Abstract
Immune checkpoint blockade-based multimodal therapy is widely used across oncology; yet drivers of resistance in most cancer types are not well understood. Here, we comprehensively characterized the tumor genome, microenvironment and microbiome in a phase 3 international randomized trial (NCT02952586) to identify factors that shape outcomes to anti-PD-L1 avelumab plus standard-of-care chemoradiotherapy versus placebo/chemoradiotherapy in individuals with locally advanced head and neck cancer. Patients receiving avelumab whose tumors contained distinct immunologic and genetic features had superior outcomes compared to those receiving placebo. By contrast, patients with increased myeloid/neutrophil activities had worse outcomes with avelumab than those treated with placebo. Strikingly, these tumors possessed telltale intratumoral bacteria, elevated tumor-associated neutrophils, high systemic neutrophil-to-lymphocyte ratios and suppressed adaptive immunity. We define tumor ecosystems associated with benefit to chemoimmunotherapy. Our data demonstrate how intratumoral bacteria affect immune checkpoint blockade response within a randomized trial. These discoveries enhance our understanding of combination immunotherapy, provide a useful multiomic resource and identify unanticipated interactions that may guide future therapeutic strategies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Any request for additional data outside of that provided in the paper by scientific and medical researchers for legitimate research purposes will be subject to Merck’s Data Sharing Policy. All requests should be submitted in writing to Merck’s data sharing portal (https://www.merckgroup.com/en/research/our-approach-to-research-and-development/healthcare/clinical-trials/commitment-responsible-data-sharing.html). When Merck has a co-research, co-development, co-marketing or co-promotion agreement or when the product has been out-licensed, the responsibility for disclosure might be dependent on the agreement between parties. Under these circumstances, Merck will endeavor to gain agreement to share data in response to requests. Processed mutation calls, gene expression matrices and TBB data are available in Supplementary Tables 1 and 2 provided in the paper. Raw RNA-seq files have been deposited in the European Genome–Phenome Archive (EGAD00001011290). WES BAM files were also deposited in the European Genome–Phenome Archive (EGAD00001011321). Sequencing data are available per Merck’s Data Sharing Policy. Source data are provided with this paper.
References
Ferris, R. L. et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 375, 1856–1867 (2016).
Burtness, B. et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet 394, 1915–1928 (2019).
Ribas, A. & Wolchok, J. D. Cancer immunotherapy using checkpoint blockade. Science 359, 1350–1355 (2018).
Tang, J. et al. Trial watch: the clinical trial landscape for PD1/PDL1 immune checkpoint inhibitors. Nat. Rev. Drug Discov. 17, 854–855 (2018).
Shamseddine, A. A., Burman, B., Lee, N. Y., Zamarin, D. & Riaz, N. Tumor Immunity and immunotherapy for HPV-related cancers. Cancer Discov. 11, 1896–1912 (2021).
Dewan, M. Z. et al. Fractionated but not single-dose radiotherapy induces an immune-mediated abscopal effect when combined with anti-CTLA-4 antibody. Clin. Cancer Res. 15, 5379–5388 (2009).
Rodriguez-Ruiz, M. E., Vanpouille-Box, C., Melero, I., Formenti, S. C. & Demaria, S. Immunological mechanisms responsible for radiation-induced abscopal effect. Trends Immunol. 39, 644–655 (2018).
McLaughlin, M. et al. Inflammatory microenvironment remodelling by tumour cells after radiotherapy. Nat. Rev. Cancer 20, 203–217 (2020).
Antonia, S. J. et al. Durvalumab after chemoradiotherapy in stage III non-small-cell lung cancer. N. Engl. J. Med. 377, 1919–1929 (2017).
Kelly, R. J. et al. Adjuvant nivolumab in resected esophageal or gastroesophageal junction cancer. N. Engl. J. Med. 384, 1191–1203 (2021).
Stone, H. B., Peters, L. J. & Milas, L. Effect of host immune capability on radiocurability and subsequent transplantability of a murine fibrosarcoma. J. Natl Cancer Inst. 63, 1229–1235 (1979).
