Abstract
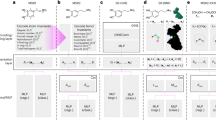
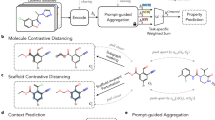
Molecular representation is a critical element in our understanding of the physical world and the foundation for modern molecular machine learning. Previous molecular machine learning models have used strings, fingerprints, global features and simple molecular graphs that are inherently information-sparse representations. However, as the complexity of prediction tasks increases, the molecular representation needs to encode higher fidelity information. This work introduces a new approach to infusing quantum-chemical-rich information into molecular graphs via stereoelectronic effects, enhancing expressivity and interpretability. Learning to predict the stereoelectronics-infused representation with a tailored double graph neural network workflow enables its application to any downstream molecular machine learning task without expensive quantum-chemical calculations. We show that the explicit addition of stereoelectronic information substantially improves the performance of message-passing two-dimensional machine learning models for molecular property prediction. We show that the learned representations trained on small molecules can accurately extrapolate to much larger molecular structures, yielding chemical insight into orbital interactions for previously intractable systems, such as entire proteins, opening new avenues of molecular design. Finally, we have developed a web application (simg.cheme.cmu.edu) where users can rapidly explore stereoelectronic information for their own molecular systems.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data and model weights are available at https://huggingface.co/gomesgroup/simg.
Code availability
The code is available at https://github.com/gomesgroup/simg (ref. 64). A web application is available at https://simg.cheme.cmu.edu where users can rapidly explore stereoelectronic information for their own molecular systems.
References
Hoffmann, R. & Laszlo, P. Representation in chemistry. Angew. Chem. Int. Ed. Engl. 30, 1–16 (1991).
Cooke, H. A historical study of structures for communication of organic chemistry information prior to 1950. Org. Biomol. Chem. 2, 3179 (2004).
Springer, M. T. Improving students’ understanding of molecular structure through broad-based use of computer models in the undergraduate organic chemistry lecture. J. Chem. Educ. 91, 1162–1168 (2014).
Gómez-Bombarelli, R. et al. Design of efficient molecular organic light-emitting diodes by a high-throughput virtual screening and experimental approach. Nat. Mater. 15, 1120–1127 (2016).
Zhavoronkov, A. et al. Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat. Biotechnol. 37, 1038–1040 (2019).
Dara, S., Dhamercherla, S., Jadav, S. S., Babu, C. M. & Ahsan, M. J. Machine learning in drug discovery: a review. Artif. Intell. Rev. 55, 1947–1999 (2022).
Gallegos, L. C., Luchini, G., St. John, P. C., Kim, S. & Paton, R. S. Importance of engineered and learned molecular representations in predicting organic reactivity, selectivity, and chemical properties. Acc. Chem. Res. 54, 827–836 (2021).
Sandfort, F., Strieth-Kalthoff, F., Kühnemund, M., Beecks, C. & Glorius, F. A structure-based platform for predicting chemical reactivity. Chem 6, 1379–1390 (2020).
Ross, J. et al. Large-scale chemical language representations capture molecular structure and properties. Nat. Mach. Intell. 4, 1256–1264 (2022).
Yang, Z., Chakraborty, M. & White, A. D. Predicting chemical shifts with graph neural networks. Chem. Sci. 12, 10802–10809 (2021).
Zhou, J. et al. Graph neural networks: a review of methods and applications. AI Open 1, 57–81 (2020).
Fang, X. et al. Geometry-enhanced molecular representation learning for property prediction. Nat. Mach. Intell. 4, 127–134 (2022).
Batzner, S. et al. E(3)-equivariant graph neural networks for data-efficient and accurate interatomic potentials. Nat. Commun. 13, 2453 (2022).
Qi, Y., Gong, W. & Yan, Q. Bridging deep learning force fields and electronic structures with a physics-informed approach. Preprint at https://doi.org/10.48550/arXiv.2403.13675 (2024).
