Abstract
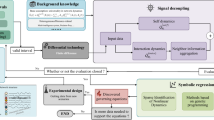
The availability of empirical data that capture the structure and behaviour of complex networked systems has been greatly increased in recent years; however, a versatile computational toolbox for unveiling a complex system’s nodal and interaction dynamics from data remains elusive. Here we develop a two-phase approach for the autonomous inference of complex network dynamics, and its effectiveness is demonstrated by the tests of inferring neuronal, genetic, social and coupled oscillator dynamics on various synthetic and real networks. Importantly, the approach is robust to incompleteness and noises, including low resolution, observational and dynamical noises, missing and spurious links, and dynamical heterogeneity. We apply the two-phase approach to infer the early spreading dynamics of influenza A flu on the worldwide airline network, and the inferred dynamical equation can also capture the spread of severe acute respiratory syndrome and coronavirus disease 2019. These findings together offer an avenue to discover the hidden microscopic mechanisms of a broad array of real networked systems.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Source data are provided with this paper. The empirical network data include C. elegans connectome54,55,56, the mushroom-body region of Drosophila57, Northern Europe power grid58, the US power grid59, Advogato social network60 retrieved from https://networkrepository.com/ and worldwide airline network data retrieved from OpenFlights (https://openflights.org/data.html). The empirical data of epidemic spreading include daily reported numbers of H1N1 and SARS cases available at Kaggle (https://www.kaggle.com/lnunes/a-brief-comparative-study-of-epidemics/data) and the daily reported numbers of COVID-19 cases61.
Code availability
All the source codes are publicly available at the Code Ocean capsule62.
Change history
29 April 2022
A Correction to this paper has been published: https://doi.org/10.1038/s43588-022-00255-8
References
Grewe, B. F., Langer, D., Kasper, H., Kampa, B. M. & Helmchen, F. High-speed in vivo calcium imaging reveals neuronal network activity with near-millisecond precision. Nat. Methods 7, 399–405 (2010).
Stetter, O., Battaglia, D., Soriano, J. & Geisel, T. Model-free reconstruction of excitatory neuronal connectivity from calcium imaging signals. PLoS Comput. Biol. 8, e1002653 (2012).
Reuter, J. A., Spacek, D. V. & Snyder, M. P. High-throughput sequencing technologies. Mol. Cell. 58, 586–597 (2015).
Levy, S. E. & Myers, R. M. Advancements in next-generation sequencing. Annu. Rev. Genom. Hum. Genet. 17, 95–115 (2016).
Colizza, V., Barrat, A., Barthélemy, M. & Vespignani, A. The role of the airline transportation network in the prediction and predictability of global epidemics. Proc. Natl Acad. Sci. USA 103, 2015–2020 (2006).
Brockmann, D. & Helbing, D. The hidden geometry of complex, network-driven contagion phenomena. Science 342, 1337–1342 (2013).
Chang, S. et al. Mobility network models of COVID-19 explain inequities and inform reopening. Nature 589, 82–87 (2021).
Newman, M., Barabási, A.-L. & Watts, D. J. The Structure and Dynamics of Networks (Princeton Univ. Press, 2011).
Barzel, B. & Barabási, A.-L. Universality in network dynamics. Nat. Phys. 9, 673–681 (2013).
Harush, U. & Barzel, B. Dynamic patterns of information flow in complex networks. Nat. Commun. 8, 2181 (2017).
Stankovski, T., Pereira, T., McClintock, P. V. & Stefanovska, A. Coupling functions: universal insights into dynamical interaction mechanisms. Rev. Mod. Phys. 89, 045001 (2017).
Breakspear, M. Dynamic models of large-scale brain activity. Nat. Neurosci. 20, 340–352 (2017).
Santolini, M. & Barabási, A.-L. Predicting perturbation patterns from the topology of biological networks. Proc. Natl Acad. Sci. USA 115, E6375–E6383 (2018).
Buldyrev, S. V., Parshani, R., Paul, G., Stanley, H. E. & Havlin, S. Catastrophic cascade of failures in interdependent networks. Nature 464, 1025–1028 (2010).
Yang, Y., Nishikawa, T. & Motter, A. E. Small vulnerable sets determine large network cascades in power grids. Science 358, eaan3184 (2017).
Pastor-Satorras, R., Castellano, C., Van Mieghem, P. & Vespignani, A. Epidemic processes in complex networks. Rev. Mod. Phys. 87, 925 (2015).
Castellano, C., Fortunato, S. & Loreto, V. Statistical physics of social dynamics. Rev. Mod. Phys. 81, 591–646 (2009).
Becker, J., Brackbill, D. & Centola, D. Network dynamics of social influence in the wisdom of crowds. Proc. Natl Acad. Sci. USA 114, E5070–E5076 (2017).