Lee, Y. et al. Therapeutic effects of ablative radiation on local tumor require CD8+ T cells: changing strategies for cancer treatment. Blood 114, 589–595 (2009).
Deng, L. et al. STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity 41, 843–852 (2014).
Weichselbaum, R. R., Liang, H., Deng, L. & Fu, Y. X. Radiotherapy and immunotherapy: a beneficial liaison?. Nat. Rev. Clin. Oncol. 14, 365–379 (2017).
Zeng, J. et al. Anti-PD-1 blockade and stereotactic radiation produce long-term survival in mice with intracranial gliomas. Int. J. Radiat. Oncol. Biol. Phys. 86, 343–349 (2013).
Deng, L. et al. Irradiation and anti-PD-L1 treatment synergistically promote antitumor immunity in mice. J. Clin. Invest. 124, 687–695 (2014).
Oweida, A. et al. Ionizing radiation sensitizes tumors to PD-L1 immune checkpoint blockade in orthotopic murine head and neck squamous cell carcinoma. Oncoimmunology 6, e1356153 (2017).
Lee, N. Y. et al. Avelumab plus standard-of-care chemoradiotherapy versus chemoradiotherapy alone in patients with locally advanced squamous cell carcinoma of the head and neck: a randomised, double-blind, placebo-controlled, multicentre, phase 3 trial. Lancet Oncol. 22, 450–462 (2021).
Mayadev, J. et al. Durvalumab in combination with chemoradiotherapy (CRT) in locally advanced cervical cancer (LACC): radiotherapy (RT) delivery and subgroup analyses from CALLA. Int. J. Radiat. Oncol. Biol. Phys. 114, 1058–1059 (2022).
Machiels, J. et al. Primary results of the phase III KEYNOTE-412 study: pembrolizumab (pembro) with chemoradiation therapy (CRT) vs placebo plus CRT for locally advanced (LA) head and neck squamous cell carcinoma (HNSCC). Ann. Oncol. 33, S1399 (2022).
Carbone, D. P. et al. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N. Engl. J. Med. 376, 2415–2426 (2017).
Si, H. et al. A blood-based assay for assessment of tumor mutational burden in first-line metastatic NSCLC treatment: results from the MYSTIC study. Clin. Cancer Res. 27, 1631–1640 (2021).
Samstein, R. M. et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 51, 202–206 (2019).
Haddad, R. I. et al. Influence of tumor mutational burden, inflammatory gene expression profile, and PD-L1 expression on response to pembrolizumab in head and neck squamous cell carcinoma. J. Immunother. Cancer 10, e003026 (2022).
Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517, 576–582 (2015).
Leemans, C. R., Snijders, P. J. F. & Brakenhoff, R. H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer 18, 269–282 (2018).
Mehanna, H. et al. Prognostic implications of p16 and HPV discordance in oropharyngeal cancer (HNCIG-EPIC-OPC): a multicentre, multinational, individual patient data analysis. Lancet Oncol. 24, 239–251 (2023).
Senbabaoglu, Y. et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol. 17, 231 (2016).
Aran, D., Hu, Z. & Butte, A. J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 18, 220 (2017).
Powles, T. et al. Avelumab maintenance in advanced urothelial carcinoma: biomarker analysis of the phase 3 JAVELIN Bladder 100 trial. Nat. Med. 27, 2200–2211 (2021).
Thorsson, V. et al. The immune landscape of cancer. Immunity 51, 411–412 (2019).
Vanhersecke, L. et al. Standardized pathology screening of mature tertiary lymphoid structures in cancers. Lab. Invest. 103, 100063 (2023).
Cabrita, R. et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature 577, 561–565 (2020).
Poropatich, K. et al. Comprehensive T-cell immunophenotyping and next-generation sequencing of human papillomavirus (HPV)-positive and HPV-negative head and neck squamous cell carcinomas. J. Pathol. 243, 354–365 (2017).
Zilionis, R. et al. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity 50, 1317–1334 (2019).
Galeano Nino, J. L. et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature 611, 810–817 (2022).
Nejman, D. et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 368, 973–980 (2020).
