Abstract
The contribution of deterministic versus stochastic processes to the spatio-temporal assembly of soil microbial communities in tropical forests requires quantification so that responses to climate change may be accurately projected. Here we report the spatio-temporal composition of soil fungal communities in a topographically homogeneous forest area in central Amazonia. Soil fungal communities have a greater turnover in space than over time. Stochastic processes are inferred to dominate in the rainy season and deterministic processes in the dry season. Our study highlights the importance of spatial heterogeneity in the absence of environmental gradients and its relationship with seasonal patterns that modulate spatial heterogeneity and contribute to environmental determinism versus stochasticity for fungal community assembly. This baseline may serve to assess deviations in community patterns caused by changes in biotic interactions with above-ground vegetation, such as those resulting from shifts in taxonomical/functional composition of trees driven by climate change.
Similar content being viewed by others
Introduction
Ecological communities are characterised by deterministic/niche-based processes, which shape species assemblages based on the differences in the selection of species in response to abiotic and biotic factors (e.g., climate, soil, vegetation, direct and soil-mediated species interactions), and by neutral1 and stochastic processes, including dispersal, ecological drift and other non-deterministic processes2,3,4. How the occurrence and/or the abundance of species within a community varies in space and time may inform about the relative importance of deterministic vs. stochastic processes for community assembly. Both processes are modulated by the interaction between spatial variability in the environment and biological processes (e.g., biotic interactions and demographic processes), which, in turn, are influenced by temporal patterns in resource availability. The rate of replacement and the relative abundance of taxa in a community can be described by the above variables/processes, more readily for organism groups with short generation times such as microorganisms5,6,7,8, and for contrasting environments, associated for example with climate9,10 or topography-related soil and hydrological properties11,12,13.
Lowland tropical rain forests are very diverse in their tree species composition. The corresponding variation in biochemical attributes (leaf chemistry, root exudates) contributes to a high degree of small-scale spatial heterogeneity of resources14 for soil microbial communities, influencing their biotic interactions and coexistence patterns. Animals also contribute to spatial heterogeneity and niche differentiation for soil bacteria15 and fungi16, by creating transient, but frequent nutrient-enriched patches by their excreta. An additional factor related to temporal variability in soil microbial community composition and assembly is attributable to seasonality of rainfall and nutrient/chemical input by litterfall17.
While recent studies have provided new insights on the links between soil fungal community structure/composition in lowland evergreen tropical rain forests and factors related to the environment18,19 and biotically mediated environmental patterns20,21,22, few studies have attempted to identify the simultaneous role of spatial and temporal variation in community composition of microbes in tropical rain forests23. In a recent study conducted across the island of Hainan, China, Wei, et al.24 have shown that spatial heterogeneity exerts a stronger influence on soil microbial community assembly than precipitation seasonality. The patterns, observed in that study conducted at the regional scale, were, however, underlain by obviously strong climatic and edaphic differences and thus masked mechanisms of community assembly associated with the intrinsic tree species diversity of lowland evergreen tropical rain forests.
The lack of knowledge on spatio-temporal community assembly in tropical rain forest soils hinders the capacity to project linked plant and soil microbial community responses to climate anomalies and extreme events, and departures from historical climatic trends. The duration, frequency and severity of drought events, for example, are projected to increase across the tropics, and particularly in some parts of the Amazon basin25,26. The consequences for soil microbial communities/processes and feedback on climate change are yet little known. Fungal communities could be affected, for example, by climate-driven changes in biotic interactions with above-ground vegetation, such as those resulting from shifts in taxonomical/functional composition of trees27. A quantitative understanding of how fungal communities vary in space and time for establishing their natural baseline variability is imperative for projecting and assessing significant departures in biogeochemical cycles and overall ecosystem function in response to future weather extremes and climate changes28.
A varying degree of precipitation seasonality in tropical lowland evergreen rain forests leads to different degrees and duration of a negative hydrological balance. The alternance between dry and wet seasons influences phenological cycles, nutrient availability29,30 and photosynthetic activity31, and, in turn, ecosystem processes. Litterfall and fine root turnover dynamics are related to seasonality, the dry season having the greatest rates of litterfall32,33,34 and new leaf flush35,36, and the rainy season the largest production of fine root biomass37,38,39. Rainy and dry seasons are also associated with different rates of soil respiration38,40,41, decomposition and mineralisation34,42, these processes being generally slowed down in the dry season.
Litterfall and fine root turnover induce changes in plant nutrient uptake and re-translocation of nutrients/carbohydrates in the dry season43, provide decomposable material, and contribute to large fluxes of dissolved organic matter (OM) early in the rainy season32. In tropical lowland rain forests in Amazonia, trees have been shown to preferentially allocate carbon to roots during the rainy season and to leaves in the dry season43. The seasonal trade-off in carbon allocation between roots and leaves affects the dynamics of the composition and quantity of root exudates, which, in a localised manner can be related to microbial community dynamics44.
Distance/environmental decay patterns of soil fungal community similarity in tropical rain forests are likely to be characterised by different processes at different spatial and temporal extents. Recent studies have shown that the relative contribution of niche-based vs. stochastic processes in determining fungal community assembly is scale-dependent45. Additionally to extent, the grain size dependence of observed distance and environmental decay patterns of soil fungi has been shown by Peguero, et al.46 along topographic gradients (plateaux, slopes and valley bottoms) in French Guyana, a landscape pattern that also characterises tropical rain forests in most of the Amazon basin. The study by Peguero, et al., however, did not investigate the temporal dynamics in spatial patterns. The selection of an obvious topography-driven soil and hydrological gradient yielded results which, while are interesting, reaffirm differences in soil and hydrological factors and associated ecological communities (trees and microbes, see e.g., de Oliveira Freitas, et al.47) that are linked to patterns determined by topography. Knowledge on the relative influence of deterministic and stochastic processes on fungal community assembly not influenced by obvious environmental gradients is lacking.
To address this knowledge gap, we followed the conceptual framework proposed by Vellend4, according to which ecological communities are controlled by four ecological processes, i.e., selection, ecological drift, dispersal and speciation, and used two complementary approaches to study the relative contribution of deterministic and stochastic processes to fungal community composition. Accordingly, we used the term ‘deterministic process’ to refer to any ecological process that involves non-aleatory niche-based mechanisms (i.e., selection and speciation), and the term ‘stochastic process’ to refer to the processes of dispersal and drift, being aware of the fact that some processes (e.g., dispersal limitation) may influence both stochastic and deterministic community assembly48.
We first characterised distance/environmental decay patterns of soil fungal communities to estimate the contribution of deterministic vs. stochastic processes in space and time to soil fungal community assembly. We then complemented the results on decay patterns by using another null model-based approach to obtain quantitative information on community assembly processes. We asked what the contribution of deterministic vs. stochastic processes to soil fungal community assembly would be like in a species-rich and diverse tropical rain forest, in the absence of topo-pedo-hydrological gradients and if these patterns could be related to seasonality in precipitation and tree phenology. We hypothesised that (H1) the turnover of fungal communities in space would be greater than in time (within-year). This is because the direct effect of fluctuations in soil moisture and soil pore water to air ratio would be counterbalanced by seasonal litter/root turnover patterns that are likely to strengthen spatial patterns related to the biogeochemical footprint of trees14, and which may alter biotic interactions between fungi and trees. We also hypothesised that (H2) the relative contribution of environmental determinism and stochastic processes to soil fungal community assembly would change with season, and that deterministic processes would dominate in the wet season. This is because in the wet season trees preferentially allocate carbon to roots, and thus impact the seasonal dynamics of rhizodeposition, and because of the increased rates of decomposition. These two factors were thus expected to result in a more deterministic environment than that exists in the dry season.
To test the above hypotheses, we characterised the fungal communities along a transect of a 1.5-km-long section in a plateau area in a central Amazonian tropical rain forest, three times during a year: in the wet season (Time 1), in the transition to the dry season (Time 2) and at the end of the dry season (Time 3). We found (i) a higher soil fungal community turnover in space than in time, (ii) a season-dependent contribution of stochastic processes vs. environmental determinism to community assembly, the first governing community assembly in the rainy season and the latter in the dry season (Table 1).