Arenas, A., Díaz-Guilera, A., Kurths, J., Moreno, Y. & Zhou, C. Synchronization in complex networks. Phys. Rep. 469, 93–153 (2008).
Barzel, B., Liu, Y.-Y. & Barabási, A.-L. Constructing minimal models for complex system dynamics. Nat. Commun. 6, 7186 (2015).
Schmidt, M. & Lipson, H. Distilling free-form natural laws from experimental data. Science 324, 81–85 (2009).
Wang, W.-X., Yang, R., Lai, Y.-C., Kovanis, V. & Grebogi, C. Predicting catastrophes in nonlinear dynamical systems by compressive sensing. Phys. Rev. Lett. 106, 154101 (2011).
Brunton, S. L., Proctor, J. L. & Kutz, J. N. Discovering governing equations from data by sparse identification of nonlinear dynamical systems. Proc. Natl Acad. Sci. USA 113, 3932–3937 (2016).
Rudy, S. H., Brunton, S. L., Proctor, J. L. & Kutz, J. N. Data-driven discovery of partial differential equations. Sci. Adv. 3, e1602614 (2017).
Udrescu, S.-M. & Tegmark, M. AI Feynman: a physics-inspired method for symbolic regression. Sci. Adv. 6, eaay2631 (2020).
Raissi, M. & Karniadakis, G. E. Hidden physics models: machine learning of nonlinear partial differential equations. J. Comput. Phys. 357, 125–141 (2018).
Iten, R., Metger, T., Wilming, H., Del Rio, L. & Renner, R. Discovering physical concepts with neural networks. Phys. Rev. Lett. 124, 010508 (2020).
Frishman, A. & Ronceray, P. Learning force fields from stochastic trajectories. Phys. Rev. X 10, 021009 (2020).
Brückner, D. B., Ronceray, P. & Broedersz, C. P. Inferring the dynamics of underdamped stochastic systems. Phys. Rev. Lett. 125, 058103 (2020).
Shandilya, S. G. & Timme, M. Inferring network topology from complex dynamics. New J. Phys. 13, 013004 (2011).
Newman, M. E. J. Network structure from rich but noisy data. Nat. Phys. 14, 542–545 (2018).
Rabinovich, M. I., Varona, P., Selverston, A. I. & Abarbanel, H. D. Dynamical principles in neuroscience. Rev. Mod. Phys. 78, 1213 (2006).
Marvel, S. A., Kleinberg, J., Kleinberg, R. D. & Strogatz, S. H. Continuous-time model of structural balance. Proc. Natl Acad. Sci. USA 108, 1771–1776 (2011).
Strogatz, S. H. Exploring complex networks. Nature 410, 268–276 (2001).
Barahona, M. & Pecora, L. M. Synchronization in small-world systems. Phys. Rev. Lett. 89, 054101 (2002).
Mangan, N. M., Kutz, J. N., Brunton, S. L. & Proctor, J. L. Model selection for dynamical systems via sparse regression and information criteria. Proc. Math. Phys. Eng. Sci. 473, 20170009 (2017).
Casadiego, J., Nitzan, M., Hallerberg, S. & Timme, M. Model-free inference of direct network interactions from nonlinear collective dynamics. Nat. Commun. 8, 2192 (2017).
Runge, J., Nowack, P., Kretschmer, M., Flaxman, S. & Sejdinovic, D. Detecting and quantifying causal associations in large nonlinear time series datasets. Sci. Adv. 5, eaau4996 (2019).
Sugihara, G. et al. Detecting causality in complex ecosystems. Science 338, 496–500 (2012).
Sun, J., Taylor, D. & Bollt, E. M. Causal network inference by optimal causation entropy. SIAM J. Appl. Dyn. Syst. 14, 73–106 (2015).
Kralemann, B., Pikovsky, A. & Rosenblum, M. Reconstructing effective phase connectivity of oscillator networks from observations. New J. Phys. 16, 085013 (2014).
Frässle, S. et al. Regression DCM for fMRI. NeuroImage 155, 406–421 (2017).
Gilson, M., Moreno-Bote, R., Ponce-Alvarez, A., Ritter, P. & Deco, G. Estimation of directed effective connectivity from fMRI functional connectivity hints at asymmetries of cortical connectome. PLoS Comput. Biol. 12, e1004762 (2016).
Deco, G., Rolls, E. T. & Romo, R. Stochastic dynamics as a principle of brain function. Prog. Neurobiol. 88, 1–16 (2009).
Genkin, M., Hughes, O. & Engel, T. A. Learning non-stationary Langevin dynamics from stochastic observations of latent trajectories. Nat. Commun. 12, 5986 (2021).
