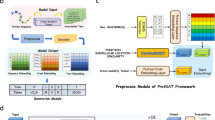
Abstract
Current AI-assisted protein design utilizes mainly protein sequential and structural information. Meanwhile, there exists tremendous knowledge curated by humans in text format describing proteins’ high-level functionalities, yet whether the incorporation of such text data can help in protein design tasks has not been explored. To bridge this gap, we propose ProteinDT, a multimodal framework that leverages textual descriptions for protein design. ProteinDT consists of three consecutive steps: ProteinCLAP, which aligns the representation of two modalities, a facilitator that generates the protein representation from the text modality and a decoder that creates the protein sequences from the representation. To train ProteinDT, we construct a large dataset, SwissProtCLAP, with 441,000 text and protein pairs. We quantitatively verify the effectiveness of ProteinDT on three challenging tasks: (1) over 90% accuracy for text-guided protein generation; (2) best hit ratio on 12 zero-shot text-guided protein editing tasks; (3) superior performance on four out of six protein property prediction benchmarks.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The dataset is available via GitHub at https://github.com/chao1224/ProteinDT. The preprocessed pretraining dataset (SwissProtCLAP) is available via HuggingFace at https://huggingface.co/datasets/chao1224/ProteinDT.
Code availability
The source code and dataset generation scripts are available via GitHub at https://github.com/chao1224/ProteinDT and via Zenodo at https://doi.org/10.5281/zenodo.14630813 (ref. 88).
References
Freschlin, C. R., Fahlberg, S. A. & Romero, P. A. Machine learning to navigate fitness landscapes for protein engineering. Curr. Opin. Biotechnol. 75, 102713 (2022).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Zhong, E. D., Lerer, A., Davis, J. H. & Berger, B. CryoDRGN2: ab initio neural reconstruction of 3D protein structures from real cryo-EM images. In 2021 IEEE/CVF International Conference on Computer Vision (ICCV), 4046–4055 (IEEE, 2021).
Hsu, C. et al. Learning inverse folding from millions of predicted structures. Proc. Mach. Learning Res. 162, 8946–8970 (2022).
Rao, R. M. et al. MSA Transformer. Proc. Mach. Learning Res. 139, 8844–8856 (2021).
Elnaggar, A. et al. ProtTrans: toward understanding the language of life through self-supervised learning. IEEE Trans. Pattern Anal. Mach. Intell. 44, 7112–7127 (2022).
Meier, J. et al. Language models enable zero-shot prediction of the effects of mutations on protein function. Adv. Neural Inf. Process. Syst. 34, 29287–29303 (2021).
Li, M. et al. SESNet: sequence–structure feature-integrated deep learning method for data-efficient protein engineering. J. Cheminformatics 15, 12 (2023).
Jing, B., Eismann, S., Suriana, P., Townshend, R. J. L. & Dror, R. Learning from protein structure with geometric vector perceptrons. In International Conference on Learning Representations (2021).
Wang, L., Liu, H., Liu, Y., Kurtin, J. & Ji, S. Learning protein representations via complete 3D graph networks. In The Eleventh International Conference on Learning Representations (2023).
Radford, A. et al. Learning transferable visual models from natural language supervision. Proc. Mach. Learning Res. 139, 8748–8763 (2021).
Nichol, A. Q. et al. GLIDE: towards photorealistic image generation and editing with text-guided diffusion models. Proc. Mach. Learning Res. 162, 16784–16804 (2022).
Ramesh, A., Dhariwal, P., Nichol, A., Chu, C. & Chen, M. Hierarchical text-conditional image generation with clip latents. Preprint at https://doi.org/10.48550/arXiv.2204.06125 (2022).
Patashnik, O., Wu, Z., Shechtman, E., Cohen-Or, D. & Lischinski, D. StyleCLIP: text-driven manipulation of StyleGAN imagery. In Proc. IEEE/CVF International Conference on Computer Vision (ICCV) 2065–2074 (IEEE, 2021).
Liu, S., Qu, M., Zhang, Z., Cai, H. & Tang, J. Structured multi-task learning for molecular property prediction. Proc. Mach. Learning Res. 151, 8906–8920 (2022).
Edwards, C., Zhai, C. & Ji, H. Text2mol: cross-modal molecule retrieval with natural language queries. In Proc. 2021 Conference on Empirical Methods in Natural Language Processing (eds Moens, M.-F. et al.) 595–607 (Association for Computational Linguistics, 2021).
Zeng, Z., Yao, Y., Liu, Z. & Sun, M. A deep-learning system bridging molecule structure and biomedical text with comprehension comparable to human professionals. Nat. Commun. 13, 862 (2022).
