Abstract
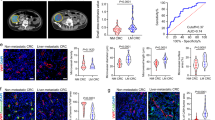
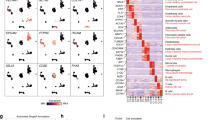
Pericytes, essential components of the tumor microenvironment, undergo phenotypic alterations that influence cancer progression, yet the molecular mechanisms governing these changes remain poorly understood. Here, we investigate the role of Notch3 signaling in pericyte phenotype and functions in colorectal cancer (CRC). Using lineage tracing approaches, we show that murine tumor pericytes originate from normal tissue-resident pericytes, which proliferate inside tumors. In vivo genetic manipulation reveals that Notch3 pathway activation promotes pericyte proliferation, while suppressing contractile protein expression, and leads to increased endothelial cell proliferation and reduced blood vessel integrity. In contrast, Notch3 deletion leads to decreased endothelial proliferation, blood vessel normalization, and a significant reduction in tumorigenesis in an advanced orthotopic mouse model. Single-cell RNA sequencing analysis uncovers significant pericyte heterogeneity in both mouse colitis-associated cancer and human CRC. It specifically identifies distinct subpopulations characterized by differential Notch3 activity, which is enriched in a synthetic subset and absent in a contractile subset, further supporting our in vivo findings. Our results establish Notch3 as a key regulator of pericyte phenotypic plasticity in CRC and suggest that targeting this pathway could represent a promising strategy for improving therapeutic outcomes through vascular normalization.

Similar content being viewed by others
Data availability
All in-house scRNA-seq data have been deposited in the Gene Expression Omnibus (GEO) database under the accession numbers GSE296872 and GSE296873. The numerical source data for the graphs can be found in Supplementary Data 2. All other data are available from the corresponding author (or other sources, as applicable) on reasonable request.
Code availability
All scripts used to generate the figures are available on GitHub [https://github.com/AthanasiaSt/Notch3-regulates-pericyte-phenotypic-plasticity-in-colorectal-cancer.git].
References
Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 73, 17–48 (2023).
Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021).
Ganesh, K. et al. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat. Rev. Gastroenterol. Hepatol. 16, 361–375 (2019).
Hanahan, D. & Coussens, L. M. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21, 309–322 (2012).
Pérez-González, A., Bévant, K. & Blanpain, C. Cancer cell plasticity during tumor progression, metastasis and response to therapy. Nat. Cancer 4, 1063–1082 (2023).
Armulik, A., Genove, G. & Betsholtz, C. Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev. Cell 21, 193–215 (2011).
Morikawa, S. et al. Abnormalities in pericytes on blood vessels and endothelial sprouts in tumors. Am. J. Pathol. 160, 985–1000 (2002).
Meng, M. B. et al. Pericytes: a double-edged sword in cancer therapy. Future Oncol. 11, 169–179 (2015).
Johansson-Percival, A. et al. Intratumoral LIGHT restores pericyte contractile properties and vessel integrity. Cell Rep. 13, 2687–2698 (2015).
Li, Z. J. et al. Pericyte phenotype switching alleviates immunosuppression and sensitizes vascularized tumors to immunotherapy in preclinical models. J. Clin. Investig. 134, e179860 (2024).
He, B. et al. Selective tubulin-binding drugs induce pericyte phenotype switching and anti-cancer immunity. EMBO Mol. Med. 17, 1071–1100 (2025).
Song, S., Ewald, A. J., Stallcup, W., Werb, Z. & Bergers, G. PDGFRbeta+ perivascular progenitor cells in tumours regulate pericyte differentiation and vascular survival. Nat. Cell Biol. 7, 870–879 (2005).
Du, R. et al. HIF1alpha induces the recruitment of bone marrow-derived vascular modulatory cells to regulate tumor angiogenesis and invasion. Cancer Cell 13, 206–220 (2008).
Clere, N., Renault, S. & Corre, I. Endothelial-to-mesenchymal transition in cancer. Front. Cell Dev. Biol. 8, 747 (2020).
Viallard, C. & Larrivée, B. Tumor angiogenesis and vascular normalization: alternative therapeutic targets. Angiogenesis 20, 409–426 (2017).
Carmeliet, P. & Jain, R. K. Principles and mechanisms of vessel normalization for cancer and other angiogenic diseases. Nat. Rev. Drug Discov. 10, 417–427 (2011).
