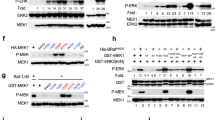
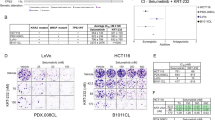
Abstract
MAPK pathway alterations are the most common oncogenic drivers. Among the approved therapies targeting this pathway are RAS, MEK and RAF inhibitors. However, therapeutic resistance and toxicities have limited their clinical success. Here, to overcome these liabilities, we developed IK-595, a potent MEK–RAF molecular glue. IK-595 traps MEK in an inactive complex with all RAF isoforms. In addition, IK-595 precludes CRAF-mediated MEK reactivation and ARAF heterodimerization, allowing for prolonged target engagement and durable MAPK pathway inhibition. This translates into superior antitumor activity across a wide range of cancer model indications harboring MAPK pathway alterations. A key advantage of IK-595 is its ability to achieve transient high plasma exposure affording a larger therapeutic window. The unique mechanism of action and improved tolerability positions IK-595 as an ideal combination partner. IK-595 is an MEK–RAF molecular glue that prolongs pathway inhibition while providing a broader therapeutic window as monotherapy and in combination.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Ryan, M. B. & Corcoran, R. B. Therapeutic strategies to target RAS-mutant cancers. Nat. Rev. Clin. Oncol. 15, 709–720 (2018).
Sugiura, R., Satoh, R. & Takasaki, T. ERK: a double-edged sword in cancer. ERK-dependent apoptosis as a potential therapeutic strategy for cancer. Cells 10, 2509 (2021).
Unal, E. B., Uhlitz, F. & Bluthgen, N. A compendium of ERK targets. FEBS Lett. 591, 2607–2615 (2017).
AACR Project GENIE Consortium AACR project GENIE: powering precision medicine through an international consortium. Cancer Discov. 7, 818–831 (2017).
Schubbert, S., Shannon, K. & Bollag, G. Hyperactive Ras in developmental disorders and cancer. Nat. Rev. Cancer 7, 295–308 (2007).
Wan, P. T. et al. Mechanism of activation of the RAF–ERK signaling pathway by oncogenic mutations of B-RAF. Cell 116, 855–867 (2004).
Deschenes-Simard, X., Kottakis, F., Meloche, S. & Ferbeyre, G. ERKs in cancer: friends or foes? Cancer Res. 74, 412–419 (2014).
Yaeger, R. & Corcoran, R. B. Targeting alterations in the RAF–MEK pathway. Cancer Discov. 9, 329–341 (2019).
Bamford, S. et al. The COSMIC (catalogue of somatic mutations in cancer) database and website. Br. J. Cancer 91, 355–358 (2004).
Jones, J. C. et al. Non-V600BRAF mutations define a clinically distinct molecular subtype of metastatic colorectal cancer. J. Clin. Oncol. 35, 2624–2630 (2017).
Owsley, J. et al. Prevalence of class I–III BRAF mutations among 114,662 cancer patients in a large genomic database. Exp. Biol. Med. (Maywood) 246, 31–39 (2021).
Lito, P. et al. Disruption of CRAF-mediated MEK activation is required for effective MEK inhibition in KRAS mutant tumors. Cancer Cell 25, 697–710 (2014).
Venkatanarayan, A. et al. CRAF dimerization with ARAF regulates KRAS-driven tumor growth. Cell Rep. 38, 110351 (2022).
Khan, Z. M. et al. Structural basis of the action of the drug trametinib at KSR-bound MEK. Nature 588, 7838 (2020).
Ryan, M. B. et al. The Pan-RAF–MEK nondegrading molecular glue NST-628 is a potent and brain-penetrant inhibitor of the RAS–MAPK pathway with activity across diverse RAS- and RAF-driven cancers. Cancer Discov. 14, 7 (2024).
Infante, J. R. et al. Safety, pharmacokinetic, pharmacodynamic, and efficacy data for the oral MEK inhibitor trametinib: a phase 1 dose-escalation trial. Lancet Oncol. 13, 773–781 (2012).
Sinkala, M., Nkhoma, P., Mulder, N. & Martin, D. P. Integrated molecular characterisation of the MAPK pathways in human cancers reveals pharmacologically vulnerable mutations and gene dependencies. Commun. Biol. 4, 9 (2021).
