Abstract
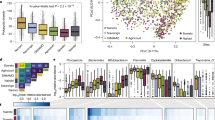
Gut microbiota composition has been extensively studied in European and North American pediatric cohorts, as well as in rural African children. Much less attention has been paid to urban African children, whose families have transitioned to a “Western” lifestyle characterized by smaller family sizes, access to perinatal care including C-section delivery, non-traditional food sources and widespread availability of antibiotics. We analyzed fecal samples from ~200 Ethiopian children aged 2-5 years from Adama, Ethiopia, using 16S rRNA gene sequencing and shotgun metagenomics. We found that well-studied factors such as delivery mode, breastfeeding and family size have only minor effects on α-diversity, whereas household crowding (single vs. multiple rooms) and consumption of the traditional fermented cereal Eragrostis tef predict higher α-diversity. Stunted growth and absence of Helicobacter pylori infection were additional factors associated with increased fecal microbial diversity. Metagenomic profiling revealed that rural African signature genera such as Segatella and Prevotella were largely absent; instead, urban Ethiopian children displayed a high Firmicutes/Bacteroidota ratio and enrichment of metabolic pathways linked to a westernized diet, resembling European rather than rural Ethiopian children. These results indicate that an urban westernized lifestyle alters gut microbiota composition, which may be partially offset by a traditional fermented diet.

Similar content being viewed by others
Data availability
The sequencing data generated in this study, including metagenomic and processed 16S rRNA reads, are publicly available in the NCBI BioProject database under accession number PRJNA1345963. For metagenomic sequencing, the raw paired-end reads have been deposited without further processing. For 16S rRNA gene sequencing, the deposited data consist of the quality-filtered, merged, and chimera-free reads as provided by the sequencing facility. Source data for all figures are provided in Supplementary Data 14. Sequencing data from external resources are available under accession numbers PRJNA504891 (rural Ethiopian children) and PRJNA716780 (Italian children).
Code availability
All custom code used for data processing and figure generation is publicly available at GitHub (https://github.com/lydiakirsche/urban-ethiopian-children-microbiome) and archived on Zenodo82 (10.5281/zenodo.18231174) to ensure long-term accessibility and reproducibility.
References
Bäckhed, F. et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 690–703 (2015).
Asnicar, F. et al. Studying vertical microbiome transmission from mothers to infants by strain-level metagenomic profiling. mSystems 2, e00164–16 (2017).
Ferretti, P. et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 24, 133–145 (2018).
Yassour, M. et al. Strain-level analysis of mother-to-child bacterial transmission during the first few months of life. Cell Host Microbe 24, 146–154 (2018).
Dominguez-Bello, M. G. et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 107, 11971–11975 (2010).
Bokulich, N. A. et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 8, 343ra82 (2016).
Shao, Y. et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 574, 117–121 (2019).
Stearns, J. C. et al. Intrapartum antibiotics for GBS prophylaxis alter colonization patterns in the early infant gut microbiome of low risk infants. Sci. Rep. 7, 1–9 (2017).
Yassour, M. et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci. Transl. Med. 8, 343ra81 (2016).
Selma-Royo, M. et al. Birthmode and environment-dependent microbiota transmission dynamics are complemented by breastfeeding during the first year. Cell Host Microbe 32, 996–1010 (2024).
Shao, Y. et al. Primary succession of Bifidobacteria drives pathogen resistance in neonatal microbiota assembly. Nat. Microbiol. 9, 2570–2582 (2024).
Schwartz, D. J., Langdon, A. E. & Dantas, G. Understanding the impact of antibiotic perturbation on the human microbiome. Genome Med. 12, 1–12 (2020).
Manara, S. et al. Maternal and food microbial sources shape the infant microbiome of a rural Ethiopian population. Curr. Biol. 33, 1939–1950. (2023).
Yatsunenko, T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012).
Pasolli, E. et al. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell 176, 649–662 (2019).
Maghini, D. G. et al. Expanding the human gut microbiome atlas of Africa. Nature 638, 718–728 (2025).
Vangay, P. et al. US immigration westernizes the human gut microbiome. Cell 175, 962–972 (2018).
De Filippo, C. et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 107, 14691–14696 (2010).
Tett, A. et al. The Prevotella copri complex comprises four distinct clades underrepresented in westernized populations. Cell Host Microbe 26, 666–679 (2019).
Smits, S. A. et al. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 357, 802–805 (2017).
Robertson, R. C. et al. The gut microbiome and early-life growth in a population with high prevalence of stunting. Nat. Commun. 14, 1–15 (2023).
Bowers, R. M. et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 35, 725–731 (2017).
Hjorth, M. F. et al. Prevotella-to-bacteroides ratio predicts body weight and fat loss success on 24-week diets varying in macronutrient composition and dietary fiber: results from a post-hoc analysis. Int. J. Obes. 43, 149–157 (2018).
Arumugam, M. et al. Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011).