Siebel, C. & Lendahl, U. Notch signaling in development, tissue homeostasis, and disease. Physiol. Rev. 97, 1235–1294 (2017).
Ntziachristos, P., Lim, J. S., Sage, J. & Aifantis, I. From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer Cell 25, 318–334 (2014).
Aster, J. C., Pear, W. S. & Blacklow, S. C. The varied roles of Notch in cancer. Annu. Rev. Pathol. 12, 245–275 (2017).
Kopan, R. & Ilagan, M. X. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216–233 (2009).
Domenga, V. et al. Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev. 18, 2730–2735 (2004).
Joutel, A. et al. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature 383, 707–710 (1996).
Liu, H., Zhang, W., Kennard, S., Caldwell, R. B. & Lilly, B. Notch3 is critical for proper angiogenesis and mural cell investment. Circ. Res. 107, 860–870 (2010).
Aburjania, Z. et al. The role of Notch3 in cancer. Oncologist 23, 900–911 (2018).
Hu, W. et al. Notch3 pathway alterations in ovarian cancer. Cancer Res. 74, 3282–3293 (2014).
Ozawa, T. et al. Nuclear Notch3 expression is associated with tumor recurrence in patients with stage II and III colorectal cancer. Ann. Surg. Oncol. 21, 2650–2658 (2014).
Varga, J. et al. AKT-dependent NOTCH3 activation drives tumor progression in a model of mesenchymal colorectal cancer. J. Exp. Med. 217, e20191515 (2020).
Serafin, V. et al. Notch3 signalling promotes tumour growth in colorectal cancer. J. Pathol. 224, 448–460 (2011).
Wang, X. W. et al. MicroRNA-206 attenuates tumor proliferation and migration involving the downregulation of NOTCH3 in colorectal cancer. Oncol. Rep. 33, 1402–1410 (2015).
Neufert, C. et al. Inducible mouse models of colon cancer for the analysis of sporadic and inflammation-driven tumor progression and lymph node metastasis. Nat. Protoc. 16, 61–85 (2021).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Pan, X. et al. Tumour vasculature at single-cell resolution. Nature 632, 429–436 (2024).
Kobayashi, H. et al. The origin and contribution of cancer-associated fibroblasts in colorectal carcinogenesis. Gastroenterology 162, 890–906 (2022).
Pelka, K. et al. Spatially organized multicellular immune hubs in human colorectal cancer. Cell 184, 4734–4752.e4720 (2021).
Lee, H. O. et al. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat. Genet. 52, 594–603 (2020).
Qi, J. et al. Single-cell and spatial analysis reveal interaction of FAP+ fibroblasts and SPP1+ macrophages in colorectal cancer. Nat. Commun. 13, 1742 (2022).
Khaliq, A. M. et al. Refining colorectal cancer classification and clinical stratification through a single-cell atlas. Genome Biol. 23, 113 (2022).
Cuervo, H. et al. PDGFRβ-P2A-CreER(T2) mice: a genetic tool to target pericytes in angiogenesis. Angiogenesis 20, 655–662 (2017).
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007).
Hong, S. P. et al. Distinct fibroblast subsets regulate lacteal integrity through YAP/TAZ-induced VEGF-C in intestinal villi. Nat. Commun. 11, 4102 (2020).
Vega, P. N. et al. Cancer-associated fibroblasts and squamous epithelial cells constitute a unique microenvironment in a mouse model of inflammation-induced colon cancer. Front. Oncol. 12, 878920 (2022).
Fre, S. et al. Notch lineages and activity in intestinal stem cells determined by a new set of knock-in mice. PLoS ONE 6, e25785 (2011).
Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12, 1088 (2021).
Oh, P. et al. In vivo mapping of notch pathway activity in normal and stress hematopoiesis. Cell Stem Cell 13, 190–204 (2013).
Ventura, A. et al. Restoration of p53 function leads to tumour regression in vivo. Nature 445, 661–665 (2007).
Hamzah, J. et al. Vascular normalization in Rgs5-deficient tumours promotes immune destruction. Nature 453, 410–414 (2008).
Krebs, L. T. et al. Characterization of Notch3-deficient mice: normal embryonic development and absence of genetic interactions with a Notch1 mutation. Genesis 37, 139–143 (2003).
