Abstract
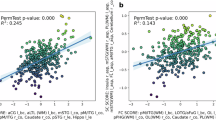
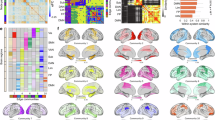
Converging evidence suggests that understanding the human brain requires more than just examining pairwise functional brain interactions. The human brain is a complex, nonlinear system, and focusing solely on linear pairwise functional connectivity often overlooks important nonlinear and higher-order interactions. Infancy is a critical period marked by significant brain development that could contribute to future learning, health, and life success. Exploring higher-order functional relationships in the brain can provide insight into brain function and development. To the best of our knowledge, there is no existing research on multiway, multiscale brain network interactions in infants. In this study, we comprehensively investigate the interactions among brain intrinsic connectivity networks (ICNs), including both pairwise (pair-FNC) and triple relationships (tri-FNC). We focused on a dataset of typically developing infants scanned during the first six months of life-a critical period for brain maturation. In total, 71 infants (aged 4-179 days) contributed 126 scans (76 from 41 males, 50 from 30 females). Our results revealed significant hierarchical, multiway, multiscale brain functional network interactions in the infant brain. These findings suggest that tri-FNC provide additional insights beyond what pairwise interactions reveal during early brain development. The tri-FNC predominantly involve the default mode, sensorimotor, visual, limbic, language, salience, and central executive domains. Notably, these triplet networks align with the classical triple network model of the human brain, which includes the default mode network, the salience network, and the central executive network. This suggests that the brain network system might already be initially established during the first six months of infancy. We also found that pair-FNC were less effective at detecting these networks. The present study suggests that exploring tri-FNC can offer additional insights beyond pair-FNC by capturing higher-order nonlinear interactions, potentially yielding more reliable biomarkers to characterize developmental trajectories.

Similar content being viewed by others
Data availability
Data collected from National Institute of Mental Health (NIMH) 2P50MH100029, R01MH118285, and R01MH119251 are available from the NIMH Data Archive (NDA). NDA is a collaborative informatics system created by the National Institutes of Health to provide a national resource to support and accelerate research in mental health. Dataset identifiers: https://doi.org/10.15154/vjcy-f58967. NeuroMark 2.1 templates are accessible on our lab website (https://trendscenter.org/data/) and GitHub (https://github.com/trendscenter/gift/tree/master/GroupICAT/icatb/icatb_templates). The UNC-BCP 4D Infant Brain Template is available at https://www.nitrc.org/projects/uncbcp_4d_atlas/.
Code availability
The codes of the GICA, and MOO-ICAR have been integrated into the group ICA Toolbox (GIFT 4.0c, https://trendscenter.org/software/gift/). The GAM model from the “mgcv" library in the R environment (https://cran.r-project.org/web/packages/mgcv/index.html).
References
Power, J. D. et al. Functional network organization of the human brain. Neuron 72, 665–678 (2011).
Park, H.-J. & Friston, K. Structural and functional brain networks: from connections to cognition. Science 342, 1238411 (2013).
Iraji, A. et al. Multi-spatial scale dynamic interactions between functional sources reveal sex-specific changes in schizophrenia. Netw. Neurosci. 6, 1–48 (2021).
Iraji, A. et al. Spatial dynamics within and between brain functional domains: A hierarchical approach to study time-varying brain function. Hum. Brain Mapp. 40, 1969–1986 (2018).
Breukelaar, I. A. et al. Cognitive ability is associated with changes in the functional organization of the cognitive control brain network. Hum. Brain Mapp. 39, 5028–5038 (2018).
Haartsen, R., Jones, E. J. H. & Johnson, M. H. Human brain development over the early years. Curr. Opin. Behav. Sci. 10, 149–154 (2016).
Tsang, T. et al. Salience network connectivity is altered in 6-week-old infants at heightened likelihood for developing autism. Commun. Biol. 7, 485 (2024).