RESULTS
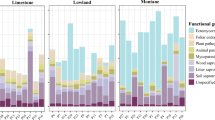
Of the total sequences, 48.3% (992 Amplicon Sequence Variants, ASVs) were assigned to a unique functional group and 98.7% (2028 ASVs) were classifiable to some taxonomic level. Fungal communities were dominated by saprotrophs (75.6%; 750 ASVs) followed by pathogens (12.6%; 125 ASVs), endophytes (1.8%; 18 ASVs) and symbionts (1.5%; 15 ASVs). Symbionts were likely to have been underrepresented due to the use of soil fungal primers that are not designed to amplify arbuscular mycorrhizal fungi that represent the dominant symbiosis type in these tropical rain forests. Significant differences in α-diversity and/or abundance of dominant functional groups between sampling times were observed for saprotrophs, pathogens, and endophytes (Fig. 1). The α-diversity of saprotrophs and pathogens was significantly higher at Time 3 when compared to Time 1. Pathogens and endophytes showed larger values of α-diversity at Time 2 when compared to Time 1. On the other hand, lower saprotroph abundance was observed at Time 3 when compared with that at Time 1, while pathogen abundance was significantly greater at Time 3 than at Time 2.
Fuchsia, rainy season; turquoise, transition into dry season; yellow, dry season. Richness, number of Amplicon Sequence Variants (ASVs) in each taxonomic group; total number for all samples per functional group in upper left corner of panels; total number per season inside plotted boxes; relative abundance, the proportion of reads. The complete pool assigned to a functional group consisted of 992 ASVs. Significant differences as determined by paired t-tests or Wilcoxon signed-rank tests, corrected for multiple comparisons using the false discovery rate method are indicated by grey arrows; * p < 0.05. Boxplots: centre line, median; empty square, mean value; box limits, 25th and 75th percentiles; whiskers, 1.5 times interquartile range; empty circles, outliers. The y-axis values for richness are number of ASVs and percent for relative abundance.
At the phylum level, the sequences were dominated by Ascomycota (67.3%), followed by Mucoromycota (21.4%) and Basidiomycota (9.9%). All other phyla including Chytridiomycota, Glomeromycota, Mortierellomycota and Rozellomycota, were represented by <1% of the total number of reads. Unidentified reads and unidentified ASVs accounted for 1.4% of the total number of reads and for 3.9% of total ASVs. The highest number of ASVs was also recorded for Ascomycota (1,406, 75,6%), followed by Basidiomycota (323, 17.4%) and Mucoromycota (32, 1.7%). Ascomycota, Basidiomycota and Mucoromycota showed significantly greater values of α-diversity at Time 2 than at Time 1; the α-diversity of Ascomycota and Basidiomycota differed significantly between Times 1 and 3 being greater at Time 3 (Supplementary Fig. 1). Significant shifts in relative abundance (RA) were observed among the three sampling times for the three main phyla. The RA of Ascomycota was lower at Time 2 than at Time 1 and greater at Time 3 than at Time 2, while the RA of Mucoromycota was higher at Time 2 than at Times 1 and 3 (Supplementary Fig. 1).
Soil analyses
Among soil properties, mineral nitrogen, soil pH, potassium (K+), cation exchange capacity (CEC) and exchangeable acidity (hydrogen, H+ plus aluminium, Al3+) differed significantly among sampling dates (Table 2). Both ammonium (NH4+) and nitrate (NO3−) were significantly lower at Time 2 than at Time 1 and higher at Time 3 than at Times 2 or 1. The values of pH increased successively from Time 1 to Time 3. Both CEC and H+ plus Al3+ decreased from Time 1 to Time 3 and from Time 2 to Time 3 while K+ showed an opposite pattern and was significantly higher at Time 3 than at Times 1 and 2. Fine root biomass was significantly higher at Time 1 than at Times 2 and 3.
Spatio-temporal turnover of fungal communities
Spatial and temporal differences in fungal communities were strongly related to the turnover of ASVs across space and over time. Such community turnover (ßsim) contributed spatially (overall plot-wise comparisons) from 91.9% (Time 1) to 94.8% (Time 2) of ß-diversity (Time 1: ßsim, 0.76, nestedness or ßsne, 0.05; Time 2: ßsim, 0.73, ßsne, 0.06; Time 3: ßsim, 0.75, ßsne, 0.04). Turnover over time (paired comparison of each plot with itself at different sampling dates) ranged from 85.8% (between Time 1 and Time 2) to 88% (between Time 1 and Time 3), with ßsim, 0.59, ßsne, 0.1 between Time 1 and Time 2; ßsim, 0.58, ßsne, 0.08 between Time 1 and Time 3; and ßsim, 0.58, ßsne, 0.08 between Time 2 and Time 3. Community composition was more similar in samples of the same plot/site. The magnitude of variability in species turnover across the full spatial extent (1.5 km; Whittaker’s ßspace, 8.1) was, on average, 3.5 times greater than the temporal variability (ßtime, 2.3; Supplementary Table 1). Significant differences in the ratio ßspace:ßtime were found between Time 1 (mean, 3.04; SD, 0.85) and Time 3 (mean, 3.56; SD, 0.61; p < 0.05).
Distance and environmental decay of fungal communities
Statistically significant relationships between community similarity and spatial distance were found at Time 1 for similarity calculated on presence/absence (pseudo-R2 0.06, p ≤ 0.05, Table 1; Supplementary Fig. 2; Supplementary Table 2) and at Time 1 and Time 2 for similarity calculated on abundance data (Time 1: pseudo-R2 0.01, p ≤ 0.01; Time 2: pseudo-R2 0.06, p ≤ 0.05; Fig. 2; Supplementary Table 3). Decay patterns were comparable when using abundance and relative abundance data. No significant decay was found for community similarity with environmental distance (Supplementary Table 2).
The similarity was calculated on abundances as 1 – Morisita index. Fuchsia, rainy season; turquoise, transition into dry season; yellow, dry season. Circles represent pairs of plots (15 soil samples). The distance decay model was fitted by using a non-linear regression with the power-law function. ∗∗p ≤ 0.01; ∗p ≤ 0.05; ns, not statistically significant.
As there were no differences between abundance and relative abundance decay patterns, differences in the rate of community similarity decay (slope) and initial similarity (intercept) among seasons were calculated only for abundance data. Slopes were significantly different between Time 1 and Time 3 (mean, −0.284; SE, 0.004 vs. mean, −0.096; SE, 0.003; z ratio, −36.61; p < 0.001) and between Time 2 (mean, −0.284; SE, 0.003) and Time 3 (z ratio, −36.72; p < 0.001). Intercepts were significantly lower at Time 3 when compared with those at Time 1 (mean, 0.27; SE, 0.005 vs. mean, 1.56; SE, 0.033; z ratio, 32.55; p < 0.001) and Time 2 (mean, 1.42; SE, 0.035; z ratio, 28.97; p < 0.001), and at Time 2 when compared with Time 1 (z ratio 3.586; p < 0.001).
Soil fungal communities showed significant distance/environmental decay over short distances (60 samples; 4 sample plots per site; grain size 1 m2; site extent ca. 25 m2; 15 sites over a linear extent of 1.5 km) with community similarity based on occurrences (Supplementary Fig. 3), as well as on abundances (Supplementary Fig. 4). Decay was stronger (larger values of pseudo-R2) when similarity was based on presence/absence matrices than for abundances. The power-law model fitted best the relationship between soil fungal community similarity and spatial distance (presence/absence: pseudo-R2 0.16, p ≤ 0.001), and the exponential decay model fitted best the relationship between community similarity and environmental distance (presence/absence: pseudo-R2 0.02, p ≤ 0.001; Supplementary Fig. 3; Supplementary Table 4). Power-law models highlighted an abrupt change in community similarity at a distance of ca. 5-6 m, corresponding to the spatial extent sampled at the site level (Supplementary Fig. 3). The environmental variables that were significantly related to community composition included pH, OM, PO43-, K+, Ca2+, Mg2+, H+ plus Al3+ and fine root biomass for presence/absence data, and pH and OM for abundance data (Supplementary Table 4).