Roper, J. et al. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat. Biotechnol. 35, 569–576 (2017).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Chhabra, Y. & Weeraratna, A. T. Fibroblasts in cancer: Unity in heterogeneity. Cell 186, 1580–1609 (2023).
De Palma, M. et al. Tie2 identifies a hematopoietic lineage of proangiogenic monocytes required for tumor vessel formation and a mesenchymal population of pericyte progenitors. Cancer Cell 8, 211–226 (2005).
Patenaude, A. et al. A novel population of local pericyte precursor cells in tumor stroma that require Notch signaling for differentiation. Microvasc. Res. 101, 38–47 (2015).
Sweeney, C. et al. Notch 1 and 3 receptor signaling modulates vascular smooth muscle cell growth, apoptosis, and migration via a CBF-1/RBP-Jk dependent pathway. FASEB J. 18, 1421–1423 (2004).
Morrow, D. et al. Notch-mediated CBF-1/RBP-J{kappa}-dependent regulation of human vascular smooth muscle cell phenotype in vitro. Am. J. Physiol. Cell Physiol. 289, C1188–C1196 (2005).
Alarcon-Martinez, L., Yemisci, M. & Dalkara, T. Pericyte morphology and function. Histol. Histopathol. 36, 633–643 (2021).
Dessalles, C. A., Babataheri, A. & Barakat, A. I. Pericyte mechanics and mechanobiology. J. Cell Sci. 134, jcs240226 (2021).
Jain, R. K. Antiangiogenesis strategies revisited: from starving tumors to alleviating hypoxia. Cancer Cell 26, 605–622 (2014).
Melissari, M. T. et al. Col6a1+/CD201+ mesenchymal cells regulate intestinal morphogenesis and homeostasis. Cell. Mol. Life Sci. 79, 1 (2021).
Chalkidi, N. et al. Activation and functions of Col6a1+ fibroblasts in colitis-associated cancer. Int. J. Mol. Sci. 25, 148 (2023).
Koliaraki, V. & Kollias, G. Isolation of intestinal mesenchymal cells from adult mice. Bio-protocol 6, e1940 (2016).
Acknowledgements
The authors would like to thank Sofia Grammenoudi for assistance in FACS sorting experiments. The authors would also like to thank Fleming’s Animal House and Flow Cytometry facilities. This work was funded by Worldwide Cancer Research (Grant No: 22-0126, to V.K.), and the project Fib3R (Grant No: 3001, to V.K.) was funded by the Hellenic Foundation of Research and Innovation (H.F.R.I.) under the “2nd Call for H.F.R.I. Research Projects to support Faculty members and Researchers.” The authors acknowledge support of this work by the project SingleOut (HFRI-FM17C3-3780) funded by the Hellenic Foundation for Research and Innovation (H.F.R.I.) under the “1st Call for H.F.R.I. Research Projects to support Faculty members and Researchers and the procurement of high-cost research equipment”. The authors also acknowledge support by the Research Infrastructure projects InfrafrontierGR (MIS 5002802) and pMedGR (MIS 5002802) funded by the Operational Programme “Competitiveness, Entrepreneurship and Innovation” (NSRF 2014-2020) and co-financed by Greece and the European Union (European Regional Development Fund), as well as by project MIS 6004752 funded by the Regional Operational Programme “ATTICA” (NSRF 2021-2027) and co-financed by Greece and the European Union (European Regional Development Fund). The graphical abstract was created using BioRender (https://BioRender.com/kvnz47d).
Author information
Authors and Affiliations
Contributions
N.C. and V.K. designed the study; N.C. V.Z.A, C.P., A.M., and M.S. performed experiments; A.S. performed the bioinformatics analysis; C.N. and V.K. supervised the bioinformatics analysis; N.C., A.S., and V.K. interpreted the experimental results, wrote the manuscript, and prepared the figures; V.K. supervised the study; all authors critically revised and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Jiha Kim, Vikneshwari Natarajan, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Kaliya Georgieva. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chalkidi, N., Stavropoulou, A., Arvaniti, VZ. et al. Notch3 regulates pericyte phenotypic plasticity in colorectal cancer. Commun Biol (2026). https://doi.org/10.1038/s42003-026-09629-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-026-09629-4