Battaglia, T. W. et al. A pan-cancer analysis of the microbiome in metastatic cancer. Cell 187, 2324–2335 (2024).
Kostic, A. D. et al. PathSeq: software to identify or discover microbes by deep sequencing of human tissue. Nat. Biotechnol. 29, 393–396 (2011).
Breitwieser, F. P., Baker, D. N. & Salzberg, S. L. KrakenUniq: confident and fast metagenomics classification using unique k-mer counts. Genome Biol. 19, 198 (2018).
Gihawi, A. et al. Major data analysis errors invalidate cancer microbiome findings. mBio 14, e0160723 (2023).
Wade, W. G. The oral microbiome in health and disease. Pharmacol. Res. 69, 137–143 (2013).
Deo, P. N. & Deshmukh, R. Oral microbiome: unveiling the fundamentals. J. Oral Maxillofac. Pathol. 23, 122–128 (2019).
Dohlman, A. B. et al. The cancer microbiome atlas: a pan-cancer comparative analysis to distinguish tissue-resident microbiota from contaminants. Cell Host Microbe 29, 281–298 (2021).
Chowell, D. et al. Improved prediction of immune checkpoint blockade efficacy across multiple cancer types. Nat. Biotechnol. 40, 499–506 (2022).
Puram, S. V. et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 171, 1611–1624 (2017).
Gong, Y. et al. The tumor ecosystem in head and neck squamous cell carcinoma and advances in ecotherapy. Mol. Cancer 22, 68 (2023).
Chu, Y. et al. Pan-cancer T cell atlas links a cellular stress response state to immunotherapy resistance. Nat. Med. 29, 1550–1562 (2023).
Chow, A., Perica, K., Klebanoff, C. A. & Wolchok, J. D. Clinical implications of T cell exhaustion for cancer immunotherapy. Nat. Rev. Clin. Oncol. 19, 775–790 (2022).
Mell, L. K. et al. Radiotherapy with durvalumab vs. cetuximab in patients with locoregionally advanced head and neck cancer and a contraindication to cisplatin: phase II results of NRG-HN004. Int. J. Radiat. Oncol. Biol. Phys. 114, 1058 (2022).
Clump, D. A. et al. A randomized phase II study evaluating concurrent or sequential fixed-dose immune therapy in combination with cisplatin and intensity-modulated radiotherapy in intermediate- or high-risk, previously untreated, locally advanced head and neck cancer (LA SCCHN). J. Clin. Oncol. 40, 6007 (2022).
Zhao, X. et al. Somatic 9p24.1 alterations in HPV− head and neck squamous cancer dictate immune microenvironment and anti-PD-1 checkpoint inhibitor activity. Proc. Natl Acad. Sci. USA 119, e2213835119 (2022).
Darragh, L. B. et al. A phase I/Ib trial and biological correlate analysis of neoadjuvant SBRT with single-dose durvalumab in HPV-unrelated locally advanced HNSCC. Nat. Cancer 3, 1300–1317 (2022).
Darragh, L. B. et al. Elective nodal irradiation mitigates local and systemic immunity generated by combination radiation and immunotherapy in head and neck tumors. Nat. Commun. 13, 7015 (2022).
Wang, L. et al. YTHDF2 inhibition potentiates radiotherapy antitumor efficacy. Cancer Cell 41, 1294–1308 (2023).
Nishiga, Y. et al. Radiotherapy in combination with CD47 blockade elicits a macrophage-mediated abscopal effect. Nat. Cancer 3, 1351–1366 (2022).
Sepich-Poore, G. D. et al. The microbiome and human cancer. Science 371, eabc4552 (2021).
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Lamont, R. J., Koo, H. & Hajishengallis, G. The oral microbiota: dynamic communities and host interactions. Nat. Rev. Microbiol. 16, 745–759 (2018).
Nagy, K. N., Sonkodi, I., Szoke, I., Nagy, E. & Newman, H. N. The microflora associated with human oral carcinomas. Oral Oncol. 34, 304–308 (1998).
Whitmore, S. E. & Lamont, R. J. Oral bacteria and cancer. PLoS Pathog. 10, e1003933 (2014).