Liu, S. et al. Multi-modal molecule structure–text model for text-based retrieval and editing. Nat. Mach. Intell. 5, 1447–1457 (2023).
Liu, S. et al. Conversational drug editing using retrieval and domain feedback. In The Twelfth International Conference on Learning Representations (2024).
The UniProt Consortium The Universal Protein Resource (UniProt). Nucleic Acids Res. 36, D190–D195 (2007).
Ashburner, M. et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
UniProt. Uniprotkg/swiss-prot (2023); https://www.uniprot.org
Boutet, E., Lieberherr, D., Tognolli, M., Schneider, M. & Bairoch, A. in Plant Bioinformatics (ed. Edwards, D.) 89–112 (Springer, 2007).
Branden, C. I. & Tooze, J. Introduction to Protein Structure (Garland, 2012).
Devlin, J., Chang, M.-W., Lee, K. & Toutanova, K. BERT: pre-training of deep bidirectional transformers for language understanding. In Proc. 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers) (eds Burstein, J. et al.) 4171–4186 (Association for Computational Linguistics, 2019).
Vaswani, A. et al. Attention is all you need. Adv. Neural Inf. Process. Syst. https://doi.org/10.48550/arXiv.1706.03762 (2017).
Steinegger, M., Mirdita, M. & Söding, J. Protein-level assembly increases protein sequence recovery from metagenomic samples manyfold. Nat. Methods 16, 603–606 (2019).
Steinegger, M. & Söding, J. Clustering huge protein sequence sets in linear time. Nat. Commun. 9, 2542 (2018).
Beltagy, I., Lo, K. & Cohan, A. SciBERT: a pretrained language model for scientific text. In Proc. 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP) (eds Inui, K. et al.) 3615–3620 (Association for Computational Linguistics, 2019).
Fricke, S. Semantic Scholar. J. Med. Libr. Assoc. 106, 145–147 (2018).
Taylor, R. et al. Galactica: a large language model for science. Preprint at https://arxiv.org/abs/2211.09085 (2022).
Li, Y., Xu, H., Zhao, H., Guo, H. & Liu, S. ChatPathway: conversational large language models for biology pathway detection. In NeurIPS 2023 AI for Science Workshop (2023).
Savage, N. Drug discovery companies are customizing ChatGPT: here’s how. Nat. Biotechnol. 41, 585–586 (2023).
Gao, Z. et al. Empowering diffusion models on the embedding space for text generation. In Proc. 2024 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies (Volume 1: Long Papers) (eds Duh, K. et al.) 4664–4683 (Association for Computational Linguistics, 2024).
Lin, Z. et al. Text generation with diffusion language models: a pre-training approach with continuous paragraph denoise. Proc. Mach. Learning Res. 202, 21051–21064 (2023).
Bar-Tal, O. et al. Lumiere: a space–time diffusion model for video generation. In SIGGRAPH Asia 2024 Conference Papers 1–11 (Association for Computing Machinery, 2024).
Rombach, R., Blattmann, A., Lorenz, D., Esser, P. & Ommer, B. High-resolution image synthesis with latent diffusion models. In Proc. IEEE/CVF Conference on Computer Vision and Pattern Recognition 10684–10695 (IEEE Computer Society, 2022).
Kabsch, W. & Sander, C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983).
Binder, J. L. et al. AlphaFold illuminates half of the dark human proteins. Curr. Opin. Struct. Biol. 74, 102372 (2022).
Rocklin, G. J. et al. Global analysis of protein folding using massively parallel design, synthesis, and testing. Science 357, 168–175 (2017).
Rohl, C. A., Strauss, C. E., Misura, K. M. & Baker, D. in Methods in Enzymology Vol. 383 (eds Brand, L. & Johnson, M. L.) 66–93 (Elsevier, 2004).
Chaudhury, S., Lyskov, S. & Gray, J. J. PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics 26, 689–691 (2010).
Park, H. et al. Simultaneous optimization of biomolecular energy functions on features from small molecules and macromolecules. J. Chem. Theory Comput. 12, 6201–6212 (2016).
Berman, H. M. et al. The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000).
Liu, S. et al. A multi-grained symmetric differential equation model for learning protein–ligand binding dynamics. Preprint at https://arxiv.org/abs/2401.15122 (2024).
McNutt, A. T. et al. gnina 1.0: molecular docking with deep learning. J. Cheminformatics 13, 43 (2021).
Salsi, E. et al. Design of O-acetylserine sulfhydrylase inhibitors by mimicking nature. J. Med. Chem. 53, 345–356 (2010).
Rao, R. et al. Evaluating protein transfer learning with TAPE. Adv. Neural Inf. Process. Syst. 32 (2019).