Roager, H. M., Licht, T. R., Poulsen, S. K., Larsen, T. M. & Bahl, M. I. Microbial enterotypes, inferred by the Prevotella-to-Bacteroides ratio, remained stable during a 6-month randomized controlled diet intervention with the new Nordic diet. Appl. Environ. Microbiol. 80, 1142–1149 (2014).
Ramne, S. et al. Gut microbiota composition in relation to intake of added sugar, sugar-sweetened beverages and artificially sweetened beverages in the Malmö offspring study. Eur. J. Nutr. 60, 2087–2097 (2021).
Angelakis, E. et al. Treponema species enrich the gut microbiota of traditional rural populations but are absent from urban individuals. New Microbes New Infect. 27, 14–21 (2019).
Victora, C. G. et al. Revisiting maternal and child undernutrition in low-income and middle-income countries: variable progress towards an unfinished agenda. Lancet 397, 1388–1399 (2021).
Ratnayani et al. Association of gut microbiota composition with stunting incidence in children under five in Jakarta slums. Nutrients 16, 3444 (2024).
Dinh, D. M. et al. Longitudinal analysis of the intestinal microbiota in persistently stunted young children in South India. PLoS ONE 11, e0155405 (2016).
Gough, E. K. et al. Linear growth faltering in infants is associated with Acidaminococcus sp. and community-level changes in the gut microbiota. Microbiome 3, 1–10 (2015).
von Schwartzenberg, R. J. et al. Caloric restriction disrupts the microbiota and colonization resistance. Nature 595, 272–277 (2021).
Hardjo, J. & Selene, N. B. Stunting and gut microbiota: a literature review. Pediatr. Gastroenterol. Hepatol. Nutr. 27, 137 (2024).
Llorca, L. et al. Characterization of the gastric microbiota in a pediatric population according to Helicobacter pylori status. Pediatr. Infect. Dis. J. 36, 173–178 (2017).
Brawner, K. M. et al. Helicobacter pylori infection is associated with an altered gastric microbiota in children. Mucosal Immunol. 10, 1169–1177 (2017).
Frost, F. et al. Helicobacter pylori infection associates with fecal microbiota composition and diversity. Sci. Rep. 9, 1–10 (2019).
Gao, J. J. et al. Association between gut microbiota and Helicobacter pylori-related gastric lesions in a high-risk population of gastric cancer. Front. Cell. Infect. Microbiol. 8, 369735 (2018).
Benavides-Ward, A. et al. Helicobacter pylori and its relationship with variations of gut microbiota in asymptomatic children between 6 and 12 years. BMC Res. Notes 11, 1–7 (2018).
Osaki, T. et al. Influence of intestinal indigenous microbiota on intrafamilial infection by Helicobacter pylori in Japan. Front. Immunol. 9, 325662 (2018).
Yang, L. et al. Helicobacter pylori infection aggravates dysbiosis of gut microbiome in children with gastritis. Front. Cell. Infect. Microbiol. 9, 473388 (2019).
Lapidot, Y., Reshef, L., Cohen, D. & Muhsen, K. Helicobacter pylori and the intestinal microbiome among healthy school-age children. Helicobacter 26, e12854 (2021).
DuPont, H. L., Jiang, Z. D., Alexander, A. S., DuPont, A. W. & Brown, E. L. Intestinal IgA-coated bacteria in healthy- and altered-microbiomes (dysbiosis) and predictive value in successful fecal microbiota transplantation. Microorganisms 11, 93 (2022).
Hsieh, C.-S. et al. Altered IgA response to gut bacteria is associated with childhood asthma in Peru. J. Immunol. 207, 398–407 (2021).
Dzidic, M. et al. Aberrant IgA responses to the gut microbiota during infancy precede asthma and allergy development. J. Allergy Clin. Immunol. 139, 1017–1025 (2017).
Eriksen, C. et al. IgG and IgM cooperate in coating of intestinal bacteria in IgA deficiency. Nat. Commun. 14, 1–12 (2023).
Catanzaro, J. R. et al. IgA-deficient humans exhibit gut microbiota dysbiosis despite secretion of compensatory IgM. Sci. Rep. 9, 1–10 (2019).
Sterlin, D. et al. Human IgA binds a diverse array of commensal bacteria. J. Exp. Med. 217, e20181635 (2020).
Van Der Waaij, L. A. et al. Immunoglobulin coating of faecal bacteria in inflammatory. Eur. J. Gastroenterol. Hepatol. 16, 669–674 (2004).
Boutard, M. et al. Functional diversity of carbohydrate-active enzymes enabling a bacterium to ferment plant biomass. PLoS Genet. 10, e1004773 (2014).
Gorvitovskaia, A., Holmes, S. P. & Huse, S. M. Interpreting prevotella and bacteroides as biomarkers of diet and lifestyle. Microbiome 4, 1–12 (2016).
Du, S. et al. Children’s home environments as reservoirs of antimicrobial resistance: divergent urban-rural risks from antibiotic resistance genes and pathogens. J. Hazard. Mater. 495, 139053 (2025).
Balachandra, S. S. et al. Antimicrobial resistance (AMR) at the community level. J. Family Med. Prim. Care 10, 1404–1411 (2021).