Orlandi, E. et al. Potential role of microbiome in oncogenesis, outcome prediction and therapeutic targeting for head and neck cancer. Oral Oncol. 99, 104453 (2019).
Hashim, D. et al. The role of oral hygiene in head and neck cancer: results from International Head and Neck Cancer Epidemiology (INHANCE) consortium. Ann. Oncol. 27, 1619–1625 (2016).
Riquelme, E. et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell 178, 795–806 (2019).
Kostic, A. D. et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215 (2013).
Cogdill, A. P., Gaudreau, P. O., Arora, R., Gopalakrishnan, V. & Wargo, J. A. The impact of intratumoral and gastrointestinal microbiota on systemic cancer therapy. Trends Immunol. 39, 900–920 (2018).
Bender, M. J. et al. Dietary tryptophan metabolite released by intratumoral Lactobacillus reuteri facilitates immune checkpoint inhibitor treatment. Cell 186, 1846–1862 (2023).
Elkrief, A. et al. Intratumoral Escherichia is associated with improved survival to single-agent immune checkpoint inhibition in patients with advanced non-small-cell lung cancer. J. Clin. Oncol. 42, 3339–3349 (2024).
Goubet, A. G. et al. Escherichia coli-specific CXCL13-producing TFH are associated with clinical efficacy of neoadjuvant PD-1 blockade against muscle-invasive bladder cancer. Cancer Discov. 12, 2280–2307 (2022).
Silver, N. et al. Intratumoral bacteria are immunosuppressive and promote immunotherapy resistance. Nat. Cancer https://doi.org/10.1038/s43018-025-01067-1 (in the press).
Sturm, G. et al. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics 35, i436–i445 (2019).
Nguyen, H., Nguyen, H., Tran, D., Draghici, S. & Nguyen, T. Fourteen years of cellular deconvolution: methodology, applications, technical evaluation and outstanding challenges. Nucleic Acids Res. 52, 4761–4783 (2024).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
Koboldt, D. C. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012).
Saunders, C. T. et al. Strelka: accurate somatic small-variant calling from sequenced tumor–normal sample pairs. Bioinformatics 28, 1811–1817 (2012).
Larson, D. E. et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics 28, 311–317 (2012).
Rimmer, A. et al. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat. Genet. 46, 912–918 (2014).
Ellrott, K. et al. Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst. 6, 271–281 (2018).
Amemiya, H. M., Kundaje, A. & Boyle, A. P. The ENCODE blacklist: identification of problematic regions of the genome. Sci. Rep. 9, 9354 (2019).
Riaz, N. et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell 171, 934–949 (2017).
Rosenthal, R., McGranahan, N., Herrero, J., Taylor, B. S. & Swanton, C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 17, 31 (2016).
Alexandrov, L. B. et al. The repertoire of mutational signatures in human cancer. Nature 578, 94–101 (2020).
Kawaguchi, S. & Matsuda, F. High-definition genomic analysis of HLA genes via comprehensive HLA allele genotyping. Methods Mol. Biol. 2131, 31–38 (2020).
Liu, P. et al. Benchmarking the human leukocyte antigen typing performance of three assays and seven next-generation sequencing-based algorithms. Front. Immunol. 12, 652258 (2021).
Shen, R. & Seshan, V. E. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 44, e131 (2016).
Leshchiner, I. et al. Comprehensive analysis of tumour initiation, spatial and temporal progression under multiple lines of treatment. Preprint at bioRxiv https://doi.org/10.1101/508127 (2019).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Katz, A. et al. CytoPro: a computational platform for accurate and robust assessment of cell contributions in bulk expression from diverse tissues and conditions. Preprint at bioRxiv https://doi.org/10.1101/2021.08.19.456930 (2021).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Walker, M. A. et al. GATK PathSeq: a customizable computational tool for the discovery and identification of microbial sequences in libraries from eukaryotic hosts. Bioinformatics 34, 4287–4289 (2018).
Oskolkov, N. KrakenUniq database based on complete microbial genomes from NCBI RefSeq, May 2020. FigShare https://doi.org/10.17044/scilifelab.21299541.v1 (2022).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
McMurdie, P. J. & Holmes, S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217 (2013).
