281/zenodo.14692589 (2026).
Acknowledgements
The authors thank Professor Jelle Hendrix (Hasselt University, Belgium) for constructive discussions on pixel-based FRET efficiency analysis, and Rik Nuyts for assistance with microscopy measurements and imaging optimization. They are grateful to the following funding sources: S.R. and S.A. were supported by KU Leuven internal funding: IDN/20/021, KA/20/026 and C14/22/085. S.D. was supported by KU Leuven internal funding: ZB/22/028. S.R. and H.V.O. received funding from the Research Foundation Flanders (FWO) through a project with grant number G0C2422N. H.V.O. and L.K. were supported by the iBOF project 21/083C. H.V.O. received the FWO infrastructure grant I009718N. In addition, S.A. and J.B.F. are recipients of FWO fellowships with grant numbers 1S95125N and 1259223 N, respectively.
Author information
Authors and Affiliations
Contributions
Experiments and data analysis were performed by S.A. and L.K. S.A. and S.V. generated the DNA constructs used in the study. The rheological characterization of the hydrogels was performed by D.L. and S.D., under the supervision of R.C. and H.V.O. The customized Matlab algorithms were developed by L.K., Q.C. and J.B.F. Q.C., M.C., H.V.O., and S.R. developed the initial concept. The work was supervised by H.V.O. and S.R. The original draft was written by S.A., under the supervision of S.R. All authors contributed to the writing of the final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Brenton Hoffman, Andrea Ravasio, Catalina Soto-Montandon and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Ophelia Bu and Christina Karlsson Rosenthal. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Aytekin, S., Kimps, L., Coucke, Q. et al. Linking molecular tension and cellular tractions: a multiscale approach to focal adhesion mechanics. Commun Biol (2026). https://doi.org/10.1038/s42003-026-09514-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-026-09514-0