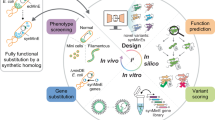
Abstract
Directed evolution, a strategy for protein engineering, optimizes protein properties (that is, fitness) by expensive and time-consuming screening or selection of a large mutational sequence space. Machine learning-assisted directed evolution (MLDE), which screens sequence properties in silico, can accelerate the optimization and reduce the experimental burden. This work introduces an MLDE framework, cluster learning-assisted directed evolution (CLADE), which combines hierarchical unsupervised clustering sampling and supervised learning to guide protein engineering. The clustering sampling selectively picks and screens variants in targeted subspaces, which guides the subsequent generation of diverse training sets. In the last stage, accurate predictions via supervised learning models improve the final outcomes. By sequentially screening 480 sequences out of 160,000 in a four-site combinatorial library with five equal experimental batches, CLADE achieves global maximal fitness hit rates of up to 91.0% and 34.0% for the GB1 and PhoQ datasets, respectively, improved from the values of 18.6% and 7.2% obtained by random sampling-based MLDE.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The GB1 dataset4 is available at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA278685/ with accession code PRJNA278685. The PhoQ dataset has been reported in the literature52. The processed version of it used in this work is owned by the Michael T. Laub laboratory and is available at https://github.com/WeilabMSU/CLADE. Source data are provided with this paper.
Code availability
All source codes and models are publicly available at https://github.com/WeilabMSU/CLADE60.
References
Tawfik, O. K. & S, D. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu. Rev. Biochem. 79, 471–505 (2010).
Wittmann, B. J., Yue, Y. & Arnold, F. H. Informed training set design enables efficient machine learning-assisted directed protein evolution. Cell Syst. 12, 1026–1045.e7 (2021).
Wu, Z., Kan, S. J., Lewis, R. D., Wittmann, B. J. & Arnold, F. H. Machine learning-assisted directed protein evolution with combinatorial libraries. Proc. Natl Acad. Sci. USA 116, 8852–8858 (2019).
Wu, N. C., Dai, L., Olson, C. A., Lloyd-Smith, J. O. & Sun, R. Adaptation in protein fitness landscapes is facilitated by indirect paths. eLife 5, e16965 (2016).
Yang, K. K., Wu, Z. & Arnold, F. H. Machine-learning-guided directed evolution for protein engineering. Nat. Methods 16, 687–694 (2019).
Siedhoff, N. E., Schwaneberg, U. & Davari, M. D. Machine learning-assisted enzyme engineering. Methods Enzymol. 643, 281–315 (2020).
Narayanan, H. et al. Machine learning for biologics: opportunities for protein engineering, developability and formulation. Trends Pharmacol. Sci. 42, 151–165 (2021).
Mazurenko, S., Prokop, Z. & Damborsky, J. Machine learning in enzyme engineering. ACS Catal. 10, 1210–1223 (2019).
Bojar, D. & Fussenegger, M. The role of protein engineering in biomedical applications of mammalian synthetic biology. Small 16, 1903093 (2020).
Kim, G. B., Kim, W. J., Kim, H. U. & Lee, S. Y. Machine learning applications in systems metabolic engineering. Curr. Opin. Biotechnol. 64, 1–9 (2020).
Tian, J., Wu, N., Chu, X. & Fan, Y. Predicting changes in protein thermostability brought about by single- or multi-site mutations. BMC Bioinformatics 11, 370 (2010).
Cang, Z. & Wei, G.-W. Analysis and prediction of protein folding energy changes upon mutation by element specific persistent homology. Bioinformatics 33, 3549–3557 (2017).
Quan, L., Lv, Q. & Zhang, Y. STRUM: structure-based prediction of protein stability changes upon single-point mutation. Bioinformatics 32, 2936–2946 (2016).
Khurana, S. et al. DeepSol: a deep learning framework for sequence-based protein solubility prediction. Bioinformatics 34, 2605–2613 (2018).
Hie, B., Bryson, B. D. & Berger, B. Leveraging uncertainty in machine learning accelerates biological discovery and design. Cell Syst. 11, 461–477 (2020).
Wang, M., Cang, Z. & Wei, G.-W. A topology-based network tree for the prediction of protein–protein binding affinity changes following mutation. Nat. Mach. Intell. 2, 116–123 (2020).
Rao, R. et al. Evaluating protein transfer learning with tape. Adv. Neural Inf. Process. Syst. 32, 9689–9701 (2019).
