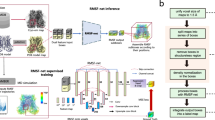
Abstract
Elucidation of both the three-dimensional structure and the dynamics of a protein is essential to understand its function. Technical breakthroughs in single-particle analysis based on cryo-electron microscopy (cryo-EM) have enabled the three-dimensional structures of numerous proteins to be solved at atomic or near-atomic resolution. However, the analysis of the dynamics of protein targets using cryo-EM is often challenging because of their large sizes and complex structural assemblies. Here, we describe DEFMap, a deep learning-based approach to directly extract the dynamics associated with the atomic fluctuations that are hidden in cryo-EM density maps. Using only cryo-EM density data, DEFMap provides dynamics that correlate well with data obtained from molecular dynamics simulations and experimental approaches. Furthermore, DEFMap successfully detects changes in dynamics that are associated with molecular recognition. This strategy combines deep learning, experimental data and molecular dynamics simulations, and may reveal a new multidisciplinary approach for protein science.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All of the datasets used in this study are publicly available in the Protein Data Bank (PDB) at https://www.rcsb.org/ and the Electron Microscopy Data Bank (EMDB) at https://www.ebi.ac.uk/pdbe/emdb/. Detailed descriptions of the datasets are provided in Supplementary Tables 1,2,3,4. The models and the preprocessed input data used in 25-fold cross-validation are available in the Zenodo public repository at https://doi.org/10.5281/zenodo.4317158.
Code availability
The Python code used to implement DEFMap is available on GitHub at https://github.com/clinfo/DEFMap (https://doi.org/10.5281/zenodo.4317158). DEFMap is available under the MIT License.
References
Boehr, D. D., Nussinov, R. & Wright, P. E. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 5, 789–796 (2009).
Kohen, A. Role of dynamics in enzyme catalysis: substantial versus semantic controversies. Acc. Chem. Res. 48, 466–473 (2015).
Cheng, Y. Single-particle cryo-EM—how did it get here and where will it go. Science 361, 876–880 (2018).
Cheng, Y., Grigorieff, N., Penczek, P. A. & Walz, T. A primer to single-particle cryo-electron microscopy. Cell 161, 438–449 (2015).
Murata, K. & Wolf, M. Cryo-electron microscopy for structural analysis of dynamic biological macromolecules. Biochim. Biophys. Acta Gen. Subj. 1862, 324–334 (2018).
Nitta, R., Imasaki, T. & Nitta, E. Recent progress in structural biology: lessons from our research history. Microscopy 67, 187–195 (2018).
Masson, G. R. et al. Recommendations for performing, interpreting and reporting hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments. Nat. Methods 16, 595–602 (2019).
Hollingsworth, S. A. & Dror, R. O. Molecular dynamics simulation for all. Neuron 99, 1129–1143 (2018).
Kühlbrandt, W. Cryo-EM enters a new era. eLife 3, e03678 (2014).
Cheng, Y. Single-particle cryo-EM at crystallographic resolution. Cell 161, 450–457 (2015).
Merk, A. et al. Breaking cryo-EM resolution barriers to facilitate drug discovery. Cell 165, 1698–1707 (2016).
Gremer, L. et al. Fibril structure of amyloid-β(1–42) by cryo-electron microscopy. Science 358, 116–119 (2017).
Kato, T., Makino, F., Miyata, T., Horváth, P. & Namba, K. Structure of the native supercoiled flagellar hook as a universal joint. Nat. Commun. 10, 1–8 (2019).
Kujirai, T. et al. Structural basis of the nucleosome transition during RNA polymerase II passage. Science 362, 595–598 (2018).
Li, X. et al. A unified mechanism for intron and exon definition and back-splicing. Nature 573, 375–380 (2019).
Vilas, J. L. et al. MonoRes: automatic and accurate estimation of local resolution for electron microscopy maps. Structure 26, 337–344.e4 (2018).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Berman, H. M. et al. The protein data bank. Nucleic Acids Res. 28, 235–242 (2000).
Hollingsworth, S. A. et al. Cryptic pocket formation underlies allosteric modulator selectivity at muscarinic GPCRs. Nat. Commun. 10, 3289 (2019).
Plattner, N., Doerr, S., De Fabritiis, G. & Noé, F. Complete protein–protein association kinetics in atomic detail revealed by molecular dynamics simulations and Markov modelling. Nat. Chem. 9, 1005–1011 (2017).