Liu, J. et al. Altered lateralization of dorsal language tracts in 6-week-old infants at risk for autism. Dev. Sci. 22 3, e12768 (2018).
Tran, X. A. et al. Functional connectivity during language processing in 3-month-old infants at familial risk for autism spectrum disorder. Eur. J. Neurosci. 53, 1621 – 1637 (2020).
Bruchhage, M. M. K., Ngo, G.-C., Schneider, N., D’Sa, V. & Deoni, S. C. L. Functional connectivity correlates of infant and early childhood cognitive development. Brain Struct. Funct. 225, 669 – 681 (2020).
Blass, E. M. & Camp, C. A. Biological bases of face preference in 6-week-old infants. Dev. Sci. 6, 524–536 (2003).
Seraji, M. et al. Spatial development of brain networks during the first six postnatal months. Commun. Biol. 8, 1514 (2025).
Fransson, P. et al. Resting-state networks in the infant brain. Proc. Natl. Acad. Sci. USA 104, 15531–15536 (2007).
Doria, V. et al. Emergence of resting state networks in the preterm human brain. Proc. Natl. Acad. Sci. USA 107, 20015–20020 (2010).
Fransson, P., Åden, U., Blennow, M. & Lagercrantz, H. The functional architecture of the infant brain as revealed by resting-state fmri. Cereb. Cortex 21, 145–154 (2011).
Gao, W. et al. Evidence on the emergence of the brain’s default network from 2-week-old to 2-year-old healthy pediatric subjects. Proc. Natl. Acad. Sci. USA 106, 6790–5 (2009).
Gao, W. et al. Functional network development during the first year: relative sequence and socioeconomic correlations. Cereb. Cortex 25, 2919–2928 (2015).
Gao, W. et al. Temporal and spatial evolution of brain network topology during the first two years of life. PloS One 6, e25278 (2011).
Gilmore, J. H., Knickmeyer, R. C. & Gao, W. Imaging structural and functional brain development in early childhood. Nat. Rev. Neurosci. 19, 123–137 (2018).
Li, Q. Functional connectivity inference from fmri data using multivariate information measures. Neural Netw. 146, 85–97 (2022).
Ashrafi, M. & Soltanian-Zadeh, H. Multivariate Gaussian copula mutual information to estimate functional connectivity with less random architecture. Entropy 24, 631 (2022).
Li, Q., Yu, S., Madsen, K. H., Calhoun, V. D. & Iraji, A. Higher-order organization in the human brain from matrix-based rényi’s entropy. In 2023 IEEE International Conference on Acoustics, Speech, and Signal Processing Workshops (ICASSPW), 1–5 (2023).
Herzog, R. et al. Genuine high-order interactions in brain networks and neurodegeneration. Neurobiol. Dis. 175, 105918 (2022).
Gatica, M. et al. High-order interdependencies in the aging brain. Brain Connect. 11, 734–744 (2021).
Xie, Q. et al. Constructing high-order functional connectivity network based on central moment features for diagnosis of autism spectrum disorder. PeerJ 9, e11692 (2021).
Li, Q., Steeg, G. V., Yu, S. & Malo, J. Functional connectome of the human brain with total correlation. Entropy 24, 1725 (2022).
Iraji, A. et al. Functional multi-connectivity: A novel approach to assess multi-way entanglement between networks and voxels. In 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), 1698–1701 (2020).
Battiston, F. et al. Networks beyond pairwise interactions: structure and dynamics. Phys. Rep. 874, 1–92 (2020).
Li, Q., Ver Steeg, G. & Malo, J. Functional connectivity via total correlation: analytical results in visual areas. Neurocomputing 571, 127143 (2023).
Li, Q. et al. Aberrant high-order dependencies in schizophrenia resting-state functional MRI networks. In NeurIPS 2023 workshop: Information-Theoretic Principles in Cognitive Systems (2023).
Iraji, A. et al. Identifying canonical and replicable multi-scale intrinsic connectivity networks in 100k+ resting-state fmri datasets. Hum. Brain Mapp. 44, 5729–5748 (2023).
Calhoun, V. D., Adalı, T., Pearlson, G. D. & Pekar, J. J. A method for making group inferences from functional mri data using independent component analysis. Hum. Brain Mapp.14 (2001).
Faghiri, A., Iraji, A., Adalı, T. & Calhoun, V. D. Analysis of high-order brain networks resolved in time and frequency using cp decomposition. ICASSP 2024 - 2024 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) (2024).
Bajracharya, P. et al. Born connected: Do infants already have adult-like multi-scale connectivity networks? bioRxiv (2024).
Watanabe, S. Information theoretical analysis of multivariate correlation. IBM J. Res. Dev. 4, 66–82 (1960).
Du, Y. et al. Artifact removal in the context of group ica: a comparison of single-subject and group approaches. Hum. Brain Mapp. 37, 1005–1025 (2015).
Torres, L., Blevins, A. S., Bassett, D. & Eliassi-Rad, T. The why, how, and when of representations for complex systems. SIAM Rev. 63, 435–485 (2021).
Benson, A. R., Gleich, D. F. & Higham, D. J. Higher-order network analysis takes off, fueled by classical ideas and new data. https://arxiv.org/abs/2103.05031 (2021).
Luppi, A. et al. Systematic evaluation of fmri data-processing pipelines for consistent functional connectomics. Nat. Commun. 15, 4745 (2024).
Zhang, Y., Lucas, M. & Battiston, F. Higher-order interactions shape collective dynamics differently in hypergraphs and simplicial complexes. Nat. Commun. 14, 1605 (2023).
Rieck, B. et al. Uncovering the topology of time-varying fmri data using cubical persistence. In NeurIPS (2020).
Han, T. S. Nonnegative entropy measures of multivariate symmetric correlations. Inf. Control 36, 133–156 (1978).
Rosas, F. E., Mediano, P. A., Gastpar, M. & Jensen, H. J. Quantifying high-order interdependencies via multivariate extensions of the mutual information. Phys. Rev. E 100, 032305 (2019).
Iraji, A., Miller, R., Adali, T. & Calhoun, V. Space: A missing piece of the dynamic puzzle. Trends Cogn. Sci.24 (2020).
Allen, E. et al. Tracking whole-brain connectivity dynamics in the resting state. Cerebral Cortex (New York, N.Y.: 1991) (2012).
Iraji, A. et al. Tools of the trade: Estimating time-varying connectivity patterns from fmri data. Soc. Cogn. Affect. Neurosci. 16, 849–874 (2020).
Vidaurre, D., Smith, S. M. & Woolrich, M. W. Brain network dynamics are hierarchically organized in time. Proc. Natl. Acad. Sci. USA 114, 12827 – 12832 (2017).
Iraji, A. et al. The spatial chronnectome reveals a dynamic interplay between functional segregation and integration. Hum. Brain Mapp. 40, 3058–3077 (2019).
Shultz, S., Klin, A. & Jones, W. Neonatal transitions in social behavior and their implications for autism. Trends Cogn. Sci. 22, 452–469 (2018).
Ford, A. et al. Caregiver greeting to infants under 6 months already reflects emerging differences in those later diagnosed with autism. Proc. B 291, 20232494 (2024).
Chen, L. et al. A 4d infant brain volumetric atlas based on the unc/umn baby connectome project (bcp) cohort. NeuroImage 253, 119097 (2022).
Du, Y. et al. Neuromark: An automated and adaptive ica based pipeline to identify reproducible fmri markers of brain disorders. NeuroImage: Clin. 28, 102375 (2020).
Meng, X. et al. Multi-model order spatially constrained ica reveals highly replicable group differences and consistent predictive results from resting data: a large n fmri schizophrenia study. NeuroImage: Clin. 38, 103434 (2023).
Calhoun, V. et al. Exploring the psychosis functional connectome: Aberrant intrinsic networks in schizophrenia and bipolar disorder. Front. Psychiatry Front. Res. Found. 2, 75 (2011).