Fabrizio, A., Briling, K. R. & Corminboeuf, C. SPAHM: the spectrum of approximated Hamiltonian matrices representations. Digital Discovery 1, 286–294 (2022).
Elton, D. C., Boukouvalas, Z., Butrico, M. S., Fuge, M. D. & Chung, P. W. Applying machine learning techniques to predict the properties of energetic materials. Sci. Rep. 8, 9059 (2018).
Rupp, M., Tkatchenko, A., Müller, K.-R. & von Lilienfeld, O. A. Fast and accurate modeling of molecular atomization energies with machine learning. Phys. Rev. Lett. 108, 058301 (2012).
Pozdnyakov, S. N. & Ceriotti, M. Smooth, exact rotational symmetrization for deep learning on point clouds. Preprint at https://doi.org/10.48550/arXiv.2305.19302 (2023).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Černý, J. & Hobza, P. Non-covalent interactions in biomacromolecules. Phys. Chem. Chem. Phys. 9, 5291 (2007).
Anighoro, A. in Quantum Mechanics in Drug Discovery (ed. Heifetz, A.) 75–86 (Humana, Springer, 2020).
Wheeler, S. E., Seguin, T. J., Guan, Y. & Doney, A. C. Noncovalent interactions in organocatalysis and the prospect of computational catalyst design. Acc. Chem. Res. 49, 1061–1069 (2016).
Weinhold, F. & Landis, C. R. Natural bond orbitals and extensions of localized bonding concepts. Chem. Educ. Res. Pract. 2, 91–104 (2001).
Llenga, S. & Gryn’ova, G. Matrix of orthogonalized atomic orbital coefficients representation for radicals and ions. J. Chem. Phys. 158, 214116 (2023).
Bartók, A. P., Kondor, R. & Csányi, G. On representing chemical environments. Phys. Rev. B 87, 184115 (2013).
Prokhorenkova, L., Gusev, G., Vorobev, A., Dorogush, A. V. & Gulin, A. Catboost: unbiased boosting with categorical features. Adv. Neur. Inf. Proc. Syst. 31, 6638–6648 (2018).
NVIDIA. MegaMolBART. GitHub https://github.com/NVIDIA/MegaMolBART (2022).
Heid, E. et al. Chemprop: a machine learning package for chemical property prediction. J. Chem. Inf. Model 64, 9–17 (2024).
Alabugin, I. V. Stereoelectronic Effects: A Bridge Between Structure and Reactivity (Wiley, 2016).
Echenique, P. & Alonso, J. L. A mathematical and computational review of Hartree–Fock SCF methods in quantum chemistry. Mol. Phys. 105, 3057–3098 (2007).
Burke, K. & Wagner, L. O. DFT in a nutshell. Int. J. Quantum. Chem. 113, 96–101 (2013).
Goerigk, L. & Grimme, S. Double-hybrid density functionals. Wiley Interdiscip. Rev. Comput. Mol. Sci. 4, 576–600 (2014).
Kneiding, H. et al. Deep learning metal complex properties with natural quantum graphs. Digital Discovery 2, 618–633 (2023).
Johnson, E. R. et al. Revealing noncovalent interactions. J. Am. Chem. Soc. 132, 6498–6506 (2010).
Axelrod, S. & Gómez-Bombarelli, R. GEOM, energy-annotated molecular conformations for property prediction and molecular generation. Sci. Data 9, 185 (2022).
Malinin, A., Prokhorenkova, L. & Ustimenko, A. Uncertainty in gradient boosting via ensembles. Preprint at https://doi.org/10.48550/arXiv.2006.10562 (2020).
Chua, K., Calandra, R., McAllister, R. & Levine, S. Deep reinforcement learning in a handful of trials using probabilistic dynamics models. In Proc. 32nd International Conference on Neural Information Processing Systems 4759–4770 (NIPS, 2018).
Goan, E. & Fookes, C. in Case Studies in Applied Bayesian Data Science (eds Mengerson, K. L. et al.) 45–87 (Springer, 2020).
