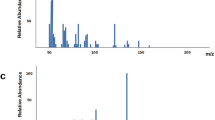
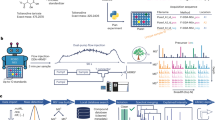
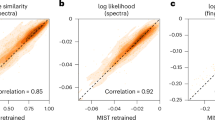
Abstract
Translating mass spectra into chemical structures is a central challenge in exposomics, making it difficult to quickly track the millions of chemicals found in humans and the environment. Unlike metabolomics, key problems in developing models for chemicals with a larger molecular space include data scarcity, model complexity and proper query strategy. Here we present a molecular structure generator (MSGo) that can generate structures directly from mass spectra and discover unknown polyfluorinated chemicals in the exposome. Trained with only virtual spectra using a transformer neural network, MSGo correctly identified 48% of structures in a validation set and was better at discovering new polyfluorinated chemicals in wastewater samples reported in the literature than experts. Applying probability-oriented masking to the virtual spectra is key to MSGo’s performance. Rapid discovery of chemicals with limited experimental mass spectral data using automated tools such as MSGo is key to tackling the current unknown polyfluorinated chemical crisis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Information, and are available via GitHub at http://github.com/aaronma2020/MSGO and via Zenodo at https://doi.org/10.5281/zenodo.17182996 (ref. 46). Source data are provided with this paper.
Code availability
MSGo was developed using Python and is available for scientific research purposes via GitHub at http://github.com/aaronma2020/MSGO and Zenodo at https://doi.org/10.5281/zenodo.17182996 (ref. 46).
References
Rappaport, S. M. & Smith, M. T. Environment and disease risks. Science 330, 460–461 (2010).
Landrigan, P. J. et al. The Lancet Commission on pollution and health. Lancet 391, 462–512 (2018).
Stein, S. E. & Scott, D. R. Optimization and testing of mass spectral library search algorithms for compound identification. J. Am. Soc. Mass. Spectrom. 5, 859–866 (1994).
Scheubert, K. et al. Significance estimation for large scale metabolomics annotations by spectral matching. Nat. Commun. 8, 1494 (2017).
Kind, T. et al. Identification of small molecules using accurate mass MS/MS search. Mass Spectrom. Rev. 37, 513–532 (2018).
Dührkop, K., Shen, H., Meusel, M., Rousu, J. & Böcker, S. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc. Natl Acad. Sci. USA 112, 12580–12585 (2015).
Ruttkies, C., Schymanski, E. L., Wolf, S., Hollender, J. & Neumann, S. MetFrag relaunched: incorporating strategies beyond in silico fragmentation. J. Cheminform. 8, 3 (2016).
Tsugawa, H. et al. Hydrogen rearrangement rules: computational MS/MS fragmentation and structure elucidation using MS-FINDER software. Anal. Chem. 88, 7946–7958 (2016).
Allen, F., Greiner, R. & Wishart, D. Competitive fragmentation modeling of ESI-MS/MS spectra for putative metabolite identification. Metabolomics 11, 98–110 (2015).
Escher, B. I., Stapleton, H. M. & Schymanski, E. L. Tracking complex mixtures of chemicals in our changing environment. Science 367, 388–392 (2020).
Vermeulen, R., Schymanski, E. L., Barabási, A.-L. & Miller, G. W. The exposome and health: where chemistry meets biology. Science 367, 392–396 (2020).
Schymanski, E. L., Meinert, C., Meringer, M. & Brack, W. The use of MS classifiers and structure generation to assist in the identification of unknowns in effect-directed analysis. Anal. Chim. Acta 615, 136–147 (2008).
Djoumbou-Feunang, Y. et al. BioTransformer: a comprehensive computational tool for small molecule metabolism prediction and metabolite identification. J. Cheminform. 11, 2 (2019).
Reymond, J.-L. The chemical space project. Acc. Chem. Res. 48, 722–730 (2015).
