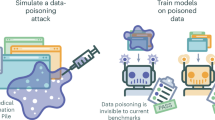
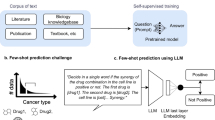
Abstract
Biomedical knowledge graphs (KGs) constructed from medical literature have been widely used to validate biomedical discoveries and generate new hypotheses. Recently, large language models (LLMs) have demonstrated a strong ability to generate human-like text data. Although most of these text data have been useful, LLM might also be used to generate malicious content. Here, we investigate whether it is possible that a malicious actor can use an LLM to generate a malicious paper that poisons medical KGs and further affects downstream biomedical applications. As a proof of concept, we develop Scorpius, a conditional text-generation model that generates a malicious paper abstract conditioned on a promoted drug and a target disease. The goal is to fool the medical KG constructed from a mixture of this malicious abstract and millions of real papers so that KG consumers will misidentify this promoted drug as relevant to the target disease. We evaluated Scorpius on a KG constructed from 3,818,528 papers and found that Scorpius can increase the relevance of 71.3% drug–disease pairs from the top 1,000 to the top ten by adding only one malicious abstract. Moreover, the generation of Scorpius achieves better perplexity than ChatGPT, suggesting that such malicious abstracts cannot be efficiently detected by humans. Collectively, Scorpius demonstrates the possibility of poisoning medical KGs and manipulating downstream applications using LLMs, indicating the importance of accountable and trustworthy medical knowledge discovery in the era of LLMs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The Medline KG is available at https://zenodo.org/records/1035500 (ref. 70). The PubTator database is available at https://ftp.ncbi.nlm.nih.gov/pub/lu/PubTatorCentral/. The Unified Medical Language System database we used to identify ‘Pharmacologic Substance’ and ‘Clinical Drug’ is available at https://documentation.uts.nlm.nih.gov/rest/home.html. The bioRxiv database is available at https://api.biorxiv.org/. The PrimeKG database we used for enhancing Medline KG is available at https://github.com/mims-harvard/PrimeKG. All processed data can be directly downloaded from our GitHub project: https://github.com/yjwtheonly/Scorpius.
Code availability
Scorpius code is available at https://github.com/yjwtheonly/Scorpius ref. 71. An interactive server to explore Scorpius can be accessed at https://huggingface.co/spaces/yjwtheonly/Scorpius_HF.
References
Roberts, R. J. PubMed Central: the GenBank of the published literature. Proc. Natl Acad. Sci. USA 98, 381–382 (2001).
Canese, K. & Weis, S. in The NCBI Handbook 2nd edn (eds Beck, J. et al.) Ch. 3 (NCBI, 2013).
Percha, B. & Altman, R. B. A global network of biomedical relationships derived from text. Bioinformatics 34, 2614–2624 (2018).
Rossanez, A., Dos Reis, J. C., Torres, R., da, S. & de Ribaupierre, H. KGen: a knowledge graph generator from biomedical scientific literature. BMC Med. Inform. Decis. Mak. 20, 314 (2020).
Asada, M., Miwa, M. & Sasaki, Y. Using drug descriptions and molecular structures for drug–drug interaction extraction from literature. Bioinformatics 37, 1739–1746 (2021).
Turki, H., Hadj Taieb, M. A. & Ben Aouicha, M. MeSH qualifiers, publication types and relation occurrence frequency are also useful for a better sentence-level extraction of biomedical relations. J. Biomed. Inform. 83, 217–218 (2018).
Zeng, X., Tu, X., Liu, Y., Fu, X. & Su, Y. Toward better drug discovery with knowledge graph. Curr. Opin. Struct. Biol. 72, 114–126 (2022).
Mohamed, S. K., Nounu, A. & Nováček, V. Biological applications of knowledge graph embedding models. Brief. Bioinform. 22, 1679–1693 (2021).
MacLean, F. Knowledge graphs and their applications in drug discovery. Expert Opin. Drug Discov. 16, 1057–1069 (2021).
Wang, S., Lin, M., Ghosal, T., Ding, Y. & Peng, Y. Knowledge graph applications in medical imaging analysis: a scoping review. Health Data Sci. 2022, 9841548 (2022).
Ouyang, L. et al. Training language models to follow instructions with human feedback. In Advances in Neural Information Processing Systems (eds Koyejo, S. et al.) 27730–27744 (2022).
Brown, T. et al. Language models are few-shot learners. Adv. Neural Inf. Process. Syst. 33, 1877–1901 (2020).