Fredriksen, S., de Warle, S., van Baarlen, P., Boekhorst, J. & Wells, J. M. Resistome expansion in disease-associated human gut microbiomes. Microbiome 11, 166 (2023).
Gudeta, A. N., Andrén Aronsson, C., Binagdie, B. B., Girma, A. & Agardh, D. Incidence of celiac disease autoimmunity and associations with maternal tuberculosis and pediatric Helicobacter pylori infections in 4-year-old Ethiopian children followed up in an HLA genotyped birth cohort. Front. Pediatr. 10, 999287 (2022).
Gudeta, A. N. et al. Distribution of HLA-DQ risk genotypes for celiac disease in Ethiopian children. HLA 96, 681–687 (2020).
Andrew, S. FastQC: a quality control tool for high throughput sequence data http://www.bioinformatics.babraham.ac.uk/projects/fastqc (2010).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Robeson II, M. S. et al. RESCRIPt: Reproducible sequence taxonomy reference database management. PLOS Computational Biology 17, e1009581 (2021).
Bokulich, N. A. et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6, 90 (2018).
Pruesse, E. et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196 (2007).
Ewels, P., Magnusson, M., Lundin, S. & Käller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357 (2012).
Nurk, S., Meleshko, D., Korobeynikov, A. & Pevzner, P. A. MetaSPAdes: a new versatile metagenomic assembler. Genome Res. 27, 824–834 (2017).
Kang, D. D. et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 2019, e7359 (2019).
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P. & Tyson, G. W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043 (2015).
Blanco-Míguez, A. et al. Extending and improving metagenomic taxonomic profiling with uncharacterized species using MetaPhlAn 4. Nat. Biotechnol. 41, 1633–1644 (2023).
Beghini, F. et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. Elife 10, e65088 (2021).
Asnicar, F. et al. Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat. Commun. 11, 1–10 (2020).
Letunic, I. & Bork, P. Interactive tree of life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 52, W78–W82 (2024).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 11, 1–11 (2010).
Buchfink, B., Reuter, K. & Drost, H. G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 18, 366–368 (2021).
Cantalapiedra, C. P., Hern̗andez-Plaza, A., Letunic, I., Bork, P. & Huerta-Cepas, J. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38, 5825–5829 (2021).
Alcock, B. P. et al. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the comprehensive antibiotic resistance database. Nucleic Acids Res. 51, D690–D699 (2023).
Oksanen, J. et al. Package vegan: Community Ecology Package https://vegandevs.github.io/vegan/, https://github.com/vegandevs/vegan (2009).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis https://ggplot2.tidyverse.org (2016).
Chen, Y., Chen, L., Lun, A. T. L., Baldoni, P. L. & Smyth, G. K. edgeR v4: powerful differential analysis of sequencing data with expanded functionality and improved support for small counts and larger datasets. Nucleic Acids Res. 53, gkaf018 (2025).
Nickols, W. A. et. al. MaAsLin 3: refining and extending generalized multivariable linear models for meta-omic association discovery. Nat. Methods (2026).
Ripley, B. Package ‘MASS’ http://www.stats.ox.ac.uk/pub/MASS4 (2025).
Koneswarakantha, B. easyalluvial: Generate Alluvial Plots with a Single Line of Code https://github.com/erblast/easyalluvial/ (2025).
Kirsche, L. Iydiakirsche/urban-ethiopian-children-microbiome: version 1.0.0. https://doi.org/10.5281/zenodo.18231174 (2026).
Acknowledgements
We are indebted to the participants of this study. We thank Lena Schultheis for help with DNA extraction. This work was supported by the Swiss National Science Foundation (Project grants 320030-236304 and 310030_192490 to A.M.), the Comprehensive Cancer Center Zürich project grant (to A.M., P.L.) the Medical Faculty of the University of Zürich (to A.M.) and the CIFAR Catalyst Fund Project CF-0533 - CP25-090 (to A.M. and M.J.B.). The sponsors had no role in the study design, data analysis or any other part of the research and manuscript submission.
Author information
Authors and Affiliations
Contributions
L.K. performed all metagenomic analyses, generated all figures, and contributed to manuscript writing and revision. P.L. performed QIIME2-based bioinformatics analyses and supported metagenomic data processing. M.J.B. and M.S. provided scientific guidance and critical review. A.N. coordinated patient recruitment and sample collection and contributed to data interpretation. A.M. conceived and supervised the study, secured funding, and led manuscript drafting and finalization. All authors approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Ching Jian, Vanja Klepac-Ceraj, Guilherme Fahur Bottino and the other anonymous reviewer(s) for their contribution to the peer review of this work. Primary handling editors: Yu-Wei Wi, Aylin Bircan, and George Inglis. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kirsche, L., Leary, P., Blaser, M.J. et al. Gut microbial signatures expose the westernized lifestyle of urban Ethiopian children. Commun Biol (2026). https://doi.org/10.1038/s42003-026-09639-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-026-09639-2