Community assembly processes
Measures of the modified Raup-Crick dissimilarity metric (RC), the standardised effect size (SES), and the taxonomic normalised stochasticity ratio index (tNST) presented similar results using Sørensen’s or Jaccard’s indices (occurrence data) and the Bray-Curtis or Ružička indices (abundance data). We therefore report here the results obtained with Jaccard’s and Ružička’s indices. Values of all metrics calculated on occurrence data at each sampling time indicated that observed dissimilarity did not differ from estimated dissimilarity, i.e., community composition was governed primarily by stochastic processes (Supplementary Table 5). Mean values of RC ranged from -0.66 (SD, 0.17; Time 1) and -0.22 (SD, 0.19; Time 3), SES between -1.76 (SD, 0.45; Time 1) and -0.65 (SD, 0.46; Time 3) and tNST between 0.69 (SD, 0.04; Time 1) and 0.82 (SD, 0.04; Time 3). Values of all indices were significantly different between Time 1 and Time 2 and between Time 1 and Time 3.
Index values calculated on abundance data indicated that there was a significant departure between observed and estimated dissimilarity (i.e., community composition was governed primarily by deterministic processes) at Time 2 and Time 3 (Fig. 3; Table 1). RC ranged from 0.6 (SD, 0.12; Time 1) and 0.95 (SD, 0.03; Time 3), and values were significantly different between Time 1 and Time 3 (p < 0.001) and between Time 2 (0.81; SD, 0.1) and Time 3 (p < 0.05). SES ranged from 1.01 (SD, 0.19; Time 1) and 2.08 (SD, 0.2; Time 3), the differences being significant between Time 1 and Time 2 (1.71; SD, 0.28; p < 0.05) and Time 1 and Time 3 (p < 0.001). The value of tNST ranged from 0.33 (SD, 0.06; Time 3) and 0.81 (SD, 0.09; Time 1) being significantly different between Time 1 and Time 2 (0.49; SD, 0.09; p < 0.05) and Time 1 and Time 3 (p < 0.01).
Different null model approaches were used to compare the observed β-diversity to the β-diversity expected by randomly sampling individuals from the regional species pool. This was done according to the probability proportional to observed species relative abundance in the regional pool and the total number of individuals in each plot. RC, modified Raup–Crick dissimilarity metric; SES, standardised effect size (SES); tNST, taxonomic normalised stochasticity ratio index; Time 1, rainy season; Time 2, transition into dry season; Time 3, dry season. RC, SES and tNST were calculated based on abundance using Ružička’s index. Differences between seasons in these metrics were tested using nst.boot (999 permutations) in ‘NST’86 and are indicated by grey arrows; *p < 0.05; ***p < 0.001. Boxplots: centre line, median; empty square, mean value; box limits, 25th and 75th percentiles; whiskers, 1.5 times interquartile range; empty circles, outliers.
Over short distances, mean values of all metrics calculated on both occurrence and abundance data indicated that observed dissimilarity did not differ from estimated dissimilarity (mean ± SD presence/absence: RC, −0.68 ± 0.05; SES, −1.92 ± 0.18; tNST, 0.73 ± 0.02; mean ± SD abundance: RC, 0.54 ± 0.06; SES, 0.97 ± 0.11; tNST, 0.99 ± 0.06).
Discussion
We assessed soil fungal community assembly, along a 1.5 km section of a plateau area in the central part of the Amazon basin, accounting for temporal changes linked to seasonality in precipitation. Our study showed that spatial distance in a highly diverse lowland rain forest in Amazonia exert a stronger influence on soil fungal community assembly than seasonal patterns linked to the precipitation regime and thus confirmed our first hypothesis. The relative magnitude of community turnover at the spatial extent studied was, on average, 3.5 times larger than that attributable to seasonality, indicating a strong relationship between spatial variability and community composition (grain size 1 m × 1 m). The ratio of ßspace:ßtime was significantly related to seasonality reflecting greater species turnover in space than in time in the dry season than in the rainy season.
Fungal communities were characterised by higher taxonomic and functional α-diversity in the dry season when compared to that in the rainy season. The same pattern has recently been observed in an eastern Amazonian lowland forest on yellow oxisol23 and could be attributable to several concomitant and interacting factors, including the reduced competitive interactions among microbial taxa due to the lack of connectivity among soil pores in the dry season49, the input of fungal species from the phyllosphere microbiome17 and/or to compositional shifts during litter decomposition that may increase overall microbial diversity50,51.
The relationship between space (physical and environmental) and time is however complex as the indirect effects that seasonality exerts on fungal community assembly through changes in litter input/quality and root turnover/exudate patterns linked to species-specific traits contribute to maintaining a high level of spatial heterogeneity. This makes it difficult to disentangle the contribution of spatial heterogeneity vs. temporal variation (seasonality) on soil fungal community assembly. A larger relative contribution of spatial over temporal variability on fungal community composition, similarly to that observed in this study has recently been reported by Wei, et al.24 for tropical forests in Hainan (18° to 20° N and 108° to 111° E), China. Their study was conducted at the regional scale, where the patterns detected could be attributable to strong climatic and edaphic differences across the extent of the study area. Here we show that, in the absence of obvious environmental gradients, a similar pattern occurs at a much smaller spatial extent (1.5 km vs. > 150 km) and is likely to be related to the intrinsic tree species diversity of tropical rain forests. Although the trees around which we sampled had been selected randomly and were not identified, a posterior identification of six focal trees and their surrounding trees in a 10-m radius showed that all focal trees belonged to different species. Using the data based on six of the 15 sites, we calculated that owing to the large diversity (120 individuals identified belonged to 80 species), the chances of randomly selecting two individuals of the same tree species would be no more than 15%.
It has previously been hypothesised that the relative importance of spatial heterogeneity in soil chemical properties in tropical lowland rain forests is expected to shift from abiotic to biotic control from regional to landscape and local scales14. This would also apply to the Amazon basin where there is a clear topography-related abiotic control at the landscape scale (dissected undulating topography associated with different soil types, water table depth and nutrient availability e.g.,47), which at the local scale is likely to shift to a biotic control, related to differences in tree species and mediated by their biochemical differences. Trees exert strong direct and indirect biotic controls on soil microbial communities through plant-derived soil resources that reflect differences in plant species composition and their spatial distribution22,52. The quality/chemistry and quantity of plant inputs (Fig. 4;53) influences soil microbial community assembly by affecting decomposers54,55, as well as via the specificity of biotic interactions between fungal symbionts/pathogens and trees56,57,58. Tree species diversity in tropical rain forests contributes therefore to both spatially and temporally uneven distribution of resources that contribute to shaping soil microbial community assemblages.
The concept was originally proposed by Townsend, et al.14. According to the authors, the diverse tree species composition that characterises lowland tropical rain forests is likely to result in a large variation in biochemical attributes among different tree species. This, in turn, would contribute to a high degree of spatio-temporal heterogeneity of resources for soil microbial communities and influences both their biotic interactions and coexistence patterns. Individual tree species might therefore create specific biogeochemical footprints. In species-rich tropical rain forests, inputs to the forest floor/soil by trees via litterfall (litter biochemistry, Lb) and root exudate (root biochemistry, Rb) create overlapping spheres of influence (viz. differently coloured ellipses) on soil microbial communities, among neighbouring tree species.