Moorthy, A. S., Wallace, W. E., Kearsley, A. J., Tchekhovskoi, D. V. & Stein, S. E. Combining fragment-ion and neutral-loss matching during mass spectral library searching: a new general purpose algorithm applicable to illicit drug identification. Anal. Chem. 89, 13261–13268 (2017).
Xing, S. et al. Retrieving and utilizing hypothetical neutral losses from tandem mass spectra for spectral similarity analysis and unknown metabolite annotation. Anal. Chem. 92, 14476–14483 (2020).
Aron, A. T. et al. Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat. Protoc. 15, 1954–1991 (2020).
Schmid, R. et al. Ion identity molecular networking for mass spectrometry-based metabolomics in the GNPS environment. Nat. Commun. 12, 3832 (2021).
Tripathi, A. et al. Chemically informed analyses of metabolomics mass spectrometry data with Qemistree. Nat. Chem. Biol. 17, 146–151 (2021).
Dührkop, K. et al. Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra. Nat. Biotechnol. 39, 462–471 (2021).
Vamathevan, J. et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 18, 463–477 (2019).
Colby, S. M., Nuñez, J. R., Hodas, N. O., Corley, C. D. & Renslow, R. R. Deep learning to generate in silico chemical property libraries and candidate molecules for small molecule identification in complex samples. Anal. Chem. 92, 1720–1729 (2019).
Skinnider, M. A. et al. A deep generative model enables automated structure elucidation of novel psychoactive substances. Nat. Mach. Intell. 3, 973–984 (2021).
Stravs, M. A., Dührkop, K., Böcker, S. & Zamboni, N. MSNovelist: de novo structure generation from mass spectra. Nat. Methods 19, 865–870 (2022).
Shrivastava, A. D. et al. MassGenie: a transformer-based deep learning method for identifying small molecules from their mass spectra. Biomolecules 11, 1793 (2021).
Litsa, E. E. et al. An end-to-end deep learning framework for translating mass spectra to de-novo molecules. Commun. Chem. 6, 132 (2023).
Butler, T. et al. MS2Mol: a transformer model for illuminating dark chemical space from mass spectra. Preprint at https://doi.org/10.26434/chemrxiv-2023-vsmpx-v4.
Madani, A. et al. Large language models generate functional protein sequences across diverse families. Nat. Biotechnol. 41, 1099–1106 (2023).
Vaswani, A. et al. Attention is all you need. In Proc. 31st Conference on Neural Information Processing Systems (NIPS) (eds von Luxburg, U. et al.) 5999–6009 (NeruIPS, 2017).
Blum, A. et al. The Madrid statement on poly-and perfluoroalkyl substances (PFASs). Environ. Health Perspect. 123, A107–A111 (2015).
Evich, M. G. et al. Per- and polyfluoroalkyl substances in the environment. Science 375, eabg9065 (2022).
Washington, J. W. et al. Nontargeted mass-spectral detection of chloroperfluoropolyether carboxylates in New Jersey soils. Science 368, 1103–1107 (2020).
Djoumbou-Feunang, Y. et al. CFM-ID 3.0: significantly improved ESI-MS/MS prediction and compound identification. Metabolites 9, 72 (2019).
Kong, F. et al. Denoising Search doubles the number of metabolite and exposome annotations in human plasma using an Orbitrap Astral mass spectrometer. Nat. Methods 22, 1008–1016 (2025).
Li, X. & Fourches, D. SMILES pair encoding: a data-driven substructure tokenization algorithm for deep learning. J. Chem. Inf. Model. 61, 1560–1569 (2021).
Dührkop, K. et al. SIRIUS 4: a rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 16, 299–302 (2019).
Wang, Y. et al. Suspect and nontarget screening of per- and polyfluoroalkyl substances in wastewater from a fluorochemical manufacturing park. Environ. Sci. Technol. 52, 11007–11016 (2018).