Acknowledgements
We thank the patients and their families, investigators, co-investigators and the study teams at each of the participating centers. We also thank A. Donahue (Pfizer) and P. Robbins (Pfizer) for their roles in establishing the research collaboration with Memorial Sloan Kettering Cancer Center and R.-Y. Tzeng (Memorial Sloan Kettering Cancer Center) for her role in data quality control. We thank colleagues at the Cleveland Clinic and Memorial Sloan Kettering core facilities, including the Cell Imaging Core (CCF), Genomics Core (CCF) and Integrated Genomics Operation (Memorial Sloan Kettering) for processing samples and providing important suggestions. We also thank colleagues at the Department of Pathology (CCF) for pathology advice and the members and alumni of the laboratory of T.A.C. at Memorial Sloan Kettering and CCF for their generous help and support of this study. Data used in this study were generated or collected by the TCGA Research Network. We acknowledge the following funding sources: NIH R01 CA205426 (T.A.C.), NIH R35 CA232097 (T.A.C.) and NIH/NCI U54 CA274513 (T.A.C.), the Sheikha Fatima bint Mubarak Chair (T.A.C.) and NIH/NCI Cancer Center Support Grant P30 CA008748. This trial was sponsored by Pfizer and was previously conducted under an alliance between Merck (CrossRef Funder ID: 10.13039/100009945) and Pfizer. Medical writing support was provided by H. Al-Ashtal of Clinical Thinking and was funded by Merck. The investigators worked with Pfizer on the trial design, collection and analysis of data and interpretation of results. Datasets were reviewed by the authors, and all authors participated fully in developing and reviewing the report for publication. All authors had full access to all data, and the first author had final responsibility for the decision to submit for publication. We vouch for the completeness and accuracy of the data and their analysis and the fidelity of the trial to the protocol and statistical analysis plan.
Author information
Authors and Affiliations
Contributions
Conceptualization: T.A.C., N.R., T.J.A. and N.Y.L. Methodology: T.J.A., N.R., R.I.H., V.M., M.S., P.H., P.B.P., A.N., M.G., D.H., I.J., D.C., A.G., J.K., D.J.M., Y.Z., N.L.S., C.B.D. and T.A.C. Investigation: T.J.A., N.R., R.I.H., C.B.D. and T.A.C. Clinical trial organization: R.I.H., N.Y.L., E.E.W.C., R.L.F., P.M.-H.C., J.-C.L. and A.P. Writing, original draft: N.R., T.J.A., C.B.D. and T.A.C. Writing, review and editing: N.R., T.J.A., M.S., R.I.H., N.Y.L. and T.A.C. Biostatistical review: M.S. Funding acquisition: T.A.C. Supervision: T.A.C., N.R. and C.B.D. All authors critically discussed and revised the paper for important intellectual content.
Corresponding authors
Ethics declarations
Competing interests
T.A.C. is a cofounder of Gritstone Oncology and holds equity. T.A.C. also holds equity in An2H. T.A.C. acknowledges grant funding from Bristol-Myers Squibb, AstraZeneca, Illumina, Pfizer, An2H and Eisai. T.A.C. has served as an advisor for Bristol-Myers, MedImmune, Squibb, Illumina, Eisai, AstraZeneca and An2H. T.A.C. is an inventor on intellectual property held by Memorial Sloan Kettering Cancer Center on using TMB to predict immunotherapy response, with pending patent, which has been licensed to PGDx. T.A.C. is on the advisory board of Cell. N.R. acknowledges research support from Pfizer, REPARE Therapeutics, Invitae and Bristol-Myers Squibb. The other authors declare no competing interests.