Ji, S., Xu, W., Yang, M. & Yu, K. 3D convolutional neural networks for human action recognition. IEEE Trans. Pattern Anal. Mach. Intell. 35, 221–231 (2013).
Maturana, D. & Scherer, S. VoxNet: a 3D convolutional neural network for real-time object recognition. In Proc. IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS) 922–928 (IEEE, 2015).
Zhirong, W. et al. 3D ShapeNets: a deep representation for volumetric shapes. In Proc. IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 1912–1920 (IEEE, 2015).
Chen, H., Dou, Q., Yu, L., Qin, J. & Heng, P.-A. VoxResNet: deep voxelwise residual networks for brain segmentation from 3D MR images. NeuroImage 170, 446–455 (2018).
Kamnitsas, K. et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 36, 61–78 (2017).
Qi, D. et al. Automatic detection of cerebral microbleeds from MR Images via 3D convolutional neural networks. IEEE Trans. Med. Imaging 35, 1182–1195 (2016).
Avramov, T. K. et al. Deep learning for validating and estimating resolution of cryo-electron microscopy density maps. Molecules 24, 1181 (2019).
Maddhuri Venkata Subramaniya, S. R., Terashi, G. & Kihara, D. Protein secondary structure detection in intermediate-resolution cryo-EM maps using deep learning. Nat. Methods 16, 911–917 (2019).
Mostosi, P., Schindelin, H., Kollmannsberger, P. & Thorn, A. Automated interpretation of cryo-EM density maps with convolutional neural networks. bioRxiv, 644476 (2019).
Xu, K., Wang, Z., Shi, J., Li, H. & Zhang, Q. C. A2-Net: molecular structure estimation from cryo-EM density volumes. In Proc. AAAI Conference on Artificial Intelligence 33, 1230–1237 (AAAI, 2019).
Lawson, C. L. et al. EMDataBank.org: unified data resource for CryoEM. Nucleic Acids Res. 39, D456–D464 (2010).
de la Rosa-Trevín, J. M. et al. Scipion: a software framework toward integration, reproducibility and validation in 3D electron microscopy. J. Struct. Biol. 195, 93–99 (2016).
Goodfellow, I., Bengio, Y. & Courville, A. Deep Learning (MIT Press, 2016).
Zhang, W., Lukoyanova, N., Miah, S., Lucas, J. & Vaughan, C. K. Insights into centromere DNA bending revealed by the Cryo-EM structure of the core centromere binding factor 3 with Ndc10. Cell Rep. 24, 744–754 (2018).
Sun, Y. et al. Molecular basis for the recognition of the human AAUAAA polyadenylation signal. Proc. Natl Acad. Sci. USA 115, E1419–E1428 (2018).
Cash, J. N. et al. Cryo–electron microscopy structure and analysis of the P-Rex1–Gβγ signaling scaffold. Sci. Adv. 5, eaax8855 (2019).
Dedden, D. et al. The architecture of Talin1 reveals an autoinhibition mechanism. Cell 179, 120–131.e13 (2019).
Wongpalee, S. P. et al. CryoEM structures of Arabidopsis DDR complexes involved in RNA-directed DNA methylation. Nat. Commun. 10, 3916 (2019).
Lan, P. et al. Structural insight into precursor tRNA processing by yeast ribonuclease P. Science 362, eaat6678 (2018).
Abascal-Palacios, G., Ramsay, E. P., Beuron, F., Morris, E. & Vannini, A. Structural basis of RNA polymerase III transcription initiation. Nature 553, 301–306 (2018).
Walls, A. C. et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181, 281–292.e6 (2020).
Walls, A. C. et al. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl Acad. Sci. USA 114, 11157–11162 (2017).
Wrapp, D. et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020).
Kostyuchenko, V. A. et al. Structure of the thermally stable Zika virus. Nature 533, 425–428 (2016).
Wu, W. et al. Expression of quasi-equivalence and capsid dimorphism in the Hepadnaviridae. PLoS Comput. Biol. 16, e1007782 (2020).
Cao, L. et al. Structural basis for neutralization of hepatitis A virus informs a rational design of highly potent inhibitors. PLoS Biol. 17, e3000229 (2019).