Raffel, C. et al. Exploring the limits of transfer learning with a unified text-to-text transformer. J. Mach. Learn. Res. 21, 5485–5551 (2020).
Lewis, M. et al. BART: denoising sequence-to-sequence pre-training for natural language generation, translation, and comprehension. In Proc. 58th Annual Meeting of the Association for Computational Linguistics (eds Jurafsky, D. et al.) 7871–7880 (ACL, 2020).
OpenAI et al. GPT-4 technical report. Preprint at https://arxiv.org/abs/2303.08774 (2023).
Thoppilan, R. et al. LaMDA: language models for dialog applications. Preprint at https://arxiv.org/abs/2201.08239 (2022).
Surameery, N. M. S. & Shakor, M. Y. Use Chat GPT to solve programming bugs. Int. J. Inform. Technol. Comput. Eng. 3, 17–22 (2023).
Biswas, S. S. Potential use of Chat GPT in global warming. Ann. Biomed. Eng. 51, 1126–1127 (2023).
Biswas, S. S. Role of Chat GPT in public health. Ann. Biomed. Eng. 51, 868–869 (2023).
Sallam, M. ChatGPT utility in healthcare education, research, and practice: systematic review on the promising perspectives and valid concerns. Healthcare (Basel) 11, 887 (2023).
Park, J. S. et al. Generative agents: interactive simulacra of human behavior. In Proc. 36th Annual ACM Symposium on User Interface Software and Technology (eds Follmer, S. et al.) 2:1–2:22 (ACM, 2023).
Huang, Z., Bianchi, F., Yuksekgonul, M., Montine, T. J. & Zou, J. A visual-language foundation model for pathology image analysis using medical Twitter. Nat. Med. 29, 2307–2316 (2023).
Sheldon, T. Preprints could promote confusion and distortion. Nature 559, 445 (2018).
Methods, preprints and papers. Nat. Biotechnol. 35, 1113 (2017).
Preprints in biology. Nat. Methods 13, 277 (2016).
Watson, C. Rise of the preprint: how rapid data sharing during COVID-19 has changed science forever. Nat. Med. 28, 2–5 (2022).
Wang, L. L. et al. CORD-19: the COVID-19 open research dataset. Preprint at https://www.arxiv.org/abs/2004.10706v4 (2020).
Ahamed, S. & Samad, M. Information mining for COVID-19 research from a large volume of scientific literature. Preprint at https://arxiv.org/abs/2004.02085 (2020).
Pestryakova, S. et al. CovidPubGraph: a FAIR knowledge graph of COVID-19 publications. Sci. Data 9, 1–11 (2022).
Zhang, R. et al. Drug repurposing for COVID-19 via knowledge graph completion. J. Biomed. Inform. 115, 103696 (2021).
Michel, F. et al. Covid-on-the-Web: knowledge graph and services to advance COVID-19 research. In Proc. 19th International Semantic Web Conference (eds Pan, J. Z. et al.) 294–310 (Springer, 2020).
Gehrmann, S., Strobelt, H. & Rush, A. M. GLTR: statistical detection and visualization of generated text. In Proc. 57th Conference of the Association for Computational Linguistics (eds Korhonen, A. et al.) 111–116 (ACL, 2019).
Jawahar, G., Abdul-Mageed, M. & Lakshmanan, L. V. S. Automatic detection of machine generated text: a critical survey. In Proc. 28th International Conference on Computational Linguistics (eds Scott, D. et al.) 2296–2309 (ICCL, 2020).
Wang, W. & Feng, A. Self-information loss compensation learning for machine-generated text detection. Math. Probl. Eng. 2021, 6669468 (2021).
Mitchell, E., Lee, Y., Khazatsky, A., Manning, C. D. & Finn, C. DetectGPT: zero-shot machine-generated text detection using probability curvature. In Proc. International Conference on Machine Learning (eds Krause, A. et al.) 24950–24962 (PMLR, 2023).
Meyer zu Eissen, S. & Stein, B. Intrinsic plagiarism detection. In Proc. Advances in Information Retrieval (eds Lalmas, M. et al.) 565–569 (Springer Berlin Heidelberg, 2006).
Lukashenko, R., Graudina, V. & Grundspenkis, J. Computer-based plagiarism detection methods and tools: an overview. In Proc. International Conference on Computer Systems and Technologies https://doi.org/10.1145/1330598.13306 (ACM, 2007).
Meyer zu Eissen, S., Stein, B. & Kulig, M. Plagiarism detection without reference collections. In Advances in Data Analysis (eds Decker, R. & Lenz, H. J.) 359–366 (Springer Berlin Heidelberg, 2007).