Yang, K. K., Wu, Z., Bedbrook, C. N. & Arnold, F. H. Learned protein embeddings for machine learning. Bioinformatics 34, 2642–2648 (2018).
Riesselman, A. J., Ingraham, J. B. & Marks, D. S. Deep generative models of genetic variation capture the effects of mutations. Nat. Methods 15, 816–822 (2018).
Alley, E. C., Khimulya, G., Biswas, S., AlQuraishi, M. & Church, G. M. Unified rational protein engineering with sequence-based deep representation learning. Nat. Methods 16, 1315–1322 (2019).
Bepler, T. & Berger, B. Learning protein sequence embeddings using information from structure. In Proc. International Conference on Learning Representations (2018).
Rao, R. et al. MSA transformer. In Proc. 38th International Conference on Machine Learning Vol. 139, 8844–8856 (PMLR, 2021).
Hamerly, G. & Elkan, C. Learning the k in k-means. Adv. Neural Inf. Process. Syst. 16, 281–288 (2004).
Frey, B. J. & Dueck, D. Clustering by passing messages between data points. Science 315, 972–976 (2007).
Blondel, V. D., Guillaume, J.-L., Lambiotte, R. & Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. 2008, P10008 (2008).
Schubert, E., Sander, J., Ester, M., Kriegel, H. P. & Xu, X. DBSCAN revisited, revisited: why and how you should (still) use DBSCAN. ACM Trans. Database Syst. 42, 1–21 (2017).
Sha, Y., Wang, S., Zhou, P. & Nie, Q. Inference and multiscale model of epithelial-to-mesenchymal transition via single-cell transcriptomic data. Nucleic Acids Res. 48, 9505–9520 (2020).
Kuang, D., Ding, C. & Park, H. Symmetric nonnegative matrix factorization for graph clustering. In Proc. 2012 SIAM International Conference on Data Mining 106–117 (SIAM, 2012).
Oller-Moreno, S., Kloiber, K., Machart, P. & Bonn, S. Algorithmic advances in machine learning for single cell expression analysis. Curr. Opin. Syst. Biol. 25, 27–33 (2021).
Saxena, A. et al. A review of clustering techniques and developments. Neurocomputing 267, 664–681 (2017).
Zhong, Y., Ma, A., Soon Ong, Y., Zhu, Z. & Zhang, L. Computational intelligence in optical remote sensing image processing. Appl. Soft Comput. 64, 75–93 (2018).
Li, G., Dong, Y. & Reetz, M. T. Can machine learning revolutionize directed evolution of selective enzymes? Adv. Synth. Catal. 361, 2377–2386 (2019).
Saito, Y. et al. Machine-learning-guided mutagenesis for directed evolution of fluorescent proteins. ACS Synth. Biol. 7, 2014–2022 (2018).
Bedbrook, C. N., Yang, K. K., Rice, A. J., Gradinaru, V. & Arnold, F. H. Machine learning to design integral membrane channelrhodopsins for efficient eukaryotic expression and plasma membrane localization. PLoS Comput. Biol. 13, e1005786 (2017).
Romero, P. A., Krause, A. & Arnold, F. H. Navigating the protein fitness landscape with Gaussian processes. Proc. Natl Acad. Sci. USA 110, E193–E201 (2013).
Mason, D. M. et al. Deep learning enables therapeutic antibody optimization in mammalian cells by deciphering high-dimensional protein sequence space. bioRxiv https://doi.org/10.1101/617860 (2019).
Hie, B. L. & Yang, K. K. Adaptive machine learning for protein engineering. Preprint at https://arxiv.org/abs/2106.05466 (2021).
Schulz, E., Speekenbrink, M. & Krause, A. A tutorial on Gaussian process regression: modelling, exploring and exploiting functions. J. Math. Psychol. 85, 1–16 (2018).
Srinivas, N., Krause, A., Kakade, S. & Seeger, M. Gaussian process optimization in the bandit setting: no regret and experimental design. In Proc. 27th International Conference on Machine Learning 1015–1022 (ACM, 2010).
Hopf, T. A. et al. Mutation effects predicted from sequence co-variation. Nat. Biotechnol. 35, 128–135 (2017).
Meier, J. et al. Language models enable zero-shot prediction of the effects of mutations on protein function. Preprint at bioRxiv https://doi.org/10.1101/2021.07.09.450648 (2021).
Yang, K. K., Chen, Y., Lee, A. & Yue, Y. Batched stochastic Bayesian optimization via combinatorial constraints design. In Proc. 22nd International Conference on Artificial Intelligence and Statistics 3410–3419 (PMLR, 2019).