Beluch, W. H., Genewein, T., Nurnberger, A. & Kohler, J. M. The power of ensembles for active learning in image classification. In Proc. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition 9368–9377 (IEEE, 2018).
León, I., Alonso, E. R., Cabezas, C., Mata, S. & Alonso, J. L. Unveiling the n→π* interactions in dipeptides. Commun Chem 2, 3 (2019).
Newberry, R. W., Bartlett, G. J., VanVeller, B., Woolfson, D. N. & Raines, R. T. Signatures of n→π* interactions in proteins. Protein Sci. 23, 284–288 (2014).
Hodges, J. A. & Raines, R. T. Energetics of an n → π* interaction that impacts protein structure. Org Lett 8, 4695–4697 (2006).
Zhou, Y., Morais-Cabral, J. H., Kaufman, A. & MacKinnon, R. Chemistry of ion coordination and hydration revealed by a K+ channel–Fab complex at 2.0 Å resolution. Nature 414, 43–48 (2001).
Bartlett, G. J., Choudhary, A., Raines, R. T. & Woolfson, D. N. n→π* interactions in proteins. Nat. Chem. Biol. 6, 615–620 (2010).
dos Passos Gomes, G. & Alabugin, I. V. Drawing catalytic power from charge separation: stereoelectronic and zwitterionic assistance in the Au(I)-catalyzed Bergman cyclization. J. Am. Chem. Soc. 139, 3406–3416 (2017).
Gomes, G. D. P., Vil’, V., Terent’ev, A. & Alabugin, I. V. Stereoelectronic source of the anomalous stability of bis-peroxides. Chem. Sci. 6, 6783–6791 (2015).
Grabowski, S. J. Tetrel bond–σ-hole bond as a preliminary stage of the SN2 reaction. Phys. Chem. Chem. Phys. 16, 1824–1834 (2014).
Sarazin, Y., Liu, B., Roisnel, T., Maron, L. & Carpentier, J.-F. Discrete, solvent-free alkaline-earth metal cations: metal···fluorine interactions and ROP catalytic activity. J. Am. Chem. Soc. 133, 9069–9087 (2011).
Ramakrishnan, R., Dral, P. O., Rupp, M. & von Lilienfeld, O. A. Quantum chemistry structures and properties of 134 kilo molecules. Sci. Data 1, 140022 (2014).
Mardirossian, N. & Head-Gordon, M. ωB97M-V: a combinatorially optimized, range-separated hybrid, meta-GGA density functional with VV10 nonlocal correlation. J. Chem. Phys. 144, 214110 (2016).
Shao, Y. et al. Advances in molecular quantum chemistry contained in the Q-Chem 4 program package. Mol. Phys. 113, 184–215 (2015).
Glendening, E. D., Landis, C. R. & Weinhold, F. NBO 7.0: new vistas in localized and delocalized chemical bonding theory. J. Comput. Chem. https://doi.org/10.1002/jcc.25873 (2019).
Ong, S. P. et al. Python Materials Genomics (pymatgen): a robust, open-source python library for materials analysis. Comput. Mater. Sci. 68, 314–319 (2013).
Blau, S., Spotte-Smith, E. W. C., Wood, B., Dwaraknath, S. & Persson, K. Accurate, automated density functional theory for complex molecules using on-the-fly error correction. Preprint at chemRxiv https://doi.org/10.26434/chemrxiv.13076030.v1 (2020).
Mathew, K. et al. Atomate: A high-level interface to generate, execute, and analyze computational materials science workflows. Comput. Mater. Sci. 139, 140–152 (2017).
Paszke, A. et al. PyTorch: an imperative style, high-performance deep learning library. In Advances in Neural Information Processing Systems Vol. 32 (eds. Wallach, H. et al.) 8024–8035 (Curran Associates, Inc., 2019).
Falcon, W. A. et al. PyTorch Lightning. GitHub https://github.com/PyTorchLightning/pytorch-lightning (2019).
