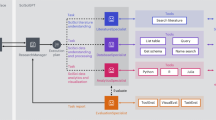
Abstract
Scientific research increasingly relies on specialized computational tools, yet effectively utilizing these tools requires substantial domain expertise. While large language models show promise in tool automation, they struggle to seamlessly integrate and orchestrate multiple tools for complex scientific workflows. Here we present SciToolAgent, a large language model-powered agent that automates hundreds of scientific tools across biology, chemistry and materials science. At its core, SciToolAgent leverages a scientific tool knowledge graph that enables intelligent tool selection and execution through graph-based retrieval-augmented generation. The agent also incorporates a comprehensive safety-checking module to ensure responsible and ethical tool usage. Extensive evaluations on a curated benchmark demonstrate that SciToolAgent outperforms existing approaches. Case studies in protein engineering, chemical reactivity prediction, chemical synthesis and metal–organic framework screening further demonstrate SciToolAgent’s capability to automate complex scientific workflows, making advanced research tools accessible to both experts and nonexperts.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The toxic compound data were obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov/#query=toxic&tab=compound), and the toxic protein data were sourced from UniProtKB (https://www.uniprot.org/uniprotkb?query=toxic&facets=reviewed%3Atrue). The SciToolEval dataset is available via Zenodo at https://doi.org/10.5281/zenodo.15691499 (ref. 44). Source data are provided with this paper.
Code availability
The source code of SciToolAgent is available via GitHub at https://github.com/hicai-zju/scitoolagent and via Zenodo at https://doi.org/10.5281/zenodo.15707181 (ref. 45).
References
Birhane, A., Kasirzadeh, A., Leslie, D. & Wachter, S. Science in the age of large language models. Nat. Rev. Phys. 5, 277–280 (2023).
Schick, T. et al. Toolformer: language models can teach themselves to use tools. Adv. Neural Inf. Process. Syst. 36, 68539–68551 (2023).
Yang, R. et al. GPT4Tools: teaching large language model to use tools via self-instruction. Adv. Neural Inf. Process. Syst. 36, 71995–72007 (2024).
Guo, T. et al. What can large language models do in chemistry? A comprehensive benchmark on eight tasks. Adv. Neural Inf. Process. Syst. 36, 59662–59688 (2023).
Zhao, W. X. et al. A survey of large language models. Preprint at https://arxiv.org/abs/2303.18223 (2023).
Min, B. et al. Recent advances in natural language processing via large pre-trained language models: a survey. ACM Comput. Surveys 56, 1–40 (2023).
Wang, L. et al. A survey on large language model based autonomous agents. Front. Comput. Sci. 18, 186345 (2024).
Ramos, M. C., Collison, C. J. & White, A. D. A review of large language models and autonomous agents in chemistry. Chem. Sci. 16, 2514–2572 (2025).
Boiko, D. A., MacKnight, R., Kline, B. & Gomes, G. Autonomous chemical research with large language models. Nature 624, 570–578 (2023).
Janakarajan, N., Erdmann, T., Swaminathan, S., Laino, T. & Born, J. Language models in molecular discovery. In Drug Development Supported by Informatics (eds Satoh, H. et al.) 121–141 (Springer, 2024).
Bran, A. M. et al. Augmenting large language models with chemistry tools. Nat. Mach. Intell. 6, 525–535 (2024).
McNaughton, A. D. et al. CACTUS: chemistry agent connecting tool usage to science. ACS Omega 9, 46563–46573 (2024).
Jin, Q., Yang, Y., Chen, Q. & Lu, Z. GeneGPT: augmenting large language models with domain tools for improved access to biomedical information. Bioinformatics 40, btae075 (2024).
Huang, K. et al. CRISPR-GPT: an LLM agent for automated design of gene-editing experiments. Preprint at https://arxiv.org/abs/2404.18021 (2024).
Liu, H. & Wang, H. GenoTEX: a benchmark for evaluating LLM-based exploration of gene expression data in alignment with bioinformaticians. Preprint at https://arxiv.org/abs/2406.15341 (2024).
Ghafarollahi, A. & Buehler, M. J. ProtAgents: protein discovery via large language model multi-agent collaborations combining physics and machine learning. Digital Discovery 3, 1389–1409 (2024).
Jia, S., Zhang, C. & Fung, V. LLMatDesign: autonomous materials discovery with large language models. Preprint at https://arxiv.org/abs/2406.13163 (2024).
Kang, Y. & Kim, J. ChatMOF: an artificial intelligence system for predicting and generating metal–organic frameworks using large language models. Nat. Commun. 15, 4705 (2024).
Wu, H. et al. ChatEDA: a large language model powered autonomous agent for EDA. IEEE Trans. Comput.-Aided Design Integr. Circuits Syst. 43, 3184–3197 (2024).
Ni, B. & Buehler, M. J. MechAgents: large language model multi-agent collaborations can solve mechanics problems, generate new data, and integrate knowledge. Extreme Mech. Lett. 67, 102131 (2024).
Yao, S. et al. ReAct: synergizing reasoning and acting in language models. In International Conference on Learning Representations https://iclr.cc/virtual/2023/oral/12647 (2023).