Liu, Y. et al. Molecular basis for the acid-initiated uncoating of human enterovirus D68. Proc. Natl Acad. Sci. USA 115, E12209–E12217 (2018).
Hamaguchi, T. et al. A new cryo-EM system for single particle analysis. J. Struct. Biol. 207, 40–48 (2019).
Kato, T. et al. CryoTEM with a cold field emission gun that moves structural biology into a new stage. Microsc. Microanal. 25, 998–999 (2019).
Ramírez-Aportela, E. et al. Automatic local resolution-based sharpening of cryo-EM maps. Bioinformatics 36, 765–772 (2019).
Abraham, M. J. et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19–25 (2015).
Lindorff-Larsen, K. et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78, 1950–1958 (2010).
Jorgensen, W. L. & Thomas, L. L. Perspective on free-energy perturbation calculations for chemical equilibria. J. Chem. Theory Comput. 4, 869–876 (2008).
Darden, T., York, D. & Pedersen, L. Particle mesh Ewald: an N⋅Log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993).
Hess, B. P-LINCS: a parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 4, 116–122 (2008).
Bussi, G., Donadio, D. & Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 (2007).
Parrinello, M. & Rahman, A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 (1981).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Doerr, S., Harvey, M. J., Noé, F. & De Fabritiis, G. HTMD: high-throughput molecular dynamics for molecular discovery. J. Chem. Theory Comput. 12, 1845–1852 (2016).
Chollet, F. Keras. https://keras.io (2015).
Schrödinger LLC. The PyMOL Molecular Graphics System (version: 2.2.0) (2018).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Acknowledgements
This work was supported by MEXT as ‘Priority Issue on the Post K computer (Building Innovative Drug Discovery Infrastructure Through Functional Control of Biomolecular Systems)’ and as ‘Program for Promoting Researches on the Supercomputer Fugaku (Application of Molecular Dynamics Simulation to Precision Medicine Using Big Data Integration System for Drug Discovery)’. S.M. was supported by JSPS KAKENHI Grant Number JP17K15106.
Author information
Authors and Affiliations
Contributions
S.M., S.I., K.T. and Y.O. designed the study. S.M. and S.I. performed the preprocessing of cryo-EM maps, designed the neural networks and carried out model training and DEFMap analyses. S.M. and M.A. performed the MD calculations. S.M. and T.K. prepared the original cryo-EM map data. K.T. and Y.O. conceived the project. S.M. and S.I. wrote the manuscript. All of the authors discussed the research, edited the manuscript and approved its final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Machine Intelligence thanks Joseph Davis, Carlos O. S. Sorzano and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Correlation plot of the cryo-EM map local resolution (a) and the given density data (b) with DynamicsMD for EMD-3948 (entry 1 in Supplementary Table 2).
Normalized residue-averaged values are shown in the correlation plot. The regression lines are coloured orange; r denotes the correlation coefficient. The average correlation coefficient for the density data of 25 cryo-EM maps used in training is 0.459 ± 0.179 (Supplementary Table 2).
Extended Data Fig. 2 Correlation plots between MD-derived and DEFMap-determined dynamics for proteins not included in the training dataset.
a,b, Normalized atomic (a, blue) and residue-averaged (b, purple) values are shown in the correlation plots; the regression lines are coloured orange; r denotes the correlation coefficient.
Extended Data Fig. 3 The map resolution dependence of the dynamics extraction accuracy for test macromolecules (Supplementary Tables 3 and 4).
a,b, Correlation coefficient values between the MD- and DEFMap-derived atomic-specific (a) and residue-specific (b) dynamics at different resolutions. The cryo-EM maps of the training and the evaluation datasets were preprocessed to identical resolutions with low-pass filters.
Extended Data Fig. 4 DEFMap performance for a map with the intermediate resolution (EMDB/PDB entry, EMD-4772/6R9T).
a, Distribution of local resolution. The local resolution is calculated using the MonoRes implementation in Scipion and mapped onto the original map using different colours as indicated in the colour bars. b,c,d, The correlation plots between DynamicsDEFMap and DynamicsMD for the intermediate-resolution map. The target cryo-EM maps are processed by low-pass filters with cutoff values of 7 Å (b), 5 Å (c), and 6 Å (d). The predictions are carried out using the models trained by the datasets preprocessed to the identical resolutions. The regression lines are coloured orange; r denotes the correlation coefficient.