Our second hypothesis that the relative contribution of environmental determinism and stochastic processes to fungal community assembly would change with seasonality was supported by our results. However, deterministic processes dominated in the dry season (Table 1), and not in the rainy season as we had hypothesised. We based our second hypothesis on the premise that during the rainy season, the concomitant availability of resources and altered soil conditions (e.g., improved moisture) increase the rate of microbial processes such as decomposition and mineralisation34,42 and would likely to auto-intensify the environmental controls exerted on microbial communities by plant-derived resources. We found a significant but weak distance decay pattern in community similarity in the rainy season (Time 1) and in the transition from the rainy into the dry season (Time 2), while no statistically significant decay was observed with environmental distance at any season (Table 1). The distance decay patterns were comparable to those found by Peguero, et al.46. However, the relative importance of a process may depend on when a community is characterised, because seasonality influences processes that govern community assembly59. Our distance decay patterns were related to seasonality and were not observable in the dry season (Table 1). In the absence of an environmental gradient, these results could indicate the dominance of stochastic processes, including ecological drift and dispersal at Times 1 and 2. Ecological drift can act alone through probabilistic factors, especially when the regional pool is very large compared with the size of local communities60. Combining the facts that turnover is a function of species pool, that fungal communities in our study area were characterised by high turnover rates, and that local communities were on average 10 times smaller than the regional pool, we can assume that ecological drift might have had a contribution to fungal community assembly. The effects of dispersal limitation are also expected to be greatest for large pools and diverse communities, because of the existence of a large proportion of rare species in the pool60 which was also the case in our study. Therefore, community assembly in the rainy season was likely governed by both ecological drift and dispersal limitation.
Estimated values of RC, SES, and tNST calculated on abundance data complemented the above findings indicating that turnover in community composition in the rainy season was governed by drift and dispersal limitation, while species selection and/or dispersal limitation (acting in concert with ecological drift) could have been the main processes governing the aggregate distribution of species in the dry season. These values were not affected by the number of samples used in the analyses (15 vs. 60). Collectively, the two different analytical approaches, support that stochastic processes could be more important for soil fungal community assembly in the rainy season, while abiotic and biotic factors (deterministic processes) would govern community assembly in the dry season.
The control exerted by plant-derived resources on fungal community assembly may be linked to the peak in litterfall that characterises tropical lowland rain forests in Amazonia late in the dry season. A study conducted in a neotropical lowland rain forest has shown that host plant taxonomic identity explained most of the variation in the phyllosphere fungal community composition61. The study has also shown that host functional traits related to leaf morphology and chemistry were significantly associated with fungal community structure. In a previous research conducted in Amazonia, in the same study area reported here, it has been shown that during the litterfall peak, in the dry season, there is an input of bacterial species from the phyllosphere (e.g. Beijerinckiaceae, see Buscardo, et al.17). This would suggest that individual tree species with their specific traits including leaf morphology, and chemistry and associated microbiome contribute to characterise the soil environment and the biotic interactions in their sphere of influence and might indeed create what Townsend, et al.14 have defined as tree-specific ‘biogeochemical footprint’. The ‘biogeochemical footprint’ extends from the canopy to the soil, influencing and being influenced by soil microbial communities (Fig. 4). Under these premises, geographic position and environmental factors would, in our case, both mirror the environmental variability linked to the presence of individual tree species, giving support to our hypothesis of a temporal alternation of the dominance of niche-based and stochastic processes, whereby niche-based processes would dominate in the dry season within the influence zone of a tree. Following the logic of the ‘biogeochemical footprint’, the environmental distance between individual neighbouring trees would overlap with their spatial distance to a degree that varies depending on the identity of neighbouring trees. This is supported by the results that included short-distance patterns, which allowed the detection of a weak albeit significant environmental distance decay co-occurring with a spatial distance decay. At the same time, high species diversity would dilute the effect of individual influence spheres (see Fig. 4; Supplementary Fig. 5) and with increasing distance it would augment the probability of the occurrence of fungal species related to stochastic effects62. Thus, a biochemically heterogeneous environment would result in emergent stochastic patterns at the large scale that would arise from small-scale deterministic processes.
Soil fungal communities in Amazonian lowland rain forests have been shown to be characterised by large turnover rates in the presence of strong environmental gradients that affect tree species diversity/structure, e.g., seasonally flooded vs. non-flooded forests21, or gradients linked to dissected topographies46. In the present study, which excluded such gradients we detected large turnover rates even at short distances (up to 6 m), reflecting the high heterogeneity that characterise the influence sphere of an individual tree and its microbiota (defined by Vandenkoornhuyse, et al.63 as the holobiont). The low initial community similarity (with very low values in the dry season) and the small amount of variation explained by geographical distance beyond the area of influence of a specific tree, might be attributable to aleatory factors other than dispersal limitation, as well as to sampling62.
A large proportion of unexplained variation in ß-diversity is expected in a tropical rain forest characterised by high diversity, due to sampling effect, local stochasticity due to ecological drift and/or unmeasured environmental variables64. Our findings indicate that soil physico-chemical explanatory variables alone are likely to be insufficient to predict community patterns in environments where obvious soil and hydrological contrasts are absent and that even a four-fold more intensive sampling does not significantly increase the amount of explained variation. We, thus, call for the inclusion of additional factors in future studies, both environmental (e.g., species-specific secondary chemicals in leaf litter) and biological (biotic interactions) for improving our understanding of the relative importance of agents and mechanisms that shape microbial assemblages in tropical rain forests.
Our study highlights the importance of spatial heterogeneity in the absence of environmental gradients and its relationship with seasonal patterns linked to the precipitation regime in determining soil fungal community assembly in a lowland tropical rain forest. Phenology linked to climate seasonality modulates spatial heterogeneity and its contribution to environmental determinism vs. stochasticity for fungal community assembly. Future studies characterising the biogeochemical footprint of target trees (and the resulting alterations of it by neighbouring trees) on fungal communities, and which consider environmental factors linked to plant—fungal biotic interactions could help to further improve our understanding of the processes that shape microbial community assembly and function in tropical rain forests. It is of great scientific and practical interest how the relative contribution of these processes may be altered by climate change. Some of the changes are likely to be mediated by shifts in taxonomical and functional composition of trees and consequent changes in biotic interactions below- and above-ground.
MATERIALS AND METHODS
Site details
The study was carried out in the Adolpho Ducke Forest Reserve (ADFR; 03°00′00″ − 03° 08′00″ S; 59°52′40″ − 59° 58′00″W), 26 km NE of Manaus, Brazil, in 2013, in a plateau area to avoid the environmental variation associated with topography (pedo-hydro-topo gradients). The region has an equatorial climate with mean annual temperature of 26 °C and mean annual precipitation of 2550 mm year−1 (period 1966–2014) with a pronounced drop in precipitation between June and October that coincides with the litterfall peak39. The year of the study was much wetter (3385 mm) than the average, however, the monthly distribution of precipitation did not deviate from the long-term pattern. There was no month with a perceived negative hydrological balance (precipitation <100 mm).
The ADFR is a 10-km2 area at an elevation of 40–140 m above sea level, characterised by a dissected and undulating topography of valley bottoms, slopes and plateaux (Supplementary Fig. 6). Soils in plateau areas are well-drained oxisols not affected by waterlogging or flooding during the rainy season; slopes and valleys are characterised by ultisols and hydromorphic spodosols65. The vegetation is lowland evergreen tropical rain forest (locally known as terra firme) and plant community composition and structure are related to topography with emergent trees up to ca. 45 m, frequent on the plateaux where average canopy height reaches 35–40 m66.
Experimental design
Initially, 20 sites were chosen along a 1.5-km-long section in a plateau area of the RFAD using random coordinates generated by the ‘Sample’ module in IDRISI Selva v.17 software67. In the field, an unidentified canopy tree closest to the predefined coordinates was selected to mark the centre of the site (reference tree). The minimum distance between any two sites was at least 40 m (Supplementary Fig. 6) to avoid autocorrelation in soil variables and associated microbial communities (Supplementary Fig. 7). The initial number of sites, which was later reduced to 15 for molecular analyses, was chosen to allow for potential losses that might occur owing to treefall, or other disturbances that may affect the sampling during the experiment. One plot of 1 m × 1 m in size and subdivided into 15 subplots was marked permanently at each site, at a distance of 1.5 m from the reference tree (Supplementary Fig. 6).