Kawashima, S., Ogata, H. & Kanehisa, M. AAindex: amino acid index database. Nucleic Acids Res. 27, 368–369 (1999).
Ofer, D. & Linial, M. ProFET: feature engineering captures high-level protein functions. Bioinformatics 31, 3429–3436 (2015).
Georgiev, A. G. Interpretable numerical descriptors of amino acid space. J. Comput. Biol. 16, 703–723 (2009).
Wittmann, B. J., Johnston, K. E., Wu, Z. & Arnold, F. H. Advances in machine learning for directed evolution. Curr. Opin. Struct. Biol. 69, 11–18 (2021).
Bubeck, S., Munos, R., Stoltz, G. & Szepesvári, C. X-armed bandits. J. Mach. Learn. Res. 12, 1655–1695 (2011).
Munos, R. Optimistic optimization of a deterministic function without the knowledge of its smoothness. Adv. Neural Inf. Process. Syst. 24, 783–791 (2011).
Pahari, S. et al. SAAMBE-3D: predicting effect of mutations on protein-protein interactions. Int. J. Mol. Sci. 21, 2563 (2020).
Rives, A. et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc. Natl Acad. Sci. USA 118, e2016239118 (2021).
Strain-Damerell, C. & Burgess-Brown, N. A. in High-Throughput Protein Production and Purification 281–296 (Springer, 2019).
Podgornaia, A. I. & Laub, M. T. Pervasive degeneracy and epistasis in a protein-protein interface. Science 347, 673–677 (2015).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Zamyatnin, A. Protein volume in solution. Progr. Biophys. Mol. Biol. 24, 107–123 (1972).
Rasmussen, C. E. in Summer School on Machine Learning 63–71 (Springer, 2003).
Hopf, T. A. et al. The EVcouplings Python framework for coevolutionary sequence analysis. Bioinformatics 35, 1582–1584 (2019).
Steinegger, M. et al. HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinformatics 20, 473 (2019).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies and species. Nat. Biotechnol. 36, 411–420 (2018).
Schmera, D., Erős, T. & Podani, J. A measure for assessing functional diversity in ecological communities. Aquatic Ecol. 43, 157–167 (2009).
YuchiQiu/CLADE: Nature Computational Science publication accompaniment (v1.0.0) (Zenodo, 2021); https://doi.org/10.5281/zenodo.5585394
Acknowledgements
This work was supported in part by NIH grants nos. GM126189 and GM129004, NSF grants nos. DMS-2052983, DMS-1761320 and IIS-1900473, NASA grant no. 80NSSC21M0023, the Michigan Economic Development Corporation, Bristol-Myers Squibb 65109, Pfizer and the MSU Foundation. We thank the IBM Thomas J. Watson Research Center, the COVID-19 High Performance Computing Consortium, NVIDIA and MSU HPCC for computational assistance. We thank F. Arnold’s laboratory for assistance with the MLDE package and M.T. Laub’s laboratory for assistance with the PhoQ dataset.
Author information
Authors and Affiliations
Contributions
All authors conceived this work and contributed to the original draft, review and editing. Y.Q. performed experiments and analyzed the data. G.-W.W. provided supervision and resources and acquired funding.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Reviewer recognition statement Nature Computational Science thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary sections 1–6, Figs. 1–11 and Tables 1–5.
Supplementary Data 1
Performance of CLADE for the GB1 and PhoQ datasets.
Supplementary Data 2
Performance of Gaussian process for the GB1 and PhoQ datasets.
Supplementary Data 3
Performance of zero-shot predictor-based CLADE and ftMLDE.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Rights and permissions
About this article
Cite this article
Qiu, Y., Hu, J. & Wei, GW. Cluster learning-assisted directed evolution. Nat Comput Sci 1, 809–818 (2021). https://doi.org/10.1038/s43588-021-00168-y
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43588-021-00168-y
This article is cited by
-
Active learning-guided optimization of cell-free biosensors for lead testing in drinking water
Nature Communications (2025)
-
Active learning-assisted directed evolution
Nature Communications (2025)
-
Integrating protein language models and automatic biofoundry for enhanced protein evolution
Nature Communications (2025)
-
Accelerating protein engineering with fitness landscape modelling and reinforcement learning
Nature Machine Intelligence (2025)
-
Computer-aided rational design strategy based on protein surface charge to improve the thermal stability of a novel esterase from Geobacillus jurassicus
Biotechnology Letters (2024)