Abstract
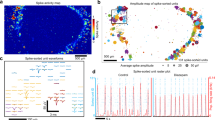
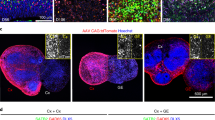
Reconstructing networks of neurons in vitro is essential for advancing our understanding of functional mechanisms and disease pathogenesis. However, neuronal culture methods including organoids are limited in network structure complexity required for their functionality and dynamics. In this study, we present modular organoid network tissues – loop connectoids – in which multiple cerebral organoids are connected via axon bundles using microfluidic devices. We compared network activity of three- and four-membered loop cerebral connectoids, two reciprocally connected organoids, and single organoids. We observed a significant trend in larger organoid networks exhibiting more complex activity, showing longer activity periods, more bursts, and richer temporal patterns. Additionally, the activity in connectoids shifts closer to a critical state, a hallmark of efficient information processing in the brain, as more organoids are connected. Pharmacological perturbation reveals prominent excitatory and inhibitory responses, supporting the physiological relevance of the observed dynamics. Furthermore, optogenetic stimulation of organoids in a specific sequence can influence their spontaneous activity propagation pattern within the network. This work represents a foundational step toward constructing more complex and physiologically relevant neural networks in vitro, offering a platform for studying neuronal network function and therapeutic intervention.

Similar content being viewed by others
Data availability
All source data of graphs and charts of the main manuscript are included in Supplementary Data 1. Any additional information about the data reported in this paper is available from the lead contact upon request.
Code availability
Source codes for processing MEA data are available on GitHub here https://github.com/utokyoIkeuchilab/loop-connectoid-activity-MEA-analysis. Calculations of network connectivity metrics using previously published32 functions can be found here http://www.brain-connectivity-toolbox.net/. The analysis on criticality metrics using previously published35 functions can be found here http://www.nicholastimme.com/software.html.
References
Cordella, F., Brighi, C., Soloperto, A. & Di Angelantonio, S. Stem cell-based 3D brain organoids for mimicking, investigating, and challenging Alzheimer’s diseases. Neural Regen. Res. 17, 330–332 (2022).
Kelava, I. & Lancaster, M. A. Stem cell models of human brain development. Cell Stem Cell 18, 736–748 (2016).
Trujillo, C. A. et al. Complex oscillatory waves emerging from cortical organoids model early human brain network development. Cell Stem Cell 25, 558–569.e7 (2019).
Sharf, T. et al. Functional neuronal circuitry and oscillatory dynamics in human brain organoids. Nat. Commun. 13, 1–20 (2022).
Jung, J. Y., Cloutman, L. L., Binney, R. J. & Lambon Ralph, M. A. The structural connectivity of higher order association cortices reflects human functional brain networks. Cortex 97, 221 (2017).
Hong, S. J. et al. Atypical functional connectome hierarchy in autism. Nat. Commun. 10, 1–13 (2019).
Cao, H., Zhou, H. & Cannon, T. D. Functional connectome-wide associations of schizophrenia polygenic risk. Mol. Psychiatry 26, 2553–2561 (2020).
Engel, J. et al. Connectomics and epilepsy. Curr. Opin. Neurol. 26, 186–194 (2013).
Birey, F. et al. Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59 (2017).
Xiang, Y. et al. hESC-derived thalamic organoids form reciprocal projections when fused with cortical organoids. Cell Stem Cell 24, 487–497.e7 (2019).
Kim, J. et al. Human assembloid model of the ascending neural sensory pathway. Nature 642, 143–153 (2025).
Miura, Y. et al. Assembloid model to study loop circuits of the human nervous system. bioRxiv https://doi.org/10.1101/2024.10.13.617729 (2024).
Kirihara, T. et al. A human induced pluripotent stem cell-derived tissue model of a cerebral tract connecting two cortical regions. iScience 14, 301–311 (2019).
Martins-Costa, C. et al. ARID1B controls transcriptional programs of axon projection in an organoid model of the human corpus callosum. Cell Stem Cell 31, 866–885.e14 (2024).
Dolgin, E. Brain tissues, assemble! Inside the push to build better brain models. Nature 641, 809–812 (2025).
Osaki, T. et al. Complex activity and short-term plasticity of human cerebral organoids reciprocally connected with axons. Nat. Commun. 15, 1–13 (2024).
Yu, C. Toward a unified analysis of the brain criticality hypothesis: reviewing several available tools. Front. Neural Circ. 16, 911245 (2022).
Duru, J. et al. Engineered biological neural networks on high density CMOS microelectrode arrays. Front. Neurosci. 16, 829884 (2022).
Hong, N. & Nam, Y. Thermoplasmonic neural chip platform for in situ manipulation of neuronal connections in vitro. Nat. Commun. 11, 1–12 (2020).
Habibey, R. et al. Engineered modular neuronal networks-on-chip represent structure-function relationship. Biosens. Bioelectron. 261, 116518 (2024).
Yamamoto, H. et al. Impact of modular organization on dynamical richness in cortical networks. Sci. Adv. 4, eaau4914 (2018).
Sato, Y. et al. Microfluidic cell engineering on high-density microelectrode arrays for assessing structure-function relationships in living neuronal networks. Front. Neurosci. 16, 943310 (2023).
Habibollahi, F., Kagan, B. J., Burkitt, A. N. & French, C. Critical dynamics arise during structured information presentation within embodied in vitro neuronal networks. Nat. Commun. 14, 1–13 (2023).
Murota, H., Yamamoto, H., Monma, N., Sato, S. & Hirano-Iwata, A. Precision microfluidic control of neuronal ensembles in cultured cortical networks. Adv. Mater. Technol. 10, 2400894 (2025).
Chow, S. Y. A. et al. Repetitive stimulation modifies network characteristics of neural organoid circuits. bioRxiv https://doi.org/10.1101/2025.01.16.633310 (2025).
Bi, G. Q. & Poo, M. M. Synaptic modifications in cultured hippocampal neurons: dependence on spike timing, synaptic strength, and postsynaptic cell type. J. Neurosci. 18, 10464–10472 (1998).
Johansen, J. P. et al. Hebbian and neuromodulatory mechanisms interact to trigger associative memory formation. Proc. Natl. Acad. Sci. USA 111, E5584–E5592 (2014).
Debanne, D. & Inglebert, Y. Spike timing-dependent plasticity and memory. Curr. Opin. Neurobiol. 80, 102707 (2023).
Ting, J. T. & Feng, G. Neurobiology of obsessive–compulsive disorder: insights into neural circuitry dysfunction through mouse genetics. Curr. Opin. Neurobiol. 21, 842–848 (2011).
Nordstrom, E. J., Bittner, K. C., McGrath, M. J., Parks, C. R. & Burton, F. H. ‘Hyperglutamatergic cortico-striato-thalamo-cortical circuit’ breaker drugs alleviate tics in a transgenic circuit model of Tourette׳s syndrome. Brain Res. 1629, 38–53 (2015).
Osaki, T. et al. Three-dimensional motor nerve organoid generation. J. Vis. Exp. 2020, 1–18 (2020).
Rubinov, M. & Sporns, O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage 52, 1059–1069 (2010).
Cutts, C. S. & Eglen, X. S. J. Detecting pairwise correlations in spike trains: an objective comparison of methods and application to the study of retinal waves. J. Neurosci. 34, 14288–14303 (2014).
Giandomenico, S. L. et al. Cerebral organoids at the air–liquid interface generate diverse nerve tracts with functional output. Nat. Neurosci. 22, 669–679 (2019).
Marshall, N. et al. Analysis of power laws, shape collapses, and neural complexity: new techniques and MATLAB support via the NCC toolbox. Front Physiol. 7, 191703 (2016).
Acknowledgements
We thank all Ikeuchi lab members for discussions and comments. We also thank past members, especially Tatsuya Osaki and Yasuhiro Ikegami, for discussions. This work was supported in part by a Grant-in-Aid for Challenging Research (Pioneering) from the JSPS (20K20643); a Grant-in-Aid for Transformative Research Areas (20H05786, 24H02307, 25H02596); AMED-CREST (JP20gm1410001, 24wm0625318, 25wm0625323); JSPS Core-to-Core Program (JPJSCCA 20190006); HFSP (RGP012/2024) and the Institute for AI and Beyond. The study was also supported by JST SPRING (JPMJSP2108), the ANRI fellowship, and a Grant-in-Aid for Research Activity Start-up from JSPS (25K23557).
Author information
Authors and Affiliations
Contributions
T.D. and Y.I. designed the experiments and wrote the manuscript. T.D performed the experiments and analyzed the data under the supervision of Y.I.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Itzy E. Morales Pantoja, Francesca Ciarpella and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Benjamin Bessieres. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Duenki, T., Ikeuchi, Y. Multi-organoid loop cerebral connectoids exhibit enhanced neuronal network dynamics and sequence-specific entrainment. Commun Biol (2026). https://doi.org/10.1038/s42003-026-09589-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-026-09589-9