Fiehn Lab—CASMI 2022—Results (ucdavis.edu) (Univ. California Davis, 2022); https://fiehnlab.ucdavis.edu/casmi/casmi-2022-results
Cai, Y., Zhou, Z. & Zhu, Z. J. Advanced analytical and informatic strategies for metabolite annotation in untargeted metabolomics. Trends Anal. Chem. 158, 116903 (2022).
Lu, S., Gao, Z., He, D., Zhang, L. & Ke, G. Data-driven quantum chemical property prediction leveraging 3D conformations with Uni-Mol+. Nat. Commun. 15, 7104 (2024).
Getzinger, G. J., Higgins, C. P. & Ferguson, P. L. Structure database and in silico spectral library for comprehensive suspect screening of per- and polyfluoroalkyl substances (PFASs) in environmental media by high-resolution mass spectrometry. Anal. Chem. 93, 2820–2827 (2021).
Koelmel, J. P. et al. FluoroMatch 2.0—making automated and comprehensive non-targeted PFAS annotation a reality. Anal. Bioanal. Chem. 414, 1201–1215 (2022).
Liu, Y., D’Agostino, L. A., Qu, G., Jiang, G. & Martin, J. W. High-resolution mass spectrometry (HRMS) methods for nontarget discovery and characterization of poly- and per-fluoroalkyl substances (PFASs) in environmental and human samples. Trends Anal. Chem. 121, 115420 (2019).
Schymanski, E. L. et al. Identifying small molecules via high resolution mass spectrometry: communicating confidence. Environ. Sci. Technol. 48, 2097–2098 (2014).
Reconciling Terminology of the Universe of Per- and Polyfluoroalkyl Substances: Recommendations and Practical Guidance (OECD, 2021); https://doi.org/10.1787/e458e796-en
Ma, Z., Yu, N., Shao, Q., Bao, Q. & Wei, S. MSGO. Zenodo https://doi.org/10.5281/zenodo.17182996 (2025).
Liu, M., Munoz, G., Duy, S. V., Sauvé, S. & Liu, J. Stability of nitrogen-containing polyfluoroalkyl substances in aerobic soils. Environ. Sci. Technol. 55, 4698–4708 (2021).
Acknowledgements
The MSGo project was supported by the National Key Research and Development Programme of China (grant no. 2024YFA0918900, X.W.), the National Natural Science Foundation of China (grant nos. 22525604, S.W., 22376092, S.W., U24A20512, S.W. and 22276090, N.Y.), the Fundamental Research Funds for the Central University (grant no. 021114380239, S.W.) and Anhui Provincial Key Research and Development Project (grant no. 2023t07020004, S.W.). We thank L. Wang, S. Yu and W. Jiang for their insights on polyfluorinated chemical synthesis; B. Zhang for constructive article feedback and Q. Bao for implementing benchmark models (Spec2Mol and MassGenie).
Author information
Authors and Affiliations
Contributions
S.W., Z.M., Q.S. and N.Y. conceived the idea and designed the algorithm and software. Q.S. and Z.M. developed the MSGo program. Q.S., N.Y. and X.W. performed data collection and data processing. S.W., Z.M., Q.S. and N.Y. verified the performance of MSGo. S.W., Q.S. and N.Y. checked the structure annotation of MSGo. B.P. and X.W. suggested and designed synthesis routes. H.Y. and S.W. designed experiments to verify the annotated structure. N.Y. and Q.S. wrote the paper. S.W., N.Y., Z.M., Q.S., L.L., X.W., B.P. and H.Y. reviewed and edited the paper. S.W. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks Cheng Wang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Text 1, Figs. 1–16 and Table 1.
Supplementary Data
Supplementary Data 1. The molecular database for polyfluorinated chemicals. Supplementary Data 2. The experimental mass spectra dataset for polyfluorinated chemicals. Supplementary Data 3. The 90 reported structures in wastewater samples from the literature.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yu, N., Ma, Z., Shao, Q. et al. Pseudodata-based molecular structure generator to reveal unknown chemicals. Nat Mach Intell 7, 1879–1887 (2025). https://doi.org/10.1038/s42256-025-01140-5
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s42256-025-01140-5