Zhao, H. Inferring the dynamics of ‘black-box’ systems using a learning machine. Sci. China Phys. Mech. Astron. 64, 270511 (2021).
Jahnke, S., Memmesheimer, R.-M. & Timme, M. Stable irregular dynamics in complex neural networks. Phys. Rev. Lett. 100, 048102 (2008).
Champion, K. P., Brunton, S. L. & Kutz, J. N. Discovery of nonlinear multiscale systems: sampling strategies and embeddings. SIAM J. Appl. Dyn. Syst. 18, 312–333 (2019).
Battiston, F. et al. The physics of higher-order interactions in complex systems. Nat. Phys. 17, 1093–1098 (2021).
Lambiotte, R., Rosvall, M. & Scholtes, I. From networks to optimal higher-order models of complex systems. Nat. Phys. 15, 313–320 (2019).
Sauer, T. Numerical solution of stochastic differential equations in finance. in Handbook of Computational Finance 529–550 (Springer, 2012).
Akaike, H. A new look at the statistical model identification. IEEE Trans. Automat. Contr. 19, 716–723 (1974).
Flores, B. E. A pragmatic view of accuracy measurement in forecasting. Omega 14, 93–98 (1986).
White, J. G., Southgate, E., Thomson, J. N. & Brenner, S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314, 1–340 (1986).
Varshney, L. R., Chen, B. L., Paniagua, E., Hall, D. H. & Chklovskii, D. B. Structural properties of the Caenorhabditis elegans neuronal network. PLoS Comput. Biol. 7, e1001066 (2011).
Yan, G. et al. Network control principles predict neuron function in the Caenorhabditis elegans connectome. Nature 550, 519–523 (2017).
Scheffer, L. K. et al. A connectome and analysis of the adult Drosophila central brain. eLife 9, e57443 (2020).
Menck, P. J., Heitzig, J., Kurths, J. & Schellnhuber, H. J. How dead ends undermine power grid stability. Nat. Commun. 5, 3969 (2014).
Kunegis, J. KONECT: the Koblenz network collection. In Proc. 22nd International Conference on World Wide Web 1343–1350 (ACM, 2013).
Rossi, R. & Ahmed, N. The network data repository with interactive graph analytics and visualization. In Twenty-Ninth AAAI Conference on Artificial Intelligence (2015).
Dong, E., Du, H. & Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 20, 533–534 (2020).
Gao, T.-T. & Yan, G. A two-phase approach for inferring complex network dynamics. Code Ocean https://doi.org/10.24433/CO.4774495.v1 (2022).
Acknowledgements
T.-T.G. and G.Y. are supported by the National Key Research and Development Program of China (grant no. 2021ZD0204500), National Natural Science Foundation of China (grant nos. 12161141016 and 11875043), Shanghai Municipal Science and Technology Major Project (grant no. 2021SHZDZX0100), Shanghai Municipal Commission of Science and Technology Project (grant nos. 18ZR1442000 and 19511132101) and Fundamental Research Funds for the Central Universities. We are also grateful for the helpful discussion with B. Barzel, J. Moore, X. Ru and T. Li.
Author information
Authors and Affiliations
Contributions
G.Y. conceived the research. G.Y. and T.-T.G. designed the research. T.-T.G. performed the research. T.-T.G. and G.Y. analysed the results. G.Y. and T.-T.G. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Computational Science thanks Matthieu Gilson and the other, anonymous reviewer(s) for their contribution to the peer review of this work. Handling editor: Jie Pan, in collaboration with the Nature Computational Science team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–20, Sections I–VI and Tables 1–5.
Source data
Source Data Fig. 1
True and inferred trajectories data.
Source Data Fig. 2
Unprocessed inferred results, time-series data and trajectories data.
Source Data Fig. 3
Unprocessed inferred results, time-series data and trajectories data.
Source Data Fig. 4
Statistical source data and trajectories data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Raw empirical data and time-series data.
Rights and permissions
About this article
Cite this article
Gao, TT., Yan, G. Autonomous inference of complex network dynamics from incomplete and noisy data. Nat Comput Sci 2, 160–168 (2022). https://doi.org/10.1038/s43588-022-00217-0
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43588-022-00217-0
This article is cited by
-
Discovering the laws behind complex networked systems
Nature Computational Science (2026)
-
Observing network dynamics through sentinel nodes
Nature Communications (2025)
-
Discovering physical laws with parallel symbolic enumeration
Nature Computational Science (2025)
-
Discovering network dynamics with neural symbolic regression
Nature Computational Science (2025)
-
Learning interpretable network dynamics via universal neural symbolic regression
Nature Communications (2025)