Soil was sampled three times during a year to characterise microbial community composition and deduce community dynamics: (i) in the wet season (Time 1, 13 April 2013), (ii) in the transition to the dry season (Time 2, 28 July) and (iii) at the end of the dry season (Time 3, 6 November). After the removal of litter, soil was collected from three randomly selected sub-plots in every plot to a depth of 5 cm using a cylindrical metal corer of 3-cm in diameter. For DNA extraction soil was collected from each sub-sample by using a sterile needle and pooled by plot at each sampling in a 2.5-ml Eppendorf vial, yielding 45 samples (3 times x 15 samples). The vials were maintained at −20 °C until molecular analyses. The remaining soil was pooled in an identical manner for soil chemical analyses.
To characterise short-distance (up to 6 m) processes/microbial community patterns (Supplementary Figs. 5, 6) within the presumed biogeochemical footprint of the reference trees, at Time 1 we undertook a complementary sampling (3 samples x 15 sites; plot size 1 m2; extent at each site ca. 25 m2). This, together with the original 15 samples at Time 1, resulted in 60 samples.
Molecular analyses
Fungal communities were characterised by analysing a total of 90 samples. DNA extractions were carried out for each sample using a PowerSoil DNA Isolation Kit (MOBIO) with 250 mg of soil. The PCR and DNA sequencing were carried out as described in detail in Geml et al.68. Briefly, primers fITS769 and ITS470 with Ion Torrent adapters were used to amplify the ITS2 region (ca. 250 bp) of the nuclear ribosomal rDNA repeat, using the following PCR conditions: one cycle of 95 °C for 5 min, then 37 cycles of 95 °C for 20 sec, 56 °C for 30 sec, and 72 °C for 1.5 min, ending with one cycle of 72 °C for 7 min. The ITS4 primer was labelled with sample-specific Multiplex Identification DNA-tags (MIDs). A negative control consisting of Milli-Q water (Merck Millipore, Burlington, MA, U.S.A.) instead of DNA were made and underwent the PCR reaction under the same experimental conditions and was shown on a gel to be amplicon-free. The PCR products were assessed for size distribution and for DNA concentration using a Bioanalyser 2100 (Agilent Technologies Inc., Santa Clara, CA, U.S.A.) and were cleaned up using 0.9x Ampure® beads (Beckman Coulter, Beverly, MA) to remove short fragments. The amplicons were diluted with Milli-Q water to achieve equal DNA concentration for each sample in the final pool to be sequenced. A quantity of 250 µl of the pool was used for emulsion PCR according to the Ion PGMTM 200XpressTM Template Kit manual. The amplicon library was sequenced using an Ion 318TM Chip by an Ion Torrent Personal Genome Machine (Life Technologies, Guilford, CT, U.S.A.) at the Naturalis Biodiversity Center, Leiden, The Netherlands.
Raw DNA sequences were processed with the ‘dada2’ package71, implemented in R v. 3.6.2 (R Development Core Team 2013), designed to resolve fine-scale DNA sequence variation with improved elimination of artificial sequences. Because ‘dada2’ does not involve clustering sequences into OTUs and is robust for removing spurious data, the output of unique ASVs capture both intra- and interspecific genetic variation of fungi found in the samples. Raw sequences were truncated to 240 bp and were denoised, with the maximum number of expected errors (maxEE) allowed in a read set to 2. Identical sequences were clustered into ASVs, while preserving information regarding their copy numbers in individual samples, and putative chimeric ASVs were eliminated. Taxonomic assignments of fungi were made with USEARCH v. 1172 based on the UNITE database (v. 8.3) of reference sequences that represents all fungal species hypotheses based on a dynamic delimitation73.
We assigned fungal ASVs to putative functional guilds using the curated FungalTraits database74. ASVs with <10 reads in a given sample were excluded from that sample to minimise the possible effects of low-abundance contaminants and tag-switching. In addition, ASVs that occurred in a single sample were deleted to minimise artefactual sequences. Normalisation of the fungal community matrix (rarefaction) was made by random subsampling to the smallest library size (11,042 reads).
Soil analyses
Soil properties were characterised for each soil sample (see Buscardo, et al.17). Soil moisture, NH4+ and NO3− were quantified within 48 h after collection in the Soil Laboratory of the National Research Institute of Amazonia, Manaus, Brazil. Soil mineral nitrogen was quantified by spectrophotometry using flow injection analysis75,76 while soil moisture content was determined by the gravimetric method by drying the soil at 105 °C to constant mass76. Soil pH, total OM, extractable PO43−, cations (K+, Ca2+ and Mg2+) and exchangeable acidity (H+ plus Al3+), were determined on air dried soil (Laboratory of Soil Analysis, University of São Paulo, Piracicaba, Brazil), according to van Raij, et al.77. Fine (≤2 mm) and coarse (>2 mm) roots were extracted from each soil sample and their mass determined after oven drying at 65 °C to constant mass.
Data analyses
All DNA samples, except one that did not amplify, were included in the analyses totalling 2055 ASVs (2,703,823 sequence reads) and an average number of 30,380 (range 11,042–69,938) sequence reads per sample. The analyses were conducted on presence/absence and abundance data. Abundance data were rarefied using the ‘GUniFrac’ R package78.
To assess the effects of seasonality on fungal functional and taxonomic groups we quantified the abundance and α-diversity (total number of ASVs) of each functional and taxonomic group at each sampling time. Between-season differences in the diversity of ASVs and soil properties were tested using pair-wise differences with the t-test or Wilcoxon’s test for non-normally distributed data. Statistical significance was corrected for multiple comparisons using the false discovery rate method79.
To test our first hypothesis that spatial heterogeneity would be more strongly related to soil fungal community assembly than seasonal patterns linked to precipitation regime, we first calculated β-diversity (βsor) and its components (spatial turnover, βsim and nestedness, βsne) at each sampling date (15 samples) using the R package ‘betapart’80 and Sørensen’s similarity index. Temporal changes in community composition between different sampling dates for each plot sampled were assessed by computing dissimilarities on presence/absence data.
We then quantified the relative magnitude of spatial (βspace, spatial variability at the stand level, i.e. full extent of the sampling transect) and temporal (βtime, temporal variability at the individual plot level) variability, by using Whittaker’s formula of ß-diversity (β = γ/α; where γ is the total number of ASVs at each sampling occasion, i.e., the total of 15 plots for space, and of three samplings per plot for time; α is the number of ASVs per plot at a given time). Between-season differences in ß-diversity were tested using pair-wise differences with the t-test or Wilcoxon’s test for non-normally distributed data.
To test our second hypothesis that the relative contribution of stochastic processes and environmental filtering in fungal community assembly would change with seasonality, and that environmental filtering would dominate in the wet season, we (1) calculated the distance decay of soil fungal community similarity with geographical and environmental distance at Times 1, 2, and 3, and then (2) used another null-model based approach to compare the observed β-diversity with the β-diversity expected from random sampling of the regional species pool (Table 1). We tested the significance of the difference using three different metrics (modified Raup-Crick dissimilarity metric, RC; standardised effect size, SES; taxonomic normalised stochasticity ratio index, tNST).
Distance decay – The geographical distance between pairs of sites was calculated using the function dist of the ‘vegan’ package. Environmental dissimilarity was calculated as the Euclidean distance between all pairs of sites considering soil physico-chemical properties quantified at each site and standardised to their z-scores ([x – mean] divided by the standard deviation) by using the function vegdist (‘vegan’). From the original environmental variable data set only continuous variables that were related significantly to soil fugal community composition were included following Martiny, et al.81. Significant explanatory variables were selected via permutation tests using the reduced model option for constrained correspondence analysis (function ANOVA in ‘vegan’). To calculate the similarity matrix, community composition was compared between each pair of sites. The similarity index was calculated using both the presence/absence and abundance matrices. Community similarity based on occurrences was determined as 1 – βsim, computed with Sørensen’s similarity index in ‘betapart’, while community similarities based on abundance (calculated as 1 – the Morisita index) and relative abundance (calculated as 1 – the Morisita-Horn index) matrices were computed in ‘vegan’. The relationships between pairwise similarities and spatial and environmental distances were modelled using the decay.model function in ‘betapart’82. Distance decay models were fitted by using non-linear regressions with negative exponential and power-law functions62,83. The goodness of fit of the decay models (i.e., explained variation) was measured as the pseudo-R2 calculated as (deviance null model – deviance distance decay model)/deviance null model, while model significance was assessed by randomising spatial or environmental distances (1000 permutations) and calculating the proportion of times in which the decay model deviance was smaller than the null model deviance84. Differences in the rate of community similarity decay and initial similarity among seasons were estimated from pairwise comparisons of the bootstrapped distributions of slopes and intercepts (1000 permutations, boot.coefs.decay in ‘betapart’), computing p-values as the proportion of times in which the bootstrapped parameters were larger (or smaller) in one season than the other82.
