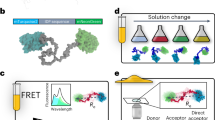
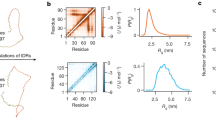
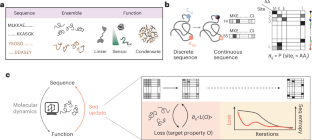
Abstract
The design of folded proteins has advanced substantially in recent years. However, many proteins and protein regions are intrinsically disordered and lack a stable fold, that is, the sequence of an intrinsically disordered protein (IDP) encodes a vast ensemble of spatial conformations that specify its biological function. This conformational plasticity and heterogeneity makes IDP design challenging. Here we introduce a computational framework for de novo design of IDPs through rational and efficient inversion of molecular simulations that approximate the underlying sequence–ensemble relationship. We highlight the versatility of this approach by designing IDPs with diverse properties and arbitrary sequence constraints. These include IDPs with target ensemble dimensions, loops and linkers, highly sensitive sensors of physicochemical stimuli, and binders to target disordered substrates with distinct conformational biases. Overall, our method provides a general framework for designing sequence–ensemble–function relationships of biological macromolecules.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All optimized sequences are provided in the Supplementary Data 1. Source data are provided with this paper.
Code availability
The complete codebase is available at the following GitHub repository: https://github.com/rkruegs123/idp-design. This repository includes a notebook containing a scaffold of a simple optimization for a custom state-level property. A snapshot of this repository, including the full source code and corresponding documentation, has also been archived on Zenodo at https://doi.org/10.5281/zenodo.15311353 (ref. 65).
References
Boehr, D. D., Nussinov, R. & Wright, P. E. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 5, 789–796 (2009).
Holehouse, A. S. & Kragelund, B. B. The molecular basis for cellular function of intrinsically disordered protein regions. Nat. Rev. Mol. Cell Biol. 25, 187–211 (2024).
van der Lee, R. et al. Classification of intrinsically disordered regions and proteins. Chem. Rev. 114, 6589–6631 (2014).
Uversky, V. N. Recent developments in the field of intrinsically disordered proteins: intrinsic disorder–based emergence in cellular biology in light of the physiological and pathological liquid–liquid phase transitions. Ann. Rev. Biophys. 50, 135–156 (2021).
Tesei, G. et al. Conformational ensembles of the human intrinsically disordered proteome. Nature 626, 897–904 (2024).
Thomasen, F. E. & Lindorff-Larsen, K. Conformational ensembles of intrinsically disordered proteins and flexible multidomain proteins. Biochem. Soc. Trans. 50, 541–554 (2022).
Mittag, T. & Forman-Kay, J. D. Atomic-level characterization of disordered protein ensembles. Cur. Opin. Struct. Biol. 17, 3–14 (2007).
Davey, N. E., Simonetti, L. & Ivarsson, Y. The next wave of interactomics: mapping the SLiM-based interactions of the intrinsically disordered proteome. Curr. Opin. Struct. Biol. 80, 102593 (2023).
Huang, Q., Li, M., Lai, L. & Liu, Z. Allostery of multidomain proteins with disordered linkers. Curr. Opin. Struct. Bio. 62, 175–182 (2020).
Moses, D., Ginell, G. M., Holehouse, A. S. & Sukenik, S. Intrinsically disordered regions are poised to act as sensors of cellular chemistry. Trends Biochem. Sci. 48, 1019–1034 (2023).
Banani, S. F. et al. Genetic variation associated with condensate dysregulation in disease. Develop. Cell 57, 1776–1788.e8 (2022).
Shrinivas, K. et al. Enhancer features that drive formation of transcriptional condensates. Mol. Cell 75, 549–561 (2019).
Sabari, B. R. Biomolecular condensates and gene activation in development and disease. Develop. Cell 55, 84–96 (2020).
Shi, M., Zhang, P., Vora, S. M. & Wu, H. Higher-order assemblies in innate immune and inflammatory signaling: a general principle in cell biology. Cur. Opin. Cell Biol. 63, 194–203 (2020).
Tsang, B., Pritišanac, I., Scherer, S. W., Moses, A. M. & Forman-Kay, J. D. Phase separation as a missing mechanism for interpretation of disease mutations. Cell 183, 1742–1756 (2020).
Dauparas, J. et al. Robust deep learning–based protein sequence design using ProteinMPNN. Science 378, 49–56 (2022).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. nature 596, 583–589 (2021).
Krishna, R. et al. Generalized biomolecular modeling and design with RoseTTAFold All-Atom. Science 384, eadl2528 (2024).
Dignon, G. L., Zheng, W., Best, R. B., Kim, Y. C. & Mittal, J. Relation between single-molecule properties and phase behavior of intrinsically disordered proteins. Proc. Natl Acad. Sci. USA 115, 9929–9934 (2018).
Joseph, J. A. et al. Physics-driven coarse-grained model for biomolecular phase separation with near-quantitative accuracy. Nat. Comput. Sci. 1, 732–743 (2021).
Tesei, G., Schulze, T. K., Crehuet, R. & Lindorff-Larsen, K. Accurate model of liquid–liquid phase behavior of intrinsically disordered proteins from optimization of single-chain properties. Proc. Natl Acad. Sci. USA 118, e2111696118 (2021).
Lotthammer, J. M., Ginell, G. M., Griffith, D., Emenecker, R. J. & Holehouse, A. S. Direct prediction of intrinsically disordered protein conformational properties from sequence. Nat. Methods 21, 465–476 (2024).
Emenecker, R. J., Guadalupe, K., Shamoon, N. M., Sukenik, S. & Holehouse, A. S. Sequence–ensemble–function relationships for disordered proteins in live cells. Preprint at bioRxiv https://doi.org/10.1101/2023.10.29.564547 (2023).
Regy, RoshanMammen, Thompson, J., Kim, Y. C. & Mittal, J. Improved coarse-grained model for studying sequence dependent phase separation of disordered proteins. Protein Sci. 30, 1371–1379 (2021).
Bradbury, J. et al. JAX: composable transformations of Python+ Numpy programs. GitHub https://github.com/jax-ml/jax (2018).
Schoenholz, S. & Cubuk, E. D. JAX MD: a framework for differentiable physics. In Proc. 34th International Conference on Neural Information Processing System Vol. 33, 11428–11441 (2020).
Thaler, