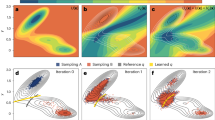
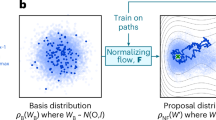
Abstract
The problem of studying rare events is central to many areas of computer simulations. We recently proposed an approach to solving this problem that involves computing the committor function, showing how it can be iteratively computed in a variational way while efficiently sampling the transition state ensemble. Here we greatly improve this procedure by combining it with a metadynamics-like enhanced sampling approach in which a logarithmic function of the committor is used as a collective variable. This procedure leads to an accurate sampling of the free energy surface in which transition states and metastable basins are studied with the same thoroughness. We show that our approach can be used in cases with the possibility of competing reactive paths and metastable intermediates. In addition, we demonstrate how physical insights can be obtained from the optimized committor model and the sampled data, thus providing a full characterization of the rare event under study.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Training and simulation data and inputs are available on GitHub56 and on Zenodo57. Source data are provided with this paper.
Code availability
The code for the training of the NN-based committor model alongside didactic tutorials is available through the open-source mlcolvar library55, which is the preferred way to access the most updated code. To obtain the results reported in the manuscript, version 1.2.2 was used and a frozen version is also available on Zenodo57. The PLUMED58,59 interface for the application of the bias is available on GitHub56 and on Zenodo57.
References
Frenkel, D. & Smit, B. Understanding Molecular Simulation: From Algorithms to Applications, Vol. 1 (Elsevier, 2001).
Hénin, J., Lelièvre, T., Shirts, M. R., Valsson, O. & Delemotte, L. Enhanced sampling methods for molecular dynamics simulations. Living J. Comput. Mol. Sci. 4, 1583 (2022).
Kang, P., Trizio, E., & Parrinello, M. Computing the committor with the committor to study the transition state ensemble. Nat. Comput. Sci. 4, 451–460 (2024).
Weinan, E. & Vanden-Eijnden, E. Transition-path theory and path-finding algorithms for the study of rare events. Annu. Rev. Phys. Chem. 61, 391–420 (2010).
Bolhuis, P. G., Dellago, C. & Chandler, D. Reaction coordinates of biomolecular isomerization. Proc. Natl Acad. Sci. USA 97, 5877–5882 (2000).
Ma, A. & Dinner, A. R. Automatic method for identifying reaction coordinates in complex systems. J. Phys. Chem. B 109, 6769–6779 (2005).
Berezhkovskii, A. & Szabo, A. One-dimensional reaction coordinates for diffusive activated rate processes in many dimensions. J. Chem. Phys. 122, 014503 (2005).
Peters, B. & Trout, B. L. Obtaining reaction coordinates by likelihood maximization. J. Chem. Phys. 125, 054108 (2006).
Vanden-Eijnden, E. Towards a theory of transition paths. J. Stat. Phys. 123, 503–523 (2006).
Rotskoff, G. M., Mitchell, A. R., & Vanden-Eijnden, E. Active importance sampling for variational objectives dominated by rare events: consequences for optimization and generalization. Proc. Mach. Learn. Res. 145, 757–780 (2022).
Chen, Y., Hoskins, J., Khoo, Y. & Lindsey, M. Committor functions via tensor networks. J. Comput. Phys. 472, 111646 (2023).
Chen, H., Roux, B. & Chipot, C. Discovering reaction pathways, slow variables, and committor probabilities with machine learning. J. Chem. Theory Comput. 19, 4414–4426 (2023).
Li, Q., Lin, B. & Ren, W. Computing committor functions for the study of rare events using deep learning. J. Chem. Phys. 151, 054112 (2019).
Jung, H. et al. Machine-guided path sampling to discover mechanisms of molecular self-organization. Nat. Comput. Sci. 3, 334–345 (2023).
Mitchell, A. R. & Rotskoff, G. M. Committor guided estimates of molecular transition rates. J. Chem. Theory Comput. 20, 9378–9393 (2024).
He, Z., Chipot, C. & Roux, B. Committor-consistent variational string method. J. Phys. Chem. Lett. 13, 9263–9271 (2022).
Khoo, Y., Lu, J. & Ying, L. Solving for high-dimensional committor functions using artificial neural networks. Res. Math. Sci. 6, 1 (2019).
Kolmogoroff, A. Über die analytischen methoden in der wahrscheinlichkeitsrechnung. Math. Ann. 104, 415–458 (1931).
Bonati, L., Piccini, G. & Parrinello, M. Deep learning the slow modes for rare events sampling. Proc. Natl Acad. Sci. USA 118, e2113533118 (2021).
Ray, D., Trizio, E. & Parrinello, M. Deep learning collective variables from transition path ensemble. J. Chem. Phys. 158, 204102 (2023).
Laio, A. & Parrinello, M. Escaping free-energy minima. Proc. Natl Acad. Sci. USA 99, 12562–12566 (2002).
Invernizzi, M. & Parrinello, M. Rethinking metadynamics: from bias potentials to probability distributions. J. Phys. Chem. Lett. 11, 2731–2736 (2020).
Trizio, E., Rizzi, A., Piaggi, P. M., Invernizzi, M. & Bonati, L. Advanced simulations with PLUMED: OPES and machine learning collective variables. Preprint at https://arxiv.org/abs/2410.18019 (2024).
Li, W. & Ma, A. Recent developments in methods for identifying reaction coordinates. Mol. Simul. 40, 784–793 (2014).
Bonati, L., Rizzi, V. & Parrinello, M. Data-driven collective variables for enhanced sampling. J. Phys. Chem. Lett. 11, 2998–3004 (2020).
Trizio, E. & Parrinello, M. From enhanced sampling to reaction profiles. J. Phys. Chem. Lett. 12, 8621–8626 (2021).
Invernizzi, M. & Parrinello, M. Exploration vs convergence speed in adaptive bias enhanced sampling. J. Chem. Theory Comput. 18, 3988–3996 (2022).
Vlugt, T. J. H. & Smit, B. On the efficient sampling of pathways in the transition path ensemble. PhysChemComm 4, 11–17 (2001).
Borrero, E. E. & Dellago, C. Avoiding traps in trajectory space: metadynamics enhanced transition path sampling. Eur. Phys. J. Spec. Top. 225, 1609–1620 (2016).
Bolhuis, P. G. & Csányi, G. Nested transition path sampling. Phys. Rev. Lett. 120, 250601 (2018).
Mandelli, D., Hirshberg, B. & Parrinello, M. Metadynamics of paths. Phys. Rev. Lett. 125, 026001 (2020).
Capelli, R., Carloni, P. & Parrinello, M. Exhaustive search of ligand binding pathways via volume-based metadynamics. J. Phys. Chem. Lett. 10, 3495–3499 (2019).
Lindorff-Larsen, K., Piana, S., Dror, R. O. & Shaw, D. E. How fast-folding proteins fold. Science 334, 517–520 (2011).
Schubert, E. & Lenssen, L. Fast k-medoids clustering in Rust and Python. J. Open Source Softw. 7, 4183 (2022).
Pizarroso, J., Portela, J. & Muñoz, A. NeuralSens: sensitivity analysis of neural networks. J. Stat. Softw. 102, 1–36 (2022).
Novelli, P., Bonati, L., Pontil, M. & Parrinello, M. Characterizing metastable states with the help of machine learning. J. Chem. Theory Comput. 18, 5195–5202 (2022).
Yin, J. et al. Overview of the SAMPL5 host–guest challenge: are we doing better? J. Comput.-Aided Mol. Des. 31, 1–19 (2017).
Rizzi, V., Bonati, L., Ansari, N. & Parrinello, M. The role of water in host–guest interaction. Nat. Commun. 12, 93 (2021).
Limongelli, V., Bonomi, M. & Parrinello, M. Funnel metadynamics as accurate binding free-energy method. Proc. Natl Acad. Sci. USA 110, 6358–6363 (2013).
Zhang, J. et al. Descriptor-free collective variables from geometric graph neural networks. J. Chem. Theory Comput. 20, 10787–10797 (2024).
Dietrich, F. M., Advincula, X. R., Gobbo, G., Bellucci, M. A. & Salvalaglio, M. Machine learning nucleation collective variables with graph neural networks. J. Chem. Theory Comput. 20, 1600–1611 (2024).
Zou, Z. & Tiwary, P. Enhanced sampling of crystal nucleation with graph representation learnt variables. J. Phys. Chem. B 128, 3037–3045 (2024).
Aristoff, D., Copperman, J., Simpson, G., Webber, R. J. & Zuckerman, D. M. Weighted ensemble: recent mathematical developments. J. Chem. Phys. 158, 014108 (2023).
Wang, Y.-G., Mei, D., Glezakou, V.-A., Li, J. & Rousseau, R. Dynamic formation of single-atom catalytic active sites on ceria-supported gold nanoparticles. Nat. Commun. 6, 6511 (2015).
Wang, Y.-G. et al. CO oxidation on Au/TiO2: condition-dependent active sites and mechanistic pathways. J. Am. Chem. Soc. 138, 10467–10476 (2016).
Yang, M., Raucci, U. & Parrinello, M. Reactant-induced dynamics of lithium imide surfaces during the ammonia decomposition process. Nat. Catal. 6, 829–836 (2023).
Tian, J. et al. Dynamically formed active sites on liquid boron oxide for selective oxidative dehydrogenation of propane. ACS Catal. 13, 8219–8236 (2023).
Perego, S., Bonati, L., Tripathi, S. & Parrinello, M. How dynamics changes ammonia cracking on iron surfaces. ACS Catal. 14, 14652–14664 (2024).
Tripathi, S., Bonati, L., Perego, S. & Parrinello, M. How poisoning is avoided in a step of relevance to the Haber–Bosch catalysis. ACS Catal. 14, 4944–4950 (2024).
Dill, K. A. & MacCallum, J. L. The protein-folding problem, 50 years on. Science 338, 1042–1046 (2012).
Sztain, T. et al. A glycan gate controls opening of the SARS-CoV-2 spike protein. Nat. Chem. 13, 963–968 (2021).
Dellago, C., Bolhuis, P. & Geissler, P. in Computer Simulations in Condensed Matter Systems: From Materials to Chemical Biology, Vol. 1 (eds Ferrario, M. et al.) 349–391 (Springer, 2006).
Bolhuis, P. G., Chandler, D., Dellago, C. & Geissler, P. L. Transition path sampling: throwing ropes over rough mountain passes, in the dark. Annu. Rev. Phys. Chem. 53, 291–318 (2002).
Lazzeri, G., Jung, H., Bolhuis, P. G. & Covino, R. Molecular free energies, rates, and mechanisms from data-efficient path sampling simulations. J. Chem. Theory Comput. 19, 9060–9076 (2023).
Bonati, L., Trizio, E., Rizzi, A. & Parrinello, M. A unified framework for machine learning collective variables for enhanced sampling simulations: mlcolvar. J. Chem. Phys. 159, 014801 (2023).
Trizio, E., Kang, P. & Parrinello, M. Supporting data for the paper: “everything everywhere all at once, a probability-based enhanced sampling approach to rare events”. GitHub https://github.com/EnricoTrizio/committor_2.0 (2025).
Trizio, E., Kang, P. & Parrinello, M. Everything everywhere all at once: a probability-based enhanced sampling approach to rare events. Zenodo https://doi.org/10.5281/zenodo.15089372 (2025).
Tribello, G. A., Bonomi, M., Branduardi, D., Camilloni, C. & Bussi, G. PLUMED 2: new feathers for an old bird. Comput. Phys. Commun. 185, 604–613 (2014).
The PLUMED Consortium. Promoting transparency and reproducibility in enhanced molecular simulations. Nat. Methods 16, 670–673 (2019).
Acknowledgements
We are grateful to L. Bonati, A. Rizzi, J. Zhang, U. Raucci, A. Triveri and F. Mambretti for discussions and feedback about this manuscript.
Author information
Authors and Affiliations
Contributions
All the authors equally contributed to the manuscript by conceptualizing the project, developing the theoretical methodologies and participating in the writing of the manuscript. Specifically, E.T. and P.K. developed the code for the training of the committor function and conducted the computational simulations.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Computational Science thanks Samik Bose, Carme Rovira, and Omar Valsson for their contribution to the peer review of this work. Primary Handling Editor: Kaitlin McCardle, in collaboration with the Nature Computational Science team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Convergence of ΔG.
Convergence with simulation time of the estimate for the folding energy of chignolin (A) and the binding energy of the G2 ligand to the OAMe octa-acid guest (B). The average estimates from independent simulations for each system (3 for chignolin, 4 for OAMe-G2) are reported as a blue solid line, whereas the uncertainty, computed as the standard deviation over the three replicas, is depicted as a shaded blue region. The reference values are provided as gray dashed lines, and the 0.5 kBT interval around the reference is marked by gray dotted lines. For chignolin, that is the unbiased estimate from ref. 33, for the calixarene, an estimate obtained using the enhanced sampling setup of ref. 38.
Supplementary information
Supplementary Information
Supplementary Figs. 1–9, Tables 1–6 and simulation computational details.
Source data
Source Data Fig. 2
Muller–Brown potential. a, Scatter plot data, reference potential isoline data, reference and learned committor data. b, Sampling distribution data. c, Free energy surface data.
Source Data Fig. 3
Alanine. a, Free energy surface isoline data, sampling distribution data. b, Free energy surface isoline data, Kolmogorov distribution data. c, Sampling distribution data. d, Free energy surface data, reference line TSE.
Source Data Fig. 4
Double-path potential. a, Potential isoline data, learned committor data. b, Potential isoline data, Kolmogorov distribution data. c, Potential isoline data, computed free energy surface data.
Source Data Extended Data Fig. 1
a, ΔG data (time, avg, s.d., ref. value) for chingolin. b, ΔG data (time, avg, s.d., ref. value) for calixarene.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Trizio, E., Kang, P. & Parrinello, M. Everything everywhere all at once: a probability-based enhanced sampling approach to rare events. Nat Comput Sci 5, 582–591 (2025). https://doi.org/10.1038/s43588-025-00799-5
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43588-025-00799-5
This article is cited by
-
Everything everywhere all at once: a probability-based enhanced sampling approach to rare events
Nature Computational Science (2025)