He, J. et al. Control risk for potential misuse of artificial intelligence in science. Preprint at https://arxiv.org/abs/2312.06632 (2023).
Liu, X. et al. ToolNet: connecting large language models with massive tools via tool graph. Preprint at https://arxiv.org/abs/2403.00839 (2024).
Hao, S., Liu, T., Wang, Z. & Hu, Z. ToolkenGPT: augmenting frozen language models with massive tools via tool embeddings. Adv. Neural Inf. Process. Syst. 36, 45870–45894 (2024).
Shinn, N., Cassano, F., Gopinath, A., Narasimhan, K. & Yao, S. Reflexion: language agents with verbal reinforcement learning. Adv. Neural Inf. Process. Syst. 36, 8634–8652 (2024).
Ingraham, J. B. et al. Illuminating protein space with a programmable generative model. Nature 623, 1070–1078 (2023).
Lin, Z. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 379, 1123–1130 (2023).
Atilgan, A. R. et al. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys. J. 80, 505–515 (2001).
Bakan, A., Meireles, L. M. & Bahar, I. Prody: protein dynamics inferred from theory and experiments. Bioinformatics 27, 1575–1577 (2011).
Cock, P. J. et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422 (2009).
Schwaller, P. et al. Molecular transformer: a model for uncertainty-calibrated chemical reaction prediction. ACS Central Science 5, 1572–1583 (2019).
Pei, Q. et al. BioT5+: towards generalized biological understanding with IUPAC integration and multi-task tuning. In Findings of the Association for Computational Linguistics: ACL 2024, 1216–1240 (Association for Computational Linguistics, 2024).
Papadatos, G. et al. SureChEMBL: a large-scale, chemically annotated patent document database. Nucleic Acids Res. 44, D1220–D1228 (2016).
Kim, S. et al. Pubchem substance and compound databases. Nucleic Acids Res. 44, D1202–D1213 (2016).
Bobbitt, N. S. et al. MOFX-DB: an online database of computational adsorption data for nanoporous materials. J. Chem. Eng. Data 68, 483–498 (2023).
Nandy, A. et al. Mofsimplify, machine learning models with extracted stability data of three thousand metal–organic frameworks. Sci. Data 9, 74 (2022).
Dubbeldam, D., Calero, S., Ellis, D. E. & Snurr, R. Q. RASPA: molecular simulation software for adsorption and diffusion in flexible nanoporous materials. Molecular Simulation 42, 81–101 (2016).
BLAST: basic local alignment search tool. NIH https://blast.ncbi.nlm.nih.gov (2024).
RDKit: open-source cheminformatics software. RDKit http://www.rdkit.org (2024).
Bajusz, D., Rácz, A. & Héberger, K. Why is tanimoto index an appropriate choice for fingerprint-based similarity calculations? J. Cheminformatics 7, 1–13 (2015).
Smith, T. F. et al. Identification of common molecular subsequences. J. Mol. Biol. 147, 195–197 (1981).
Hu, E. J. et al. LoRA: low-rank adaptation of large language models. In International Conference on Learning Representations https://iclr.cc/virtual/2022/poster/6319 (2022).
Vaswani, A. et al. Attention is all you need. Adv. Neural Inf. Process. Syst. 30, 5998–6008 (2017).
Yu, J. Dataset for the paper “SciToolAgent: A knowledge graph-driven scientific agent for multi-tool integration”. Zenodo https://doi.org/10.5281/zenodo.15691499 (2025).
Yu, J. & Ding, K. HICAI-ZJU/SciToolAgent: V1.0.1. Zenodo https://doi.org/10.5281/zenodo.15707182 (2025).
Acknowledgements
This work is funded by National Science and Technology Major Project (2023ZD0120802, Q.Z.), Zhejiang Provincial ‘Jianbing’ ‘Lingyan’ Research and Development Program of China (2025C01097, K.D. and Q.Z.), NSFC62301480 (K.D.), NSFC62302433 (Q.Z.), NSFCU23A20496 (Q.Z.), NSFCU23B2055 (H.C.), the Fundamental Research Funds for the Central Universities (226-2023-00138, H.C.), Zhejiang Provincial Natural Science Foundation of China (LQ24F020007, Q.Z.) and Hangzhou West Lake Pearl Project Leading Innovative Youth Team Project (TD2023017, K.D.). The artificial-intelligence-driven experiments, simulations and model training were performed on the robotic AI-Scientist platform of the Chinese Academy of Sciences.
Author information
Authors and Affiliations
Contributions
K.D. conceived the study and designed the method. J.Y. and J.H. implemented the method and experiments. K.D. and J.Y. performed the result analyses. Y.Y. developed the online service. K.D., J.Y., Q.Z. and H.C. wrote and revised the paper. Q.Z. and H.C. supervised the entire project. All authors wrote the paper, reviewed it and approved the final paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Computational Science thanks Jihan Kim, Lei Shi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Kaitlin McCardle, in collaboration with the Nature Computational Science team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Sections 1–5, Fig. 1 and Tables 1–3.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ding, K., Yu, J., Huang, J. et al. SciToolAgent: a knowledge-graph-driven scientific agent for multitool integration. Nat Comput Sci 5, 962–972 (2025). https://doi.org/10.1038/s43588-025-00849-y
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43588-025-00849-y