Extended Data Fig. 5 The spatial distribution of the fragment-specific dynamics in the atomic models (EMDB/PDB entry, EMD-20308/6PCV).
The representative fragments-specific dynamics for Rac exchanger 1 complexed with G protein beta gamma subunits are mapped onto the atomic model using different colours as indicated in the colour bar (HDX-MS, left; MD, middle; DEFMap, right). The fragments not detected in the HDX-MS experiments are coloured grey.
Extended Data Fig. 6 Residue-specific dynamics profiles for apo/holo proteins.
a,b, DynamicsMD (black) and DynamicsDEFMap (magenta) datasets for 6OIS/EMD-20080 and 6OIT/EMD-20081 (a), 6AGB/EMD-9616 and 6AH3/EMD-9622 (b), and 6EU2/EMD-3957 and 6EU1/EMD-3956 (c), respectively, plotted against the residue IDs, numbered according to their order in the coordinate files. r denotes the correlation coefficient between DynamicsMD and DynamicsDEFMap.
Extended Data Fig. 7 Atomic models of residues showing allosteric dynamics changes induced by ligand binding in the DMS3-RDM1 complex with DRD1 peptide.
a,b, The main chains of the hinge domain in DMS3 dimer 1 (a) and in the RDM1-DMS3 dimer 2 binding interface (b) are coloured according to the extent of ligand-induced changes in dynamics using the indicated colour bar. The side chains of residues showing the dynamics changes of less than -0.5 are represented as sticks (apo, magenta; holo, cyan).
Extended Data Fig. 8 Conformational changes of the spike protein of SARS-CoV-2.
a, Overall architecture of the spike protein of SARS-CoV-2. The spike protein mediates the entry of the virus into host cells. The protein is composed of two subunits: S1 subunit is involved in the recognition of the host cell receptor and S2 subunit is involved in the viral and membrane fusion process. The receptor binding domain in the S1 subunit exhibits a hinge-like motion and the ‘down’ and ‘up’ conformations are shown in grey and magenta, respectively. The conformational transition from the ‘down’ to ‘up’ states is indicated by a red arrow in the right-hand panel. The structural components associated with the structural transition from prefusion to post-fusion forms, the central helix, β-rich region, and the heptad-repeat 1 (HR1) motif, are indicated. b, Conformational rearrangement of the HR1 motif in the state transition from the prefusion to post-fusion forms. The post-fusion form of the spike protein in SARS-CoV-2 is shown in green by superimposing with the prefusion form. The conformational rearrangement of the HR1 motif is indicated by a yellow arrow. The prefusion form is coloured according to the indicated colour bar.
Extended Data Fig. 9 A correlation plot between DynamicsDEFMap calculated using EMD21375 and EMD-21375.
The regression lines are coloured orange; r denotes the correlation coefficient.
Extended Data Fig. 10 DEFMap-based dynamics visualization of virus particles.
The DEFMap-derived dynamics data are mapped onto the cryo-EM maps of the viral particles by applying the icosahedral symmetry. The colour range is defined by the minimum and maximum values in each inference. The scale bar represents 50 Å and is indicated by black lines. EMDB/PDB entries are indicated. The atomic model corresponding to EMD-9053 (right) is not available.
Supplementary information
Supplementary Information
Supplementary Figs. 1–3 and Tables 1–5.
Rights and permissions
About this article
Cite this article
Matsumoto, S., Ishida, S., Araki, M. et al. Extraction of protein dynamics information from cryo-EM maps using deep learning. Nat Mach Intell 3, 153–160 (2021). https://doi.org/10.1038/s42256-020-00290-y
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s42256-020-00290-y
This article is cited by
-
Visualization of RIM-BP2’s crane-like function in neuronal vesicle transport using FRET
Communications Biology (2025)
-
Super-resolution microscopy for structural biology
Nature Methods (2025)
-
Non-averaged single-molecule tertiary structures reveal RNA self-folding through individual-particle cryo-electron tomography
Nature Communications (2024)
-
3D variability analysis reveals a hidden conformational change controlling ammonia transport in human asparagine synthetase
Nature Communications (2024)
-
Accurate Prediction of Protein Structural Flexibility by Deep Learning Integrating Intricate Atomic Structures and Cryo-EM Density Information
Nature Communications (2024)