Comparing observed β-diversity with expected β-diversity – For comparing the observed β-diversity to the β-diversity expected from a random sampling of the regional species pool, the null model regional ‘species’ pool was defined as the total number of ASVs observed at the stand level (i.e., across all plots). Observed β-diversity (i.e., dissimilarity between each pair of plots within the stand) was measured using both presence/absence data (1 - Sørensen’s and 1 - Jaccard’s index) and abundance data (Bray-Curtis and Ružička). The null model to simulate species assemblages in each plot was applied by randomly sampling individuals from the regional pool, according to the probability proportional to observed species occurrence/relative abundance in the regional pool and the total number of individuals in each plot. Three metrics were used to estimate the contribution of stochastic processes and environmental filtering in fungal community assembly based on the null model analysis of dissimilarity (999 randomised matrices): (i) RC85 that calculates the proportion of observed dissimilarities across samples that are higher than those estimated from the null model. This metric ranges from –1 to 1; values between –0.95 and –1 indicate mass effect, values between 0.95 and 1 indicate environmental filtering and values between –0.95 and 0.95 indicate drift; (ii) SES as the difference between the observed and mean expected dissimilarity, divided by the standard deviation of expected values, where values around zero indicate that observed dissimilarity does not differ from estimated dissimilarity, values greater than zero indicate a higher observed dissimilarity than expected by chance and values lower than zero indicate a lower dissimilarity than expected by chance; (iii) tNST, a measure of the relative position of observed dissimilarity values between extreme values (0 to 1) under pure deterministic and pure stochastic assembly86. RC and SES reflect the contribution to community assembly of deterministic processes (RC > |0.95|; SES > |2|) based on the significance of the difference between observed and null expectation, while tNST reflects the contribution of stochastic assembly (tNST > 0.5) based on magnitude of this difference. RC, SES and tNST were calculated using the tNST function, while differences between seasons in these metrics were tested using nst.boot (999 permutations), both in ‘NST’86.
Phylogenetic diversity analyses were not made as the ITS region is not phylogenetically conserved87, and as such ITS-based analyses are not suitable for multiple sequence alignments required for phylogenetic diversity analyses88.
Data availability
Sequences were deposited at DDBJ/EMBL/GenBank under the BioProject accession number PRJNA579099. Data supporting the findings of this study are available at https://doi.org/10.6084/m9.figshare.25008662.
References
Hubbell, S. P. The unified neutral theory of biodiversity and biogeography. (Princeton University Press, 2001).
Mori, A. S., Isbell, F. & Seidl, R. β-Diversity, community assembly, and ecosystem functioning. Trends Ecol. Evol. 33, 549–564, https://doi.org/10.1016/j.tree.2018.04.012 (2018).
Weiher, E. et al. Advances, challenges and a developing synthesis of ecological community assembly theory. Philos. Trans. Royal Soc. B: Biol. Sci. 366, 2403–2413, https://doi.org/10.1098/rstb.2011.0056 (2011).
Vellend, M. Conceptual synthesis in community ecology. Q. R. Biol. 85, 183–206, https://doi.org/10.1086/652373 (2010).
Wang, K. et al. Community assembly of bacteria and archaea in coastal waters governed by contrasting mechanisms: a seasonal perspective. Mol. Ecol. 29, 3762–3776, https://doi.org/10.1111/mec.15600 (2020).
Stegen, J. C., Lin, X., Konopka, A. E. & Fredrickson, J. K. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 6, 1653–1664, https://doi.org/10.1038/ismej.2012.22 (2012).
Liu, J. et al. Spatiotemporal dynamics of the archaeal community in coastal sediments: assembly process and co-occurrence relationship. ISME J. 14, 1463–1478, https://doi.org/10.1038/s41396-020-0621-7 (2020).
Zhang, K., Delgado-Baquerizo, M., Zhu, Y.-G. & Chu, H. Space is more important than season when shaping soil microbial communities at a large spatial scale. mSystems 5, e00783–00719, https://doi.org/10.1128/mSystems.00783-19 (2020).
Bañares-de-Dios, G. et al. Linking patterns and processes of tree community assembly across spatial scales in tropical montane forests. Ecology 101, e03058, https://doi.org/10.1002/ecy.3058 (2020).
Srivastava, D. S. et al. Geographical variation in the trait-based assembly patterns of multitrophic invertebrate communities. Funct. Ecol. 37, 73–86, https://doi.org/10.1111/1365-2435.14096 (2023).
Kraft, N. J. B. & Ackerly, D. D. Functional trait and phylogenetic tests of community assembly across spatial scales in an Amazonian forest. Ecol. Monogr. 80, 401–422, https://doi.org/10.1890/09-1672.1 (2010).
Peguero, G. et al. Nutrient-based species selection is a prevalent driver of community assembly and functional trait space in tropical forests. J. Ecol. 111, 1218–1230, https://doi.org/10.1111/1365-2745.14089 (2023).
Bernard-Verdier, M. et al. Community assembly along a soil depth gradient: contrasting patterns of plant trait convergence and divergence in a Mediterranean rangeland. J. Ecol. 100, 1422–1433, https://doi.org/10.1111/1365-2745.12003 (2012).
Townsend, A. R., Asner, G. P. & Cleveland, C. C. The biogeochemical heterogeneity of tropical forests. Trends Ecol. Evol. 23, 424–431, https://doi.org/10.1016/j.tree.2008.04.009 (2008).
Buscardo, E. et al. Of mammals and bacteria in a rainforest: temporal dynamics of soil bacteria in response to simulated N pulse from mammalian urine. Funct. Ecol. 32, 773–784, https://doi.org/10.1111/1365-2435.12998 (2018).
Buscardo, E. et al. Nitrogen pulses increase fungal pathogens in Amazonian lowland tropical rain forests. J. Ecol. 110, 1775–1789, https://doi.org/10.1111/1365-2745.13904 (2022).
Buscardo, E. et al. Spatio-temporal dynamics of soil bacterial communities as a function of Amazon forest phenology. Sci. Rep. 8, 4382, https://doi.org/10.1038/s41598-018-22380-z (2018).
Vasco-Palacios, A. M., Bahram, M., Boekhout, T. & Tedersoo, L. Carbon content and pH as important drivers of fungal community structure in three Amazon forests. Plant Soil 450, 111–131, https://doi.org/10.1007/s11104-019-04218-3 (2020).
Adamo, I. et al. Soil fungal community composition correlates with site-specific abiotic factors, tree community structure, and forest age in regenerating tropical rainforests. Biology 10, 1120, https://doi.org/10.3390/biology10111120 (2021).
McGuire, K. L., Fierer, N., Bateman, C., Treseder, K. K. & Turner, B. L. Fungal community composition in neotropical rain forests: the influence of tree diversity and precipitation. Microb. Ecol. 63, 804–812, https://doi.org/10.1007/s00248-011-9973-x (2012).
Peay, K. G., Baraloto, C. & Fine, P. V. A. Strong coupling of plant and fungal community structure across western Amazonian rainforests. ISME J. 7, 1852–1861, https://doi.org/10.1038/ismej.2013.66 (2013).
Barberán, A. et al. Relating belowground microbial composition to the taxonomic, phylogenetic, and functional trait distributions of trees in a tropical forest. Ecol. Lett. 18, 1397–1405, https://doi.org/10.1111/ele.12536 (2015).