Erhardt, E. et al. Comparison of multi-subject ica methods for analysis of fmri data. Hum. Brain Mapp. 32, 2075–95 (2011).
Lin, Q.-H., Liu, J., Zheng, Y.-R., Liang, H. & Calhoun, V. Semiblind spatial ICA of fmri using spatial constraints. Hum. Brain Mapp. 31, 1076–88 (2009).
Lu, W. & Rajapakse, J. Approach and applications of constrained ica. IEEE Trans. Neural Netw. 16, 203–212 (2005).
T. M., C. & J. A., T. Elements of Information Theory 2 edn (Wiley-Interscience, Hoboken, NJ, USA, 2006).
Hlinka, J., Paluš, M., Vejmelka, M., Mantini, D. & Corbetta, M. Functional connectivity in resting-state fmri: is linear correlation sufficient?. Neuroimage 54, 2218–2225 (2011).
Liégeois, R., Yeo, B. T. & Van De Ville, D. Interpreting null models of resting-state functional mri dynamics: not throwing the model out with the hypothesis. NeuroImage 243, 118518 (2021).
Bell, A. J. & Sejnowski, T. J. An information-maximization approach to blind separation and blind deconvolution. Neural Comput. 7, 1129–1159 (1995).
Abou Elseoud, A. et al. The effect of model order selection in group pica. Hum. Brain Mapp. 31, 1207–16 (2009).
Ray, K. et al. ICA model order selection of task co-activation networks. Front. Neurosci. 7, 237 (2013).
Nelder, J. A. & Wedderburn, R. W. M. Generalized linear models. J. R. Stat. Soc. Ser. A (Gen.) 135, 370–384 (1972).
Hastie, T. & Tibshirani, R. Generalized Additive Models (Wiley Online Library, 1990).
Cavanaugh, J. E. & Neath, A. A. The akaike information criterion: Background, derivation, properties, application, interpretation, and refinements. WIREs Comput. Stat. 11, e1460 (2019).
Seraji, M. et al. Spatial development of brain networks during the first six postnatal months. [Data set]. NIMH Data Archive https://doi.org/10.15154/vjcy-f589 (2026).
Acknowledgements
We are deeply grateful to the families and their infants who generously volunteered to participate in this research. We also extend our sincere thanks to the research coordinators, assistants, and fellows at the Marcus Autism Center-Brittney Sholar, Carly Reineri, Joanna Beugnon, Lindsey Evans, Jordan Pincus, Jennifer Gutierrez, Tristan Ponzo, and Adriana Mendez-for their invaluable efforts in data collection. We further thank the MRI technologists at the Emory Center for Systems Imaging Core-Michael White, Sarah Basadre, and Samira Yeboah-for their skilled technical support. Finally, we acknowledge Dr.Lei Zhou and Michael Valente for their contributions to equipment development and data acquisition protocols.
This work was supported by the National Institutes of Health (NIH) grant number R01MH119251 to Sarah Shultz and Armin Iraji, R01EB027147 to Sarah Shultz and Vince Calhoun, P50MH100029 to Sarah Shultz, and National Science Foundation (NSF) grant number 2112455 to Vince Calhoun.
Author information
Authors and Affiliations
Contributions
Q.L. served as the first author, with primary responsibility for the conception, design, and overall execution of the study, as well as for drafting the manuscript. S.S. contributed critically to data collection and manuscript preparation, ensuring the accuracy and integrity of the collected data. Z.F., H.W., M.S., and P.B. were responsible for data preprocessing and made significant contributions to manuscript drafting. A.I., S.S., and V.C. provided expert supervision throughout the research process, offering essential guidance, critical feedback, and substantive contributions to the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Michel Thiebaut de Schotten and Jasmine Pan.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, Q., Fu, Z., Walum, H. et al. Deciphering multiway multiscale brain network connectivity from birth to 6 months. Commun Biol (2026). https://doi.org/10.1038/s42003-026-09549-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-026-09549-3