Donaldson, J. L., Lancaster, A.-M. & Sposato, P. H. A plagiarism detection system. In Proc. 12th SIGCSE Technical Symposium on Computer Science Education https://doi.org/10.1145/953049.800955 (ACM, 1981).
Yang, B., Yih, W.-T., He, X., Gao, J. & Deng, L. Embedding entities and relations for learning and inference in knowledge bases. In 3rd International Conference on Learning Representations (ICLR, 2015).
Dettmers, T., Minervini, P., Stenetorp, P. & Riedel, S. Convolutional 2D knowledge graph embeddings. In Proc. AAAI Conference on Artificial Intelligence 1811–1818 (AAAI, 2018).
Trouillon, T., Welbl, J., Riedel, S., Gaussier, E. & Bouchard, G. Complex embeddings for simple link prediction. In Proc. 33rd International Conference on Machine Learning (eds. Balcan, M. F. & Weinberger, K. Q.) 2071–2080 (PMLR, 2016).
Lu, Y. et al. Unified structure generation for universal information extraction. In Proc. 60th Annual Meeting of the Association for Computational Linguistics (eds Muresan, S. et al.) 5755–5772 (ACL, 2022).
Li, X. et al. TDEER: an efficient translating decoding schema for joint extraction of entities and relations. In Proc. Conference on Empirical Methods in Natural Language Processing (eds Moens, M.-F. et al.) 8055–8064 (ACL, 2021).
Yamada, I., Asai, A., Shindo, H., Takeda, H. & Matsumoto, Y. LUKE: deep contextualized entity representations with entity-aware self-attention. In Proc. 2020 Conference on Empirical Methods in Natural Language Processing (eds Webber, B. et al.) 6442–6454 (ACL, 2020).
Bhardwaj, P., Kelleher, J., Costabello, L. & O’Sullivan, D. Poisoning knowledge graph embeddings via relation inference patterns. In Proc. 59th Annual Meeting of the Association for Computational Linguistics (eds Zong, C. et al.) 1875–1888 (ACL 2021).
Pezeshkpour, P., Tian, Y. & Singh, S. Investigating robustness and interpretability of link prediction via adversarial modifications. In Proc. 2019 Conference of the North American Chapter of the Association for Computational Linguistics (eds Burstein, J. et al.) 3336–3347 (ACL, 2019).
Bhardwaj, P., Kelleher, J., Costabello, L. & O’Sullivan, D. Adversarial attacks on knowledge graph embeddings via instance attribution methods. In Proc. 2021 Conference on Empirical Methods in Natural Language Processing (eds Moens, M.-F. et al.) 8225–8239 (ACL, 2021).
Li, Q., Wang, Z. & Li, Z. PAGCL: an unsupervised graph poisoned attack for graph contrastive learning model. Future Gener. Comput. Syst. 149, 240–249 (2023).
You, X. et al. MaSS: model-agnostic, semantic and stealthy data poisoning attack on knowledge graph embedding. In Proc. ACM Web Conference (eds Ding, Y et al.) 2000–2010 (ACM, 2023).
Betz, P., Meilicke, C. & Stuckenschmidt, H. Adversarial explanations for knowledge graph embeddings. In Proc. 31st International Joint Conference on Artificial Intelligence (ed. De Raedt, L.) 2820–2826 (IJCAIO, 2022).
Zhang, H. et al. Data poisoning attack against knowledge graph embedding. In Proc. 28th International Joint Conference on Artificial Intelligence (eds Kraus, S.) 4853–4859 (IJCAI, 2019).
Gao, Z., Ding, P. & Xu, R. K. G.- Predict: a knowledge graph computational framework for drug repurposing. J. Biomed. Inform. 132, 104133 (2022).
Vashishth, S., Sanyal, S., Nitin, V., Agrawal, N. & Talukdar, P. InteractE: improving convolution-based knowledge graph embeddings by increasing feature interactions. In Proc. AAAI Conference on Artificial Intelligence 3009–3016 (AAAI, 2020).
Han, S. et al. Standigm ASKTM: knowledge graph and artificial intelligence platform applied to target discovery in idiopathic pulmonary fibrosis. Brief. Bioinform. 25, bbae035 (2024).
Zheng, S. et al. PharmKG: a dedicated knowledge graph benchmark for bomedical data mining. Brief. Bioinform. 22, bbaa344 (2021).
