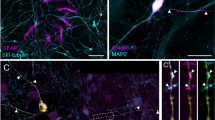
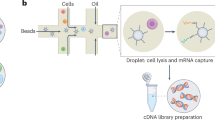
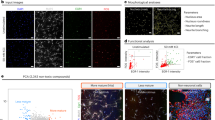
Abstract
High-throughput phenotypic screening has historically relied on manually selected features, limiting our ability to capture complex cellular processes, particularly neuronal activity dynamics. While recent advances in self-supervised learning have revolutionized the study of cellular morphology and transcriptomics, dynamic cellular processes remain challenging to phenotypically profile. To address this, we developed Plexus, a self-supervised model designed to capture and quantify network-level neuronal activity. Unlike existing tools that focus on static readouts, Plexus leverages a network-level cell encoding method, efficiently encoding dynamic neuronal activity into rich representational embeddings. In turn, Plexus achieves state-of-the-art performance in detecting phenotypic changes in neuronal activity. Here we validated Plexus using a comprehensive GCaMP6m simulation framework and demonstrated its ability to classify distinct phenotypes compared with traditional signal-processing approaches. To enable practical application, we integrated Plexus with a scalable experimental system using human induced pluripotent stem cell-derived neurons expressing the GCaMP6m calcium indicator and CRISPR interference machinery. This platform successfully identified nearly 17 times as many phenotypic changes in response to genetic perturbations compared with conventional methods, as demonstrated in a 52-gene CRISPR interference screen across multiple induced pluripotent stem cell lines. Using this framework, we identified potential genetic modifiers of aberrant neuronal activity in frontotemporal dementia, illustrating its utility for understanding complex neurological disorders.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The processed files containing the time-series data, the model checkpoints, model embeddings in the form of h5ad files and the raw dataset metadata files are available via Zenodo at https://zenodo.org/records/14714574 (ref. 43).
Code availability
All code for plexus model training and inference is available via GitHub at https://github.com/pgrosjean/plexus/tree/main (https://zenodo.org/records/15811302) (ref. 44). All code for time-series extraction from microscopy is available via GitHub at https://github.com/pgrosjean/plexus-extract/tree/main (https://zenodo.org/records/15811338) (ref. 45). All code for the multivariate Hawkes process simulation is available via GitHub at https://github.com/pgrosjean/plexus-simulate/tree/main (https://zenodo.org/records/15811345) (ref. 42).
References
Kelley, M. E. et al. High-content microscopy reveals a morphological signature of bortezomib resistance. eLife 12, e91362 (2023).
Yu, S. et al. Integrating inflammatory biomarker analysis and artificial-intelligence-enabled image-based profiling to identify drug targets for intestinal fibrosis. Cell Chem. Biol. 30, 1169–1182.e8 (2023).
Tegtmeyer, M. et al. High-dimensional phenotyping to define the genetic basis of cellular morphology. Nat. Commun. 15, 347 (2024).
Chandrasekaran, S. N. et al. Three million images and morphological profiles of cells treated with matched chemical and genetic perturbations. Nat. Methods 21, 1114–1121 (2024).
Reisen, F. et al. Linking phenotypes and modes of action through high-content screen fingerprints. Assay Drug Dev. Technol. 13, 415–427 (2015).
Ziegler, S., Sievers, S. & Waldmann, H. Morphological profiling of small molecules. Cell Chem. Biol. 28, 300–319 (2021).
MacDonald, M. L. et al. Identifying off-target effects and hidden phenotypes of drugs in human cells. Nat. Chem. Biol. 2, 329–337 (2006).
Atmaramani, R. et al. Deep learning analysis on images of IPSC-derived motor neurons carrying fals-genetics reveals disease-relevant phenotypes. Preprint at bioRxiv https://doi.org/10.1101/2024.01.04.574270 (2024).
Lee, B. R. et al. Scaled, high fidelity electrophysiological, morphological, and transcriptomic cell characterization. eLife 10, e65482 (2021).
Boivin, B. et al. A multiparametric activity profiling platform for neuron disease phenotyping and drug screening. Mol. Biol. Cell 33, ar54 (2021).
Huang, X. et al. Human amyotrophic lateral sclerosis excitability phenotype screen: target discovery and validation. Cell Rep. 35, 109224 (2021).
Williams, L. A. et al. Discovery of novel compounds and target mechanisms using a high throughput, multiparametric phenotypic screen in a human neuronal model of tuberous sclerosis. Preprint at bioRxiv https://doi.org/10.1101/2024.02.22.581652 (2024).
Chen, T.-W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013).
Singh, S., Carpenter, A. E. & Genovesio, A. Increasing the content of high-content screening: an overview. SLAS Discov. 19, 640–650 (2014).
Kim, V. et al. Self-supervision advances morphological profiling by unlocking powerful image representations. Sci Rep 15, 4876 (2025).