Fey, M. & Lenssen, J. E. Fast graph representation learning with PyTorch Geometric. Preprint at https://doi.org/10.48550/arXiv.1903.02428 (2019).
Corso, G., Cavalleri, L., Beaini, D., Liò, P. & Veličković, P. Principal neighbourhood aggregation for graph nets. Preprint at https://doi.org/10.48550/arXiv.2004.05718 (2020).
Li, G., Müller, M., Thabet, A. & Ghanem, B. DeepGCNs: can GCNs go as deep as CNNs? Preprint at https://doi.org/10.48550/arXiv.1904.03751 (2019).
Godwin, J. et al. Simple GNN regularisation for 3D molecular property prediction & beyond. Preprint at https://doi.org/10.48550/arXiv.2106.07971 (2021).
Veličković, P. et al. Graph attention networks. Preprint at https://doi.org/10.48550/arXiv.1710.10903 (2017).
Cai, C. & Wang, Y. A note on over-smoothing for graph neural networks. Preprint at https://doi.org/10.48550/arXiv.2006.13318 (2020).
Boiko, D. et al. Advancing molecular machine learned representations with stereoelectronics-infused molecular graphs. Zenodo https://doi.org/10.5281/zenodo.14393496 (2024).
Acknowledgements
We thank NSF ACCESS (project no. CHE220012), Google Cloud Platform, NVIDIA Academic Hardware Grant Program (project titled ‘New molecular graph representations in joint feature spaces’) for computational resources. G.G. and D.B. acknowledge the financial support by the National Science Foundation Center for Computer-Assisted Synthesis (grant no. 2202693) and a supporting seed grant from X, the moonshot factory (an Alphabet company). G.G. thanks Carnegie Mellon University (CMU) and the departments of chemistry and chemical engineering for the startup support. G.G. thanks F. Weinhold (University of Wisconsin, Madison) for the development of NBO and the many discussions about the theory and software. S.M.B. acknowledges financial support by the Laboratory Directed Research and Development Program of Lawrence Berkeley National Laboratory under US Department of Energy Contract No. DE-AC02-05CH11231. Computational resources for the high-throughput virtual screening and datasets development were provided by the National Energy Research Scientific Computing Center (NERSC), a US Department of Energy Office of Science User Facility under contract no. DE-AC02-05CH11231 and by the Lawrencium computational cluster resource provided by the IT Division at the Lawrence Berkeley National Laboratory (Supported by the Director, Office of Science, Office of Basic Energy Sciences, of the US Department of Energy under contract no. DE-AC02-05CH11231). We thank J. Kitchin (CMU Chemical Engineering) and O. Isayev (CMU Chemistry) for their constructive feedback. Any opinions, findings and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation, the US Department of Energy, Alphabet (and its subsidiaries) or any of the other funding sources.
Author information
Authors and Affiliations
Contributions
D.A.B. designed the computational pipeline and implemented SIMG* prediction, active learning process, downstream task analysis and the first version of large molecule analysis. T.R. implemented the lone pair prediction model and performed analysis of large molecule predictions. B.S.-L. advised on the development of machine learning pipeline and software development. S.M.B. performed quantum-chemistry calculations and advised on analysis of NBO data. G.G. designed the concept and performed preliminary studies. S.M.B. and G.G. supervised the project. D.A.B., T.R. and G.G. wrote this manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–12.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Boiko, D.A., Reschützegger, T., Sanchez-Lengeling, B. et al. Advancing molecular machine learning representations with stereoelectronics-infused molecular graphs. Nat Mach Intell 7, 771–781 (2025). https://doi.org/10.1038/s42256-025-01031-9
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s42256-025-01031-9
This article is cited by
-
Machine learning interatomic potentials at the centennial crossroads of quantum mechanics
Nature Computational Science (2025)
-
Prediction of Key Properties in Multiple Resonance Thermally Activated Delayed Fluorescence Materials Using Lightweight Feature Encoding
Chemical Research in Chinese Universities (2025)