Buscardo, E. et al. Effects of natural and experimental drought on soil fungi and biogeochemistry in an Amazon rain forest. Commun. Earth Environ. 2, 55, https://doi.org/10.1038/s43247-021-00124-8 (2021).
Wei, Y., Quan, F., Lan, G., Wu, Z. & Yang, C. Space rather than seasonal changes explained more of the spatiotemporal variation of tropical soil microbial communities. Microbiol. Spectr. 10, e01846-01822, https://doi.org/10.1128/spectrum.01846-22 (2022).
Chadwick, R., Good, P., Martin, G. & Rowell, D. P. Large rainfall changes consistently projected over substantial areas of tropical land. Nat. Clim. Change 6, 177, https://doi.org/10.1038/nclimate2805 (2015).
Neelin, J. D., Münnich, M., Su, H., Meyerson, J. E. & Holloway, C. E. Tropical drying trends in global warming models and observations. Proc. Natl Acad. Sci. 103, 6110–6115, https://doi.org/10.1073/pnas.0601798103 (2006).
Esquivel-Muelbert, A. et al. Compositional response of Amazon forests to climate change. Glob. Change Biol. 25, 39–56, https://doi.org/10.1111/gcb.14413 (2019).
Buscardo, E., Forkuor, G., Rubino, A. & Storozum, M. Land and people. Commun. Earth Environ. 2, 178, https://doi.org/10.1038/s43247-021-00240-5 (2021).
Lodge, D. J., McDowell, W. H. & McSwiney, C. P. The importance of nutrient pulses in tropical forests. Trends Ecol. Evol. 9, 384–387, https://doi.org/10.1016/0169-5347(94)90060-4 (1994).
Buscardo, E. et al. in Interactions between biosphere, atmosphere and human land use in the Amazon basin (eds Laszlo Nagy, Bruce R. Forsberg, & Paulo Artaxo) 225-266 (Springer Berlin Heidelberg, 2016).
Guan, K. et al. Photosynthetic seasonality of global tropical forests constrained by hydroclimate. Nat. Geosci. 8, 284–289, https://doi.org/10.1038/ngeo2382 (2015).
Cleveland, C. C., Reed, S. C. & Townsend, A. R. Nutrient regulation of organic matter decomposition in a tropical rain forest. Ecology 87, 492–503, https://doi.org/10.1890/05-0525 (2006).
Schaap, K. J. et al. Litter inputs and phosphatase activity affect the temporal variability of organic phosphorus in a tropical forest soil in the Central Amazon. Plant Soil 469, 423–441, https://doi.org/10.1007/s11104-021-05146-x (2021).
Luizão, F. J. & Schubart, H. O. R. Litter production and decomposition in a terra-firme forest of Central Amazonia. Experientia 43, 259–265, https://doi.org/10.1007/BF01945549 (1987).
Wu, J. et al. Leaf development and demography explain photosynthetic seasonality in Amazon evergreen forests. Science 351, 972–976, https://doi.org/10.1126/science.aad5068 (2016).
Lopes, A. P. et al. Leaf flush drives dry season green-up of the Central Amazon. Remote Sens. Environ. 182, 90–98, https://doi.org/10.1016/j.rse.2016.05.009 (2016).
Luizão, F., Luizão, R. & Chauvel, A. Premiers résultats sur la dynamique des biomasses racinaires et microbiennes dans un latosol d’Amazonie centrale (Brésil) sous forêt et sous pâturage. Cahiers ORSTOM. Série Pédologie 27, 69–79 (1992).
Malhi, Y. et al. The productivity, metabolism and carbon cycle of two lowland tropical forest plots in south-western Amazonia, Peru. Plant Ecol. Divers. 7, 85–105, https://doi.org/10.1080/17550874.2013.820805 (2014).
Luizão, F. J. Litter production and mineral element input to the forest floor in a Central Amazonian forest. GeoJournal 19, 407–417, https://doi.org/10.1007/BF00176910 (1989).
Luizão, R. C. C., Bonde, T. A. & Rosswall, T. Seasonal variation of soil microbial biomass—The effects of clearfelling a tropical rainforest and establishment of pasture in the central Amazon. Soil Biol. Biochem. 24, 805–813, https://doi.org/10.1016/0038-0717(92)90256-W (1992).
Cleveland, C. C. & Townsend, A. R. Nutrient additions to a tropical rain forest drive substantial soil carbon dioxide losses to the atmosphere. Proc. Natl Acad. Sci. 103, 10316–10321, https://doi.org/10.1073/pnas.0600989103 (2006).
Cornejo, F. H., Varela, A. & Wright, S. J. Tropical forest litter decomposition under seasonal drought: nutrient release, fungi and bacteria. Oikos 70, 183–190, https://doi.org/10.2307/3545629 (1994).
Girardin, C. A. J. et al. Seasonal trends of Amazonian rainforest phenology, net primary productivity, and carbon allocation. Glob. Biogeochem. Cycles 30, 700–715, https://doi.org/10.1002/2015GB005270 (2016).
Paterson, E., Gebbing, T., Abel, C., Sim, A. & Telfer, G. Rhizodeposition shapes rhizosphere microbial community structure in organic soil. New Phytol. 173, 600–610, https://doi.org/10.1111/j.1469-8137.2006.01931.x (2007).
Boraks, A. et al. Scale-dependent influences of distance and vegetation on the composition of aboveground and belowground tropical fungal communities. Microb. Ecol. 81, 874–883, https://doi.org/10.1007/s00248-020-01608-4 (2021).
Peguero, G. et al. Decay of similarity across tropical forest communities: integrating spatial distance with soil nutrients. Ecology 103, e03599, https://doi.org/10.1002/ecy.3599 (2022).
de Oliveira Freitas, R. et al. Arbuscular mycorrhizal fungal communities along a pedo-hydrological gradient in a Central Amazonian terra firme forest. Mycorrhiza 24, 21–32, https://doi.org/10.1007/s00572-013-0507-x (2014).
Vellend, M. et al. Assessing the relative importance of neutral stochasticity in ecological communities. Oikos 123, 1420–1430, https://doi.org/10.1111/oik.01493 (2014).
Carson, J. K. et al. Low pore connectivity increases bacterial diversity in soil. App. Environ. Microbiol. 76, 3936–3942, https://doi.org/10.1128/aem.03085-09 (2010).
Hudson, H. J. The ecology of fungi on plant remains above the soil. New Phytol. 67, 837–874, https://doi.org/10.1111/j.1469-8137.1968.tb06399.x (1968).
Voříšková, J. & Baldrian, P. Fungal community on decomposing leaf litter undergoes rapid successional changes. ISME J. 7, 477–486, https://doi.org/10.1038/ismej.2012.116 (2013).
Wolf, J. A. Geospatial observations on biodiversity and biogeochemistry of a tropical forest rhizosphere PhD thesis, University of California, (2015).
Hättenschwiler, S., Coq, S., Barantal, S. & Handa, I. T. Leaf traits and decomposition in tropical rainforests: revisiting some commonly held views and towards a new hypothesis. New Phytol. 189, 950–965, https://doi.org/10.1111/j.1469-8137.2010.03483.x (2011).
Aneja, M. K. et al. Microbial colonization of beech and spruce litter: influence of decomposition site and plant litter species on the diversity of microbial community. Microb. Ecol. 52, 127–135, https://doi.org/10.1007/s00248-006-9006-3 (2006).
Fanin, N., Hättenschwiler, S. & Fromin, N. Litter fingerprint on microbial biomass, activity, and community structure in the underlying soil. Plant Soil 379, 79–91, https://doi.org/10.1007/s11104-014-2051-7 (2014).
Sarmiento, C. et al. Soilborne fungi have host affinity and host-specific effects on seed germination and survival in a lowland tropical forest. Proc. Natl Acad. Sci. 114, 11458–11463, https://doi.org/10.1073/pnas.1706324114 (2017).