Doron, M. et al. Unbiased single-cell morphology with self-supervised vision transformers. Preprint at bioRxiv https://doi.org/10.1101/2023.06.16.545359 (2023).
Kobayashi, H., Cheveralls, K. C., Leonetti, M. D. & Royer, L. A. Self-supervised deep learning encodes high-resolution features of protein subcellular localization. Nat. Methods 19, 995–1003 (2022).
Sivanandan, S. et al. A pooled cell painting CRISPR screening platform enables de novo inference of gene function by self-supervised deep learning. Preprint at bioRxiv https://doi.org/10.1101/2023.08.13.553051 (2023).
Wang, C. et al. Scalable production of iPSC-derived human neurons to identify tau-lowering compounds by high-content screening. Stem Cell Rep. 9, 1221–1233 (2017).
Tian, R. et al. CRISPR interference-based platform for multimodal genetic screens in human iPSC-derived neurons. Neuron 104, 239–255 (2019).
Serio, A. et al. Astrocyte pathology and the absence of non-cell autonomy in an induced pluripotent stem cell model of TDP-43 proteinopathy. Proc. Natl Acad. Sci. USA 110, 4697–4702 (2013).
He, K. et al. Masked autoencoders are scalable vision learners. Preprint at https://doi.org/10.48550/arXiv.2111.06377 (2021).
Vaswani, A. et al. Attention is all you need. Preprint at https://doi.org/10.48550/arXiv.1706.03762 (2023).
Goncalves, B. & Gresland, P. Perfect simulation for interacting Hawkes processes with variable length memory. Preprint at https://doi.org/10.48550/arXiv.2209.09143 (2022).
Watts, D. J. & Strogatz, S. H. Collective dynamics of ‘small-world’ networks. Nature 393, 440–442 (1998).
Song, A., Gauthier, J. L., Pillow, J. W., Tank, D. W. & Charles, A. S. Neural anatomy and optical microscopy (NAOMi) simulation for evaluating calcium imaging methods. J. Neurosci. Methods 358, 109173 (2021).
Ansari, A. F. et al. Chronos: learning the language of time series. Preprint at https://doi.org/10.48550/arXiv.2403.07815 (2024).
Heo, S.-J. et al. Compact CRISPR genetic screens enabled by improved guide RNA library cloning. Genome Biol. 25, 25 (2024).
Kreitzer, F. R. et al. A robust method to derive functional neural crest cells from human pluripotent stem cells. Am. J. Stem Cells 2, 119–131 (2013).
Karch, C. M. et al. A Comprehensive resource for induced pluripotent stem cells from patients with primary tauopathies. Stem Cell Rep. 13, 939–955 (2019).
Pachitariu, M. & Stringer, C. Cellpose 2.0: how to train your own model. Nat. Methods 19, 1634–1641 (2022).
Ando, D. M., McLean, C. Y. & Berndl, M. Improving phenotypic measurements in high-content imaging screens. Preprint at bioRxiv https://doi.org/10.1101/161422 (2017).
Celik, S. et al. Building, benchmarking, and exploring perturbative maps of transcriptional and morphological data. PLoS Comput. Biol. 20, e1012463 (2024).
Kraus, O. et al. Masked autoencoders for microscopy are scalable learners of cellular biology. In 2024 IEEE/CVF Conference on Computer Vision and Pattern Recognition 11757–11768 (IEEE, 2024).
Shahidullah, M., Santarelli, L. C., Wen, H. & Levitan, I. B. Expression of a calmodulin-binding KCNQ2 potassium channel fragment modulates neuronal M-current and membrane excitability. Proc. Natl Acad. Sci. USA 102, 16454–16459 (2005).
Biervert, C. et al. A potassium channel mutation in neonatal human epilepsy. Science 279, 403–406 (1998).
Lee, I.-C., Yang, J.-J., Wong, S.-H., Liou, Y.-M. & Li, S.-Y. Heteromeric Kv7.2 current changes caused by loss-of-function of KCNQ2 mutations are correlated with long-term neurodevelopmental outcomes. Sci. Rep. 10, 13375 (2020).
Trygg, J. & Wold, S. Orthogonal projections to latent structures (O-PLS). J. Chemom. 16, 119–128 (2002).
Sohn, P. D. et al. Pathogenic tau impairs axon initial segment plasticity and excitability homeostasis. Neuron 104, 458–470.e5 (2019).
Mohl, G. A. et al. Multi-omic phenotyping of iPSC-derived neurons harboring the MAPT V337M mutation reveals tau hypophosphorylation and perturbed axon morphology pathways. Preprint at bioRxiv https://doi.org/10.1101/2024.06.04.597496 (2025).
Bardy, C. et al. Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro. Proc. Natl Acad. Sci. USA 112, E2725–E2734 (2015).