Garutti, M. et al. BRAF and MEK inhibitors and their toxicities: a meta-analysis. Cancers (Basel) 15, 141 (2022).
Rasco, D. W. et al. Phase 1 study of the pan-RAF inhibitor tovorafenib in patients with advanced solid tumors followed by dose expansion in patients with metastatic melanoma. Cancer Chemother. Pharmacol. 92, 15–28 (2023).
Awada, G. et al. A phase 2 clinical trial of trametinib and low-dose dabrafenib in patients with advanced pretreated NRASQ61R/K/L mutant melanoma (TraMel-WT). Cancers (Basel) 13, 2010 (2021).
Yen, I. et al. ARAF mutations confer resistance to the RAF inhibitor belvarafenib in melanoma. Nature 594, 418–423 (2021).
Carlino, M. S. et al. Antiproliferative effects of continued mitogen-activated protein kinase pathway inhibition following acquired resistance to BRAF and/or MEK inhibition in melanoma. Mol. Cancer Ther. 12, 1332–1342 (2013).
Long, G. V. et al. Increased MAPK reactivation in early resistance to dabrafenib/trametinib combination therapy of BRAF-mutant metastatic melanoma. Nat. Commun. 5, 5694 (2014).
Center for Drug Evaluation and Research. Clinical Pharmacology and Biotherapeutics Review. Application number: 204114Orig1s000. FDA https://www.accessdata.fda.gov/drugsatfda_docs/nda/2013/204114orig1s000clinpharmr.pdf (2013).
Barbato, M. I. et al. FDA approval summary: dabrafenib in combination with trametinib for BRAFV600E mutation-positive low-grade glioma. Clin. Cancer Res. 30, 263–268 (2024).
Casey, D. et al. FDA approval summary: selumetinib for plexiform neurofibroma. Clin. Cancer Res. 27, 4142–4146 (2021).
Salama, A. K. S. et al. Dabrafenib and trametinib in patients with tumors with BRAFV600E mutations: results of the NCI-MATCH trial subprotocol H. J. Clin. Oncol. 38, 3895–3904 (2020).
Guo, S., Jiang, X., Mao, B. & Li, Q. X. The design, analysis and application of mouse clinical trials in oncology drug development. BMC Cancer 19, 718 (2019).
Ryan, M. B. et al. KRASG12C-independent feedback activation of wild-type RAS constrains KRASG12C inhibitor efficacy. Cell Rep. 39, 110993 (2022).
Hagenbeek, T. J. et al. An allosteric pan-TEAD inhibitor blocks oncogenic YAP/TAZ signaling and overcomes KRAS G12C inhibitor resistance. Nat. Cancer 4, 812–828 (2023).
Monaco, K. A. et al. LXH254, a potent and selective ARAF-sparing inhibitor of BRAF and CRAF for the treatment of MAPK-driven tumors. Clin. Cancer Res. 27, 2061–2073 (2021).
Martinez-Garcia, M. et al. First-in-human, phase I dose-escalation study of the safety, pharmacokinetics, and pharmacodynamics of RO5126766, a first-in-class dual MEK/RAF inhibitor in patients with solid tumors. Clin. Cancer Res. 18, 4806–4819 (2012).
Garnett, M. J., Rana, S., Paterson, H., Barford, D. & Marais, R. Wild-type and mutant B-RAF activate C-RAF through distinct mechanisms involving heterodimerization. Mol. Cell 20, 963–969 (2005).
Yao, Z. et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature 548, 234–238 (2017).
Dahlman, K. B. et al. BRAFL597 mutations in melanoma are associated with sensitivity to MEK inhibitors. Cancer Discov. 2, 791–797 (2012).
Kim, K. B. et al. Phase II study of the MEK1/MEK2 inhibitor trametinib in patients with metastatic BRAF-mutant cutaneous melanoma previously treated with or without a BRAF inhibitor. J. Clin. Oncol. 31, 482–489 (2013).
Dumaz, N. et al. In melanoma, RAS mutations are accompanied by switching signaling from BRAF to CRAF and disrupted cyclic AMP signaling. Cancer Res. 66, 9483–9491 (2006).
Awad, M. M. et al. Acquired resistance to KRASG12C inhibition in cancer. N. Engl. J. Med. 384, 2382–2393 (2021).
Acknowledgements
We received no specific funding for this work.
Author information
Authors and Affiliations
Contributions
E.H. conceptualized the study. M.B. was the medicinal chemistry lead. E.H., R.C., B.L., A.Y., M.S.-M. and V.D.J. designed the experiments. R.C., S.R.W., D.H. and B.L. performed the in vitro assays and analyzed the data. A.Y. led the structural biology efforts. V.D.J. and J.C. conducted the in vivo studies with pharmacokinetic assistance from J.D.M. and W.B. A.I. carried out the bioinformatics analysis. S.S. and W.B. provided essential scientific guidance. S.K.R., J.E. and M.X.Z. supervised the project. E.H. and J.E. wrote the manuscript with input from all authors. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
All authors are previous employees and shareholders of Ikena Oncology.
Peer review
Peer review information
Nature Cancer thanks Evripidis Gavathiotis, Sandra Misale and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Medicinal chemistry optimization schematic for the discovery of IK-595.
IK-595 was discovered through structure-activity relationship (SAR) optimization and was structurally enabled by x-ray analysis of MEK1 and BRAF co-crystals. Multiple known MEK binding chemical scaffolds including that of trametinib were used as starting points and informed the ultimate selection of IK-595. The SAR optimization focused on three key objectives; 1) identifying molecules that bound tightly within the MEK allosteric pocket, 2) identifying molecules that made tight interactions with the RAF isoforms, and 3) identifying molecules that had moderate pharmacokinetic clearance. The internal fused biaryl ring system of a trametinib-like chemical series that led to IK-595 was opened in order to enhance the pharmacokinetic clearance relative to closed ring molecules. Multiple heteroatom, alkyl, fluoroalkyl, nitrile, and ether substitutions in the core as well as heteroatom and ring size adjustments in the terminal ring system were explored to maximize MEK binding and to further optimize the pharmacokinetic profile. The addition of the cloro atom in the terminal ring system was key to achieving the optimal balance of clearance and off-rate binding. In addition, based on the binding orientation, both oxygens of the sulfamide group created hydrogen bonds with RAF thereby enabling IK-595’s ability to function as a tight MEK-RAF molecular glue.
Extended Data Fig. 2 Synthetic scheme for the synthesis of IK-595.
Step 1: 5-(2-chloro-3-nitrophenoxy)-3-cyclopropyl-1-(2-jluoro-4-iodophenyl)-6,8-dimethylpyridof 2,3-d]pyrimidine-2,4,7 (1H,3H, 8H)-trione. To a stirred mixture of 3-cyclopropyl-l-(2-fluoro-4-iodophenyl)-6,8-dimethyl-2,4,7-trioxopyrido[2,3-d]pyrimidin-5-yl trifluoromethanesulfonate (1.00 g, 1.62 mmol, 1.0 equiv) and 2-chloro-5-nitrophenol (0.85 g, 4.87 mmol, 3.0 equiv) in THF, cesium fluoride (0.74 g, 4.87 mmol, 3.0 equiv) was added in portions at 60 °C over 30 min. The reaction mixture was stirred for an additional 30 min at 60 °C. The resulting mixture was diluted with water (30 mL) and extracted with EA (3 ×30 mL). The combined organic layers were washed with brine (50 mL) and dried over anhydrous Na2SO4. After filtration, the filtrate was concentrated under reduced pressure. The residue was purified by silica gel column chromatography, eluted with PE/THF (1:1) resulting in 5-(2-chloro-3-nitrophenoxy)-3-cyclopropyl-1-(2-fluoro-4-iodophenyl)-6,8-dimethylpyrido[2,3-d]pyrimidine-2,4,7-trione (540 mg, 52%) as a light yellow solid; ES-LCMS m/z 639 [M + H]. Step 2: 5-(3-amino-2-chlorophenoxy)-3-cyclopropyl-1-(2-jluoro-4-iodophenyl)-6,8- dimethylpyridof 2,3-d]pyrimidine-2,4, 7(1H,3H,8H)-trione. To a stirred mixture of 5-(2-chloro-3-nitrophenoxy)-3-cyclopropyl-l-(2-fluoro-4-iodophenyl)-6,8-dimethylpyrido[2,3-d]pyrimidine-2,4,7-trione (540 mg, 0.84 mmol, 1.0 equiv) in AcOH, iron (472 mg, 8.45 mmol, 10.0 equiv) was added in portions at 50 °C. The resulting mixture was stirred for additional 30 min at 50 °C. The mixture was diluted with water (50 mL) and slowly neutralized to pH 7 with saturated NaHCO3 (aq.), then extracted with EA (3 ×20 mL). The combined organic layers were washed with brine (50 mL) and dried over anhydrous Na2SO4. After filtration, the filtrate was concentrated under reduced pressure. This resulted in 5-(3-amino-2-chlorophenoxy)-3-cyclopropy 1-1-(2-fluoro-4-iodophenyl)-6,8-dimethylpyrido[2,3-d]pyrimidine-2,4,7-trione (195 mg, crude, 37%) as a light brown solid; ES-LCMS m/z 609 [M + H]. Step 3: 5-{2-chloro-3-[(methylsulfamoyl)amino]phenoxy}-3-cyclopropyl-1-(2-fluoro-4-iodophenyl)-6,8-dimethylpyrido[2,3-d]pyrimidine-2,4,7-trione. To a stirred mixture of 5-(3-amino-2-chlorophenoxy)-3-cyclopropy 1-1-(2-fluoro-4-iodophenyl)-6,8-dimethylpyrido[2,3-d]pyrimidine-2,4,7-trione (195 mg, crude, 0.32 mmol, 1.0 equiv) in DCM, triethylamine (64 mg, 0.64 mmol, 2.0 equiv) was added at 0 °C, then Nmethylsulfamoyl chloride (83 mg, 0.64 mmol, 2.0 equiv) was added dropwise over 2 min at 0 °C. The reaction mixture was stirred for additional 30 min at 0 °C. The reaction was quenched with water (10 mL) and then extracted with EtOAc (3×10 mL). The organic layer was dried over anhydrous Na2SO4, filtered and the filtrate was concentrated under reduced pressure to afford 5-{2-chloro-3-[(methylsulfamoyl)amino]phenoxy}-3-cyclopropyl-1-(2-fluoro-4-iodophenyl)-6,8-dimethylpyrido[2,3-d]pyrimidine-2,4,7-trione (130 mg, crude, 57%). The crude product mixture was used in the next step directly without further purification; ES-LCMS m/z 702 [M + H]. Step 4: 4-(2-chloro-3-((N-methylsulfamoyl)amino)phenoxy)-N-cyclopropyl-2-((2-fluoro-4-iodophenyl)amino)-1,5-dimethyl-6-oxo-1,6-dihydropyridine-3-carboxamide. To a stirred solution of 5-{2-chloro-3-[(methylsulfamoyl)amino]phenoxy}-3-cyclopropyl-1-(2-fluoro-4-iodophenyl)-6,8-dimethylpyrido[2,3-d]pyrimidine-2,4,7-trione (130 mg, 0.18 mmol, 1.0 equiv) in THF (1.0 mL) and water (1.0 mL), LiOH (44 mg, 1.85 mmol, 10.0 equiv) was added in portions at room temperature. The reaction mixture was stirred for an additional 30 min at room temperature. The resulting mixture was diluted with water (10 mL) and extracted with EA (3 ×10 mL). The combined organic layers were washed with brine (1 ×20 mL), dried over anhydrous Na2SO4. After filtration, the filtrate was concentrated under reduced pressure. The crude product was purified by Prep-HPLC [Column: YMC C18 30*150 mm, 5 uM, Mobile Phase A: Water(0.05%NH3.H2O), Mobile Phase B: ACN; Flow rate: 35 mL/min; Gradient: 40% B to 75% B in 8 min, Wave Length: 254/220 nm] to afford 4-{2-chloro-3-[(methylsulfamoyl)amino]phenoxy}-N-cyclopropyl-2-[(2-fluoro-4-iodophenyl)amino]-1,5-dimethyl-6-oxopyridine-3-carboxamide (51 mg, 41 %) as an off-white solid; ES-LCMS m/z 676.1 [M + H]. 1HNMR (400 MHz, DMSO-d6) 8 9.01 (br, s, lH), 8.41 (br, s, lH), 7.98 (d, J = 3.6 Hz, 1H), 7.59-7.51 (m, 1H), 7.39-7.27 (m, 2H), 7.23-7.12 (m, 2H), 6.63-6.55 (m, 1H), 6.55-6.48 (m, 1H), 3.36 (s, 3H), 2.53 (d, J = 2.6 Hz, 3H), 2.21-2.11 (m, 1H), 1.76 (2, 3H), 0.42-0.33 (m, 2H), -0.01—0/05 (m, 2H).
Extended Data Fig. 3 IK-595 has prolonged inhibition of MEK and ERK1/2 phosphorylation following compound washout.
A. Western blot analysis of phosphorylation MEK and ERK1/2 and total protein levels of MEK and ERK1/2 from HCT-116 cell lysates taken a 0, 0.5, 1, 2, 4, 6, and 24 hours after washout following treatment of HCT-116 cells with DMSO, trametinib (10 nM), or IK-595 (3 nM) for 1 hour. Representative of 2 independent experiments B. Quantification of MEK and ERK1/2 phosphorylation from (A). All phosphorylation values were first normalized to total MEK or ERK1/2 protein expression, then normalized to DMSO control.
Extended Data Fig. 4 Crystal structural determination of IK-595 interactions with MEK1 and BRAF.
A. The 1-fluoro-3-iodobenzene moiety of IK-595 interacts with residues I99, V127, M143, I141, F129, L118, F209, L141, L115, V211, L215, I216 and M219. This network of hydrophobic contacts provides IK-595 with a high binding affinity to MEK. In addition to these hydrophobic contacts, IK-595 establishes several polar interactions with the backbones of residues from N78, G79, V127, His188, D190, V211 and the side chains of residues K97, M143 and R189. These interactions contribute further to the compound’s specificity and stability within the allosteric pocket. B. At the MEK-BRAF interface, the sulfonamide group of IK-595 extends out from MEK to interact directly with residues N660, N661 and R662 of BRAF. Notably, IK-595 forms polar interactions and hydrogen bonds with MEK1 arginine 234, and BRAF asparagine 660 and arginine 662, acting as a “molecular glue” that enhances the stability of the MEK-BRAF complex. C. Avutometinib occupies a similar allosteric site to that of IK-595. Given the significant difference in the chemical scaffold, avutometinib establishes fewer contacts with surrounding residues leading to a less stable binding mode compared to IK-595. D. Structural analysis by Molecular Operating Environment (MOE) demonstrates that IK-595, due to its increased number of rotatable bonds and extended side chains makes more interactions than avutometinib with residues within the allosteric pocket. These features enable IK-595’s enhanced filling and binding to MEK.
Extended Data Fig. 5 IK-595 demonstrates differentiated dose-dependent tumor growth inhibition and tumor pharmacodynamic modulation.
A. Mean tumor volumes ± SD in AsPC-1 tumor-bearing mice (n = 8 mice/group) treated with vehicle (5% DMSO/95% PEG400), or IK-595 at 3, 1, 0.3, or 0.1 mg/kg PO, QD. One-way ANOVA with a Tukey’s multiple comparison test. B. Mean tumor volumes ± SD in AsPC-1 tumor-bearing mice (n = 8 mice/group) treated with vehicle (5% DMSO/95% PEG400), or IK-595 at 6, 3, 1, or 0.3 mg/kg PO, QOD. One-way ANOVA with a Tukey’s multiple comparison test. C. Mean ERK1/2 phosphorylation levels normalized to total ERK1/2 protein levels ± SD measured by an MSD assay in AsPC-1 tumors taken at 4 or 24 hours (n = 4 tumors/group) following a single dose of vehicle (5% DMSO/95% PEG400), or IK-595 at 3, 1, 0.3, or 0.1 mg/kg PO. Significance was determined by a One-Way ANOVA with a Dunnett’s multiple comparisons test. D. Mean DUSP6 mRNA levels normalized to GAPDH mRNA levels ± SD measured by RT-PCR in AsPC-1 tumors described in (A). Significance was determined using a Two-way ANOVA with a Tukey’s multiple comparison test. E. Mean tumor volumes ± SD (left panel) and percent body weight change ± SD (right panel) in NCI-H441 tumor-bearing mice (n = 10 mice/group) treated with vehicle (5% DMSO/95% PEG400), or IK-595 at 2 mg/kg QD, 6 mg/kg QOD, or 6 mg/kg Q3D. One-way ANOVA with a Tukey’s multiple comparison test.
Extended Data Fig. 6 IK-595 demonstrates improved efficacy and tolerability compared to avutometinib.