Spear, E. R. & Broders, K. D. Host-generalist fungal pathogens of seedlings may maintain forest diversity via host-specific impacts and differential susceptibility among tree species. New Phytol. 231, 460–474, https://doi.org/10.1111/nph.17379 (2021).
Hoeksema, J. D. et al. Evolutionary history of plant hosts and fungal symbionts predicts the strength of mycorrhizal mutualism. Commun. Biol. 1, 116, https://doi.org/10.1038/s42003-018-0120-9 (2018).
Hanson, C. A., Fuhrman, J. A., Horner-Devine, M. C. & Martiny, J. B. H. Beyond biogeographic patterns: processes shaping the microbial landscape. Nat. Rev. Microbiol. 10, 497–506, https://doi.org/10.1038/nrmicro2795 (2012).
Chase, J. M. & Myers, J. A. Disentangling the importance of ecological niches from stochastic processes across scales. Philos. Trans. Royal Soc. B: Biol. Sci. 366, 2351–2363, https://doi.org/10.1098/rstb.2011.0063 (2011).
Kembel, S. W. & Mueller, R. C. Plant traits and taxonomy drive host associations in tropical phyllosphere fungal communities. Botany 92, 303–311, https://doi.org/10.1139/cjb-2013-0194 (2014).
Nekola, J. C. & McGill, B. J. Scale dependency in the functional form of the distance decay relationship. Ecography 37, 309–320, https://doi.org/10.1111/j.1600-0587.2013.00407.x (2014).
Vandenkoornhuyse, P., Quaiser, A., Duhamel, M., Le Van, A. & Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 206, 1196–1206, https://doi.org/10.1111/nph.13312 (2015).
Myers, J. A. et al. Beta-diversity in temperate and tropical forests reflects dissimilar mechanisms of community assembly. Ecol. Lett. 16, 151–157, https://doi.org/10.1111/ele.12021 (2013).
Chauvel, A., Lucas, Y. & Boulet, R. On the genesis of the soil mantle of the region of Manaus, Central Amazonia, Brazil. Experientia 43, 234–241, https://doi.org/10.1007/bf01945546 (1987).
Ribeiro, J. E. L. et al. Flora da Reserva Ducke. Guia de identificacao das plantas vasculares de uma floresta de terra-firme na Amazonia Central. 799 (INPA-DFID, 1999).
IDRISI selva manual (Clark Labs, Clark University, Worcester, MA, USA, 2012).
Geml, J. et al. Large-scale fungal diversity assessment in the Andean Yungas forests reveals strong community turnover among forest types along an altitudinal gradient. Mol. Ecol. 23, 2452–2472, https://doi.org/10.1111/mec.12765 (2014).
Ihrmark, K. et al. New primers to amplify the fungal ITS2 region – evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 82, 666–677, https://doi.org/10.1111/j.1574-6941.2012.01437.x (2012).
White, T. J., Bruns, T. D., Lee, S. B. & Taylor, J. W. in PCR - Protocols and applications - A laboratory manual (eds N. Innis, D. Gelfand, J. Sninsky, & T. White) 315-322 (Academic Press, 1990).
Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583, https://doi.org/10.1038/nmeth.3869 (2016).
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461, https://doi.org/10.1093/bioinformatics/btq461 (2010).
Kõljalg, U. et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277, https://doi.org/10.1111/mec.12481 (2013).
Põlme, S. et al. FungalTraits: a user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers. 105, 1–16, https://doi.org/10.1007/s13225-020-00466-2 (2020).
Mackereth, F. J. H., Heron, J. & Talling, J. F. Water analysis: some revised methods for limnologists. 120 (Freshwater Biological Association, 1978).
Allen, S. E. Chemical analysis of ecological materials. (Blackwell Scientific Publications, 1989).
van Raij, B., Andrade, J. C., Cantarella, H. & Quaggio, J. A. Análise química para avaliação da fertilidade de solos tropicais. 285 (Instituto Agronomico, 2001).
GUniFrac: Generalized UniFrac Distances. R package version 1.1. https://CRAN.R-project.org/package=GUniFrac (2018).
Benjamini, Y. & Hochberg, Y. Controlling the false diiscovery rate: a practical and powerful approach to multiple testing. J. Royal Stat. Soc. Ser. B (Methodol.) 57, 289–300 (1995).
Baselga, A. & Orme, C. D. L. betapart: an R package for the study of beta diversity. Methods Ecol. Evol. 3, 808–812, https://doi.org/10.1111/j.2041-210X.2012.00224.x (2012).
Martiny, J. B. H. et al. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4, 102, https://doi.org/10.1038/nrmicro1341 (2006).
Gómez-Rodríguez, C. & Baselga, A. Variation among European beetle taxa in patterns of distance decay of similarity suggests a major role of dispersal processes. Ecography 41, 1825–1834, https://doi.org/10.1111/ecog.03693 (2018).
Nekola, J. C. & White, P. S. The distance decay of similarity in biogeography and ecology. J. Biogeogr. 26, 867–878, https://doi.org/10.1046/j.1365-2699.1999.00305.x (1999).
Martínez-Santalla, S., Martín-Devasa, R., Gómez-Rodríguez, C., Crujeiras, R. M. & Baselga, A. Assessing the nonlinear decay of community similarity: permutation and site-block resampling significance tests. J. Biogeogr. 49, 968–978, https://doi.org/10.1111/jbi.14351 (2022).
Chase, J. M., Kraft, N. J. B., Smith, K. G., Vellend, M. & Inouye, B. D. Using null models to disentangle variation in community dissimilarity from variation in α-diversity. Ecosphere 2, art24, https://doi.org/10.1890/ES10-00117.1 (2011).
Ning, D., Deng, Y., Tiedje, J. M. & Zhou, J. A general framework for quantitatively assessing ecological stochasticity. Proc. Natl Acad. Sci. 116, 16892–16898, https://doi.org/10.1073/pnas.1904623116 (2019).
Nilsson, R. H., Kristiansson, E., Ryberg, M., Hallenberg, N. & Larsson, K.-H. Intraspecific ITS variability in the kingdom fungi as expressed in the international sequence databases and its implications for molecular species identification. Evol. Bioinform. 4, EBO.S653, https://doi.org/10.4137/ebo.S653 (2008).
Brown, S. P. & Jumpponen, A. Phylogenetic diversity analyses reveal disparity between fungal and bacterial communities during microbial primary succession. Soil Biol. Biochem. 89, 52–60, https://doi.org/10.1016/j.soilbio.2015.06.025 (2015).
Acknowledgements
EB and LN acknowledge support of the Fundação para a Ciência e a Tecnologia, Portugal (Grant/Award Number: SFRH/BPD/77795/2011) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil (Grant/Award Number: 480568/2011). The authors thank Adalberto Luis Val, coordinator of INCT ADAPTA (INPA) for financial support. Symbols used in Fig. 1 and Supplementary Fig. 5 are courtesy of the Integration and Application Network (ian.umces.edu/symbols/), Center for Environmental Science, University of Maryland. Technical staff at Naturalis, Leiden, the Netherlands carried out the PCRs and the sequencing of DNA.
Author information
Authors and Affiliations
Contributions
LN planned the research; LN and EB established the experiment and undertook the fieldwork; EB extracted the DNA and made the statistical analyses; JG conducted the bioinformatical analyses; EB and LN led the writing of the manuscript with input from J.G. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
Erika Buscardo is an Editorial Board Member for Communications Earth & Environment but was not involved in the editorial review of, nor the decision to publish this article. None of the other authors have competing interests to declare.
Peer review
Peer review information
Communications Earth & Environment thanks Christopher Barnes, Tessa Camenzind and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Clare Davis. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Buscardo, E., Geml, J. & Nagy, L. Seasonal dependence of deterministic versus stochastic processes influencing soil fungal community composition in a lowland Amazonian rain forest. Commun Earth Environ 5, 334 (2024). https://doi.org/10.1038/s43247-024-01273-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s43247-024-01273-2
This article is cited by
-
Seasonal dynamics of soil bacterial and fungal communities in Colombian tropical forests and páramo ecosystems
Soil Ecology Letters (2025)