Grosjean, P. pgrosjean/plexus-simulate: Plexus-simulate version 1.0.0. Zenodo. https://doi.org/10.5281/zenodo.15811345 (2025).
Grosjean, P. et al. Network-aware self-supervised learning enables high-content phenotypic screening for genetic modifiers of neuronal activity dynamics: v2. Zenodo. https://doi.org/10.5281/zenodo.15809433 (2025).
Grosjean, P. pgrosjean/plexus: Plexus version 1.0.0. Zenodo. https://doi.org/10.5281/zenodo.15811302 (2025).
Grosjean, P. pgrosjean/plexus-extract: Plexus-extract version 1.0.0. Zenodo. https://doi.org/10.5281/zenodo.15811338 (2025).
Acknowledgements
We thank the following members of the Laboratory for Genomics Research for their technical support on the work presented in this paper: P. Nguyen, S. Federman, B. Cunningham and B. Kwan-Leong. We also thank members of the GSK novel human genetics research unit for their helpful discussion. We thank J. Dinis for his help managing the unique collaboration that gave rise to this research. We acknowledge D. Muir, N. Teyssier, U. Khan and A. Lee for their scientific feedback while writing this paper. This work is supported by the Laboratory for Genomics Research established by GSK, UCSF and UC Berkeley and by grant no. DAF2018-191905 (https://doi.org/10.37921/550142lkcjzw) from the Chan Zuckerberg Initiative DAF, an advised fund of the Silicon Valley Community Foundation (funder https://doi.org/10.13039/100014989) (M.J.K.).
Author information
Authors and Affiliations
Contributions
P.G., M.K., M.J.K. and J.I. conceived of the project. P.G. designed the machine-learning methodologies, simulation framework and performed all downstream analysis. C.N. and A.Y. provided critical input on the methodological framework for SSL and subsequent technical analysis. E.U., S.B. and A.D. provided input on the methodology related to the cellular model and assay development. K.S., K.M., I.F. and P.G. performed the arrayed CRISPRi screens. P.G. and A.D. generated the astrocytes and developed the co-culture model. S.A. and P.G. analysed the imaging data. D.Z. and S.-J.H. generated the single-guide RNA virus for use in the CRISPRi screen. G.L., K.S. and P.G. performed the experimental design for the CRISPRi screen. B.T., A.L., S.S. and L.P. all provided input on the CRISPRi screening workflow and maintained the organization in which the CRISPRi screens were performed. P.G. wrote the paper. A.Y., M.J.K., J.I. and M.K. supervised the research. All authors contributed to the paper.
Corresponding authors
Ethics declarations
Competing interests
M.K. is a coscientific founder of Montara Therapeutics and serves on the Scientific Advisory Boards of Engine Biosciences, Casma Therapeutics, Alector and Montara Therapeutics, and is an advisor to Modulo Bio and Recursion Therapeutics. M.K. is an inventor on US Patent 11,254,933 related to CRISPRi and CRISPRa screening, and on a US Patent application on in vivo screening methods. J.I., C.N., I.F., D.Z. and S.S. are employees of GSK. The other authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks Anthony Zannas and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Activity measured by multi-electrode array in iNeuron monoculture vs astrocyte and iNeuron co-culture measured throughout culture.
(n = 12 wells of cultured astrocytes and iNeurons; Bar height represents mean; Error bars represent 95% confidence interval).
Extended Data Fig. 2 Plexus reconstructs masked neuronal activity dynamics.
Nine representative examples of the reconstruction task with 50% masking. The original GCaMP6m data are shown in black, normalized between 0 and 1 for visualization purposes. The red traces depict the model’s reconstruction, specifically at the locations where masking was applied, highlighting Plexus’s ability to infer activity in masked regions.
Extended Data Fig. 3
Simulated and experimental activity phenotypes. Representative traces of the eight simulated activity phenotypes (left traces) used for the linear probing classification task and the closest matching in vitro activity data (right traces) by distance in the Plexus embedding space.
Extended Data Fig. 4 Neuron counts and transduction efficiency in the arrayed CRISPRi screen.
(a) Boxplot (center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; points, outliers) of the transduction efficiency per well (n = 32 wells of co-cultures for non-targeting guides, n = 16 wells of co-cultures for all other guides). (b) The number of total neurons per field of view (center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; points, outliers). (c) The number of transduced neurons per field of view (center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; points, outliers).
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2, Figs. 1–7 and Methods.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Grosjean, P., Shevade, K., Nguyen, C. et al. Network-aware self-supervised learning enables high-content phenotypic screening for genetic modifiers of neuronal activity dynamics. Nat Mach Intell 7, 2009–2025 (2025). https://doi.org/10.1038/s42256-025-01156-x
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s42256-025-01156-x