Klausen, M. S. et al. NetSurfP-2.0: improved prediction of protein structural features by integrated deep learning. Proteins 87, 520–527 (2019).
Hou, J., Adhikari, B. & Cheng, J. DeepSF: deep convolutional neural network for mapping protein sequences to folds. Bioinformatics 34, 1295–1303 (2018).
Fox, N. K., Brenner, S. E. & Chandonia, J.-M. SCOPe: Structural Classification of Proteins—extended, integrating SCOP and ASTRAL data and classification of new structures. Nucleic Acids Res. 42, D304–D309 (2013).
AlQuraishi, M. ProteinNet: a standardized data set for machine learning of protein structure. BMC Bioinform. 20, 311 (2019).
Moult, J., Fidelis, K., Kryshtafovych, A., Schwede, T. & Tramontano, A. Critical assessment of methods of protein structure prediction (CASP)—Round XII. Proteins 86, 7–15 (2018).
Sarkisyan, K. S. et al. Local fitness landscape of the green fluorescent protein. Nature 533, 397–401 (2016).
Hochreiter, S. & Schmidhuber, J. Long short-term memory. Neural Comput. 9, 1735–1780 (1997).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 770–778 (IEEE, 2016).
Zhang, N. et al. OntoProtein: protein pretraining with Gene Ontology embedding. In International Conference on Learning Representations (2022).
Ingraham, J. et al. Illuminating protein space with a programmable generative model. Nature 623, 1070–1078 (2023).
Wei, C.-H., Allot, A., Leaman, R. & Lu, Z. PubTator Central: automated concept annotation for biomedical full text articles. Nucleic Acids Res. 47, W587–W593 (2019).
Angermueller, C. et al. Model-based reinforcement learning for biological sequence design. In International Conference on Learning Representations (2020).
Gelman, S., Fahlberg, S. A., Heinzelman, P., Romero, P. A. & Gitter, A. Neural networks to learn protein sequence–function relationships from deep mutational scanning data. Proc. Natl Acad. Sci. USA 118, e2104878118 (2021).
Luo, Y. et al. ECNet is an evolutionary context-integrated deep learning framework for protein engineering. Nat. Commun. 12, 5743 (2021).
Biswas, S., Khimulya, G., Alley, E. C., Esvelt, K. M. & Church, G. M. Low-N protein engineering with data-efficient deep learning. Nat. Methods 18, 389–396 (2021).
Notin, P. et al. Tranception: protein fitness prediction with autoregressive transformers and inference-time retrieval. Proc. Mach. Learning Res. 162, 16990–17017 (2022).
Radford, A. et al. Language models are unsupervised multitask learners. OpenAI Blog 1, 9 (2019).
Lewis, M. et al. BART: denoising sequence-to-sequence pre-training for natural language generation, translation, and comprehension. In Proc. 58th Annual Meeting of the Association for Computational Linguistics (eds Jurafsky, D. et al.) 7871–7880 (Association for Computational Linguistics, 2020).
Raffel, C. et al. Exploring the limits of transfer learning with a unified text-to-text transformer. J. Mach. Learning Res. 21, 1–67 (2020).
Ho, J., Jain, A. & Abbeel, P. Denoising diffusion probabilistic models. Adv. Neural Inf. Process. Syst. 33, 6840–6851 (2020).
Vincent, P. A connection between score matching and denoising autoencoders. Neural Comput. 23, 1661–1674 (2011).
Song, Y. & Ermon, S. Generative modeling by estimating gradients of the data distribution. Adv. Neural Inf. Process. Syst. 32 (2019).
Song, Y. et al. Score-based generative modeling through stochastic differential equations. In International Conference on Learning Representations (2021).
Hjelm, R. D. et al. Learning deep representations by mutual information estimation and maximization. In International Conference on Learning Representations (2019).
Bachman, P., Hjelm, R. D. & Buchwalter, W. Learning representations by maximizing mutual information across views. Adv. Neural Inf. Process. Syst. 32 (2019).
Oord, A. v. d., Li, Y. & Vinyals, O. Representation learning with contrastive predictive coding. Preprint at https://arxiv.org/abs/1807.03748 (2018).
He, K., Fan, H., Wu, Y., Xie, S. & Girshick, R. Momentum contrast for unsupervised visual representation learning. In Proc. IEEE/CVF Conference on Computer Vision and Pattern Recognition 9729–9738 (IEEE, 2020).
Liu, S. et al. Pre-training molecular graph representation with 3D geometry. In International Conference on Learning Representations (2022).
LeCun, Y., Chopra, S., Hadsell, R., Ranzato, M. & Huang, F. in Predicting Structured Data Vol. 1 (eds Bakir, G. et al.) (MIT Press, 2006).