A. Mean tumor volumes ± SD in NCI-H2122 tumor-bearing mice (n = 10 mice/group) treated with vehicle (5% DMSO/95% PEG400), avutometinib 0.3 or 3 mg/kg PO, QD, or IK-595 at 3 mg/kg PO, QD. One-way ANOVA with a Tukey’s multiple comparison test. B. Mean body weight measurements ± SD from mice in (A).
Extended Data Fig. 7 IK-595-sotorasib combination attenuates the RAS/MAPK pathway.
A. KRAS-GTP pulldown assay evaluating expression of activated KRAS levels in NCI-H358 (n = 3, in a single experiment) (left panel) and SW837 (n = 3, in a single experiment) (right panel) treated with DMSO, sotorasib alone (1000 nM), IK-595 alone (3 nM), or sotorasib +IK-595 for 48 hours. B. Heatmap depicting the alterations in mRNA expression of RAS/MAPK pathway genes in parental NCI-H358 and NCI-H358-R cell lines untreated or exposed to constant sotorasib (250 nM). C. KRAS-GTP pulldown assay evaluating expression of activated KRAS levels in NCI-H358, and NCI-H358-R cell lines treated with DMSO, IK-595 (3 nM), sotorasib (250 nM and 1000 nM), or IK-595 + sotorasib (250 and 1000 nM) combinations for 48 hours. (n = 3, in a single experiment) D. Western blot analysis measuring KRAS. Vinculin, Total ERK1/2 and phospho-ERK1/2 expression levels in NCI-H358 parental and NCI-H358R cell lines. All treatments were for 24 hours. Data is from a single experiment.
Extended Data Fig. 8 IK-595 combinations with inhibitors within the MAPK pathway, mediators of resistance, and standard of care chemotherapy.
Loewe Sum of Synergy Scores of IK-595 combinations with BI-3406, inavolisib, and RMC-4550 (NCI-H1373, HPAF, and NCI-H2122 cell lines), LX-254 (HPAF, NCI-H2122, and AsPC-1 cell lines), IK-930 (HCT-116 and SK-MEL-2 cell lines), everolimus (NCI-H1373, NCI-H2122, and Mia-PACA-2 cell lines), lapatinib and tucatinib (SKCO-1, LS180, HCT-116 cell lines), cetuximab (HCA7, DLD-1, and HT-29 cell lines), paclitaxel (HPAC, HPAF-II, and DANG cell lines), and gemcitabine (PSN-1, DANG, and AsPC-1 cell lines). All synergy scores were calculated from data obtained from a 5-day CTG assay using the integrated synergy score algorithm from the online tool ComBenefit. An integrated score of over 10 is considered synergistic.
Supplementary information
Supplementary Table 1
Cell lines were obtained from commercial vendors as indicated and were cultured under conditions recommended by the supplier. All cell lines were confirmed to be free of Mycoplasma contamination before use.
Supplementary Table 2
X-ray crystallographic data collected for the MEK–BRAF–IK-595 ternary complex. Data collection and refinement statistics are summarized, including resolution, space group, unit-cell dimensions, R factors and model validation metrics.
Source data
Source Data Fig. 1
AlphaLisa statistical source data.
Source Data Fig. 2
Mass spectrometry protein quantification.
Source Data Fig. 3
Graph raw values.
Source Data Fig. 4
Graph raw values.
Source Data Fig. 5
Graph raw values and statistical source data.
Source Data Fig. 6
Graph raw values and statistical source data.
Source Data Fig. 7
Graph raw values and statistical source data.
Source Data Fig. 8
Graph raw values and statistical source data.
Source Data Extended Data Fig. 3
Western blot quantifications.
Source Data Extended Data Fig. 5
Graph raw values.
Source Data Extended Data Fig. 6
Graph raw values.
Source Data Extended Data Fig. 7
Graph raw values.
Source Data Extended Data Fig. 8
Graph raw values.
Source Data Raw Blots
Images of unedited western blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Haines, E., Burke, M., Catterall, R. et al. The MEK–RAF molecular glue IK-595 has potent antitumor activity across RAS/MAPK pathway-altered cancers. Nat Cancer 7, 116–130 (2026). https://doi.org/10.1038/s43018-025-01081-3
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43018-025-01081-3
This article is cited by
-
A molecular glue halts RAS–MAPK signaling
Nature Cancer (2026)