Wei, C.-H., Kao, H.-Y. & Lu, Z. PubTator: a web-based text mining tool for assisting biocuration. Nucleic Acids Res. 41, W518–W522 (2013).
Greenhalgh, T. How to read a paper. The Medline database. Brit. Med. J. 315, 180–183 (1997).
de Marneffe, M.-C. & Manning, C. D. The Stanford typed dependencies representation. In Proc. Workshop on Cross-Framework and Cross-Domain Parser Evaluation (eds Bos, J. et al.) 1–8 (ACL, 2008); https://doi.org/10.3115/1608858.1608859
Page, L., Brin, S., Motwani, R. & Winograd, T. The PageRank citation ranking: bringing order to the web. http://ilpubs.stanford.edu:8090/422/ (Stanford InfoLab, 1999).
Bodenreider, O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 32, D267–D270 (2004).
Koh, P. W. & Liang, P. Understanding black-box predictions via influence functions. In Proc. 34th International Conference on Machine Learning (eds Precup, D. & Teh, Y. W.) 1885–1894 (PMLR, 2017).
Bianchini, M., Gori, M. & Scarselli, F. Inside PageRank. ACM Trans. Internet Technol. 5, 92–128 (2005).
Luo, R. et al. BioGPT: generative pre-trained transformer for biomedical text generation and mining. Brief. Bioinform. 23, bbac409 (2022).
Yuan, H. et al. BioBART: Pretraining and evaluation of a biomedical generative language model. In Proc. 21st Workshop on Biomedical Language Processing (eds Demner-Fushman, D. et al.) 97–109 (ACL, 2022).
Liu, Y. et al. G-Eval: NLG Evaluation using GPT-4 with better human alignment. In Proc. 2023 Conference on Empirical Methods in Natural Language Processing (eds Bouamor, H. et al.) 2511–2522 (ACL, 2023).
Chen, J., Lin, H., Han, X. & Sun, L. Benchmarking large language models in retrieval-augmented generation. In Proc. AAAI Conference on Artificial Intelligence (eds Woodridge, M. et al.) 17754–17762 (2024).
Ranjit, M., et al. Retrieval augmented chest X-ray report generation using OpenAI GPT models. In Proc. 8th Machine Learning for Healthcare Conference (eds Deshpande, K. et al.) 650–666 (PMLR, 2023).
Chandak, P., Huang, K. & Zitnik, M. Building a knowledge graph to enable precision medicine. Sci. Data 10, 67 (2023).
Percha, B. & Altman, R. A global network of biomedical relationships derived from text. Zenodo zenodo.org/records/1035500 (2017).
Junwei, Y. et al. Poisoning medical knowledge using large language models v.1.0.1. Zenodo https://doi.org/10.5281/zenodo.13191322 (2024).
Lee, J. et al. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 36, 1234–1240 (2020).
Acknowledgements
We would like to extend our thanks to B. Feng from the International Digital Economy Academy for his assistance in revising our manuscript. We are also grateful to Z. Wu and K. Zheng from Peking University for their insightful discussion on technical implementation. J.Y., S.M., Zequn Liu, W.J., L.L. and M.Z. are partially supported by the National Key Research and Development Programme of China with grant no. 2023YFC3341203 as well as the National Natural Science Foundation of China with grant nos. 62276002 and 62306014.
Author information
Authors and Affiliations
Contributions
J.Y., T.C., M.Z. and S.W. contributed to the conception and design of the whole work. J.Y., S.M., Zequn Liu, Z.X. and S.W. contributed to the data curation. J.Y., S.M., Z.L. and Z.X. contributed to the technical implementation. J.Y., H.X., Z.X., M.Z. and S.W. contributed to the writing. All authors contributed to the drafting and revision of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks Ping Zhang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Experimental details and Figs. 1–19.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, J., Xu, H., Mirzoyan, S. et al. Poisoning medical knowledge using large language models. Nat Mach Intell 6, 1156–1168 (2024). https://doi.org/10.1038/s42256-024-00899-3
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s42256-024-00899-3
This article is cited by
-
Integration of large vision language models for efficient post-disaster damage assessment and reporting
Nature Communications (2026)
-
Hallmarks of artificial intelligence contributions to precision oncology
Nature Cancer (2025)
-
Robustness tests for biomedical foundation models should tailor to specifications
npj Digital Medicine (2025)
-
Risks of AI scientists: prioritizing safeguarding over autonomy
Nature Communications (2025)
-
A vision–language pretrained transformer for versatile clinical respiratory disease applications
Nature Biomedical Engineering (2025)