Khosla, P. et al. Supervised contrastive learning. Adv. Neural Inf. Process. Syst. 33, 18661–18673 (2020).
Liu, S., Guo, H. & Tang, J. Molecular geometry pretraining with SE(3)-invariant denoising distance matching. In International Conference on Learning Representations (2023).
Huang, W., Hayashi, T., Wu, Y., Kameoka, H. & Toda, T. Voice transformer network: sequence-to-sequence voice conversion using Transformer with text-to-speech pretraining. In Interspeech 2020, 21st Annual Conference of the International Speech Communication Association, Virtual Event, Shanghai, China, 25–29 October 2020 (eds Meng, H. et al.) 4676–4680 (ISCA, 2020).
Karita, S. et al. A comparative study on Transformer vs RNN in speech applications. In IEEE Automatic Speech Recognition and Understanding Workshop, ASRU 2019, Singapore, December 14–18, 2019 449–456 (IEEE, 2019).
Chang, H. et al. Muse: text-to-image generation via masked generative transformers. Proc. Mach. Learning Res. 202, 4055–4075 (2023).
Song, Y. & Kingma, D. P. How to train your energy-based models. Preprint at https://arxiv.org/abs/2101.03288 (2021).
Hoogeboom, E., Nielsen, D., Jaini, P., Forré, P. & Welling, M. Argmax flows and multinomial diffusion: learning categorical distributions. Adv. Neural Inf. Process. Syst. 34, 12454–12465 (2021).
Austin, J., Johnson, D. D., Ho, J., Tarlow, D. & van den Berg, R. Structured denoising diffusion models in discrete state-spaces. Adv. Neural Inf. Process. Syst. 34, 17981–17993 (2021).
Li, X., Thickstun, J., Gulrajani, I., Liang, P. S. & Hashimoto, T. B. Diffusion-LM improves controllable text generation. Adv. Neural Inf. Process Syst. 35, 4328–4343 (2022).
Bond-Taylor, S., Hessey, P., Sasaki, H., Breckon, T. P. & Willcocks, C. G. Unleashing Transformers: parallel token prediction with discrete absorbing diffusion for fast high-resolution image generation from vector-quantized codes. In Computer Vision - ECCV 2022: 17th European Conference, Tel Aviv, Israel, October 23–27, 2022, Proc., Part XXIII (eds Avidan, S.) 170–188 (Springer, 2022).
Liu, S. et al. A text-guided protein design framework. Zenodo https://doi.org/10.5281/zenodo.14630813 (2025).
Acknowledgements
This project was partly done during S.L.’s internship at Nvidia and PhD programme at Mila-UdeM, and was supported in part by the Natural Sciences and Engineering Research Council (NSERC) Discovery Grant, the Canada CIFAR AI Chair Program, collaboration grants between Microsoft Research and Mila, Samsung Electronics Co., Ltd., Amazon Faculty Research Award, Tencent AI Lab Rhino-Bird Gift Fund, two NRC Collaborative R&D Projects, IVADO Fundamental Research Project grant PRF-2019-3583139727 and NSF award CHE 2226451.
Author information
Authors and Affiliations
Contributions
S.L., Y.L., A.G., Y.Z., Z.X., W.N., A.R., C.X., J.T., H.G. and A.A. conceived and designed the experiments. S.L., Z.X. and J.L. contributed to the first round of editing tasks (dataset, prompt and evaluation). S.L., Y.L., A.G. and Z.X. fixed and finalized the editing tasks (dataset, prompt and evaluation). S.L. and Y.L. performed the experiments. S.L., Y.L. and A.G. analysed the data. S.L., Y.L. and Z.L. contributed analysis tools. S.L., Y.L., Z.L., A.G., C.X., H.G. and A.A. wrote the paper. C.X., J.T., H.G. and A.A. contributed equally to advising this project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks Sergio Romero-Romero and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information
Supplementary Sections A–D, Tables 1–21, Figs. 1–7 and References.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, S., Li, Y., Li, Z. et al. A text-guided protein design framework. Nat Mach Intell 7, 580–591 (2025). https://doi.org/10.1038/s42256-025-01011-z
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s42256-025-01011-z
This article is cited by
-
Protein foundation models: a comprehensive survey
Science China Life Sciences (2026)
-
Boosting the predictive power of protein representations with a corpus of text annotations
Nature Machine Intelligence (2025)
-
A trimodal protein language model enables advanced protein searches
Nature Biotechnology (2025)
-
Advancing biomolecular understanding and design following human instructions
Nature Machine Intelligence (2025)
-
Ab-initio amino acid sequence design from protein text description with ProtDAT
Nature Communications (2025)