Peer review
Peer review information
Nature Cancer thanks N. Gopalakrishna Iyer, Bertrand Routy, Daniel Spakowicz and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Characterization of cohorts and association of TMB, PD-L1, and CD8 infiltrate with outcomes.
a. Comparable PFS outcomes are seen in the biomarker analysis cohorts relative to the full dataset. Cox proportional hazards model used to estimate hazard ratio (and 95% CI) between placebo and avelumab in subgroups with genomic data. b. Tumor mutation burden (TMB) is higher in HPV negative (median=5.37 Mut/Mb, IQR=3.95-7.23, N=272) vs. HPV positive (median=4.26 Mut/Mb, IQR=2.53-6.53, N=149) (p < 0.001; Wilcoxon-rank sum test). c. Associations between PFS, treatment, and classical biomarkers were comparable in HPV-positive and HPV-negative patients (Cox proportional hazards model and 95% CI). d. Association between PFS in placebo/CRT arm and expression of PD-L1 by tumor cells (left) vs immune cells (right). PD-L1 high vs low populations (log-rank test).
Extended Data Fig. 2 HPV-related features and genetic associations with outcomes.
a. Volcano plot showing mutations associated with HPV positive vs HPV negative cancers (Fisher’s Exact Test; dotted line represents p-value adjusted for false discovery). Recurrently mutated genes in SCCHN occur at markedly different frequencies between HPV+ (N=149) and HPV- (N=272) tumors. b. Oncoplot of patients identified as HPV-positive (N=149) by p16 status reveals that cases of low coverage by NGS of viral genomes (left side of oncoplot) are more likely to have mutations or copy number changes in TP53 and CDKN2A, suggesting these cases may not be driven by HPV.
Extended Data Fig. 3 Alternative immune deconvolution methods reveal consistent TME findings and T-cell repertoire identifies clonal differences in responders.
a. (left) Concordance between CytoPro and ssGSEA methods for selected signatures related to tertiary lymphoid structures (*** indicates p-value < 0.001) (right) Illustration of relationship between CytoPro signature (Z-normalized) and raw ssGSEA score for Neutrophils (linear regression, shaded region corresponds to 95% CI, N=342). b. Correlation between IHC CD8 staining (positive cells per total area) with ssGSEA CD8 T cell staining as a validation for ssGSEA measurement of CD8 T cells (r=0.61; p < 0.001, Pearson Correlation; shaded region corresponds to 95% CI, N=342). c. Hierarchical clustering of cell-type contributions estimated by ssGSEA. d. Associations between ssGSEA signatures and PFS (Cox proportional hazards model, 95% CI, and log-rank test, n=344 patients). Note significant association with Tfh (T follicular helper) cells and improved outcomes in Avelumab (N=176, p < 0.001; log-rank test) which is not significant in Placebo and has a significant interaction by treatment arm (N=166, p=0.02). In contrast, Neutrophils are associated with worse outcomes for patients receiving avelumab (p = 0.003; log-rank test).
Extended Data Fig. 4 Validation of WES derived TBB.
a. Spearman correlation of TBB between 2 analytic pipelines, PathSeq and KrakenUniq, demonstrates good correlation of bacterial burden from tumors (rho=0.93, N=421, p < 2.2*10−16) and normal tissue (rho=0.45; N=421, p < 2.2*10−16). b. Genera counts for each sample in cohort, ordered from highest tumor bacterial abundance (N=421). c. Frequency of species identified in this cohort and HNSC from TCGA are highly correlated (rho = 0.53; p < 0.001; Spearman Correlation). d. LogTBB derived from head and neck cancer cases in TCGA with both WGS and WES (n=155 cancers, with strong linear correlation r=0.88 p < 1*10−32). e Total bacterial read counts aligned to genera in tumor and PBMC samples (split by high vs. low TBB in tumor samples; Wilcoxon test, n=421patients) illustrates markedly higher bacterial reads in tumor compared to blood (p < 0.001; Wilcoxon test). f. Evaluation of log-scale abundance of bacterial reads from WES with T Stage for entire cohort (left) shows a trend towards an association with larger tumors (p=0.064; Wilcoxon Test, n=421 patients) but not N Stage (right, n=421 patients) (p=0.38). g. Evaluation of log-scale abundance of bacterial reads for each Phyla across n=18 samples in 16S compared to WES abundance, shows a strong linear relationship (r=0.97; p=0.00035; Pearson Correlation) h. Correlation of genera abundances between 16S and WES illustrates that samples from same tumor correlate strongly (N=16 tumors). i. Pearson correlation between 16S determined bacterial species and those identified from WES is significantly higher between corresponding samples, than from unrelated samples (Wilcox test, p=0.00016, n=8 cases). Boxplots show IQR (Q1–Q3); line = median; whiskers = most extreme points within 1.5×IQR).
Extended Data Fig. 5 RNAScope 16S IHC Validation.
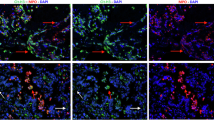
16S IHC staining by RNAscope of selected TBB high (n=4; left) and TBB low (n=4; right) tumors (red: 16S probe; blue DAPI). Note increased bacterial staining in TBB high tumors. Within each cohort, a scrambled 16S probe (red) was utilized as a negative control illustrating specificity of staining.
Extended Data Fig. 6 Relationship between TBB, gene expression, and cellular composition of the TME.
a. Expression of individual immune related genes and TBB. Note strong inverse relationship (linear regression) with gene markers of adaptive immune cells (CD8A, CD3E, PDCD1, CD19, CD79A) compared with neutrophil markers (NEAT1, NAMPT1, CXCL8) (n=342 cases). b. CD8 T cell abundance determined by IHC and split by TBB status. Increased CD8 T cell staining is noted in the TBB low samples (p < 0.001, N=323, Wilcoxon rank-sum test,). c. Total number of TCR clonotypes per sample is ordered from low to high and colored by TBB status, highlighting expansion of clones in TBB low samples. Boxplot shows IQR (Q1–Q3); line = median; whiskers = most extreme points within 1.5×IQR, N=304 tumors) d. Relative abundance of clones by size shows expansion of small clones in TBB high samples (avelumab high, placebo high), compared to low samples (n=310 patients, avelumab low, placebo low; all comparisons by Wilcoxon test, Yellow NA:NA are those samples for which TCR sequencing was available but TBB was not measured due to lack of WES for that sample). e. Patient TBB is associated with pre-treatment peripheral neutrophil count (R=0.21, p=0.001, N=421) in patients with high TBB but not in patients with low TBB (R=−0.06, p=0.43, Pearson Correlation, shaded region corresponds to 95% CI).
Extended Data Fig. 7 Association of TBB with outcomes of ICB.
a. Associations between TBB and PFS in patients treated with avelumab in HPV negative patients (left; p = 0.011) and HPV positive patients (right; p=0.077; both log-rank test) b. Multivariate cox proportional hazard model (and 95% CI) in Avelumab treated patients, identifies TBB as associated with PFS after correcting for stratification variables (HR = 2.03 p = 0.002, n=421 patients) c. Comprehensive cut-point analysis illustrates a wide range of TBB cutpoints associate with outcomes, including both the median or 99th% in patients treated with Avelumab (left). Each dot represents a log rank test at a specific cut point, with the y axis showing the p-value for the log-rank test. Every possible cutpoint was tested through the dataset and the summary is plotted. In (right) Placebo + SoC CRT arm TBB is not associated with outcomes regardless of the cutpoint. (N=421 pts) d. Histograms of BLAST E-values for remaining unaligned reads after alignment to GCHR38. Note unaligned reads have significant lower E-values aligning to bacterial genomes rather than alignment to a human genome. e. PFS in Avelumab/CRT arm split by TBB considering only Top 5 genera by abundance and known to be members of the oral microbiome (p=0.012; log-rank test).
Extended Data Fig. 8
Schematic showing workflow of elastic net modeling with nested cross-validation.
Supplementary information
Supplementary Table
Supplementary Tables 1 and 2.
Source data
Source Data Figs. 1–6
Statistical source data.
Source Data Extended Data Figs. 1–4, 6 and 7
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Riaz, N., Alban, T.J., Haddad, R.I. et al. Tumor ecosystem and microbiome features associated with efficacy and resistance to avelumab plus chemoradiotherapy in head and neck cancer. Nat Cancer 7, 98–115 (2026). https://doi.org/10.1038/s43018-025-01068-0
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43018-025-01068-0
This article is cited by
-
Tumor bacterial burden dictates immunotherapy fate in head and neck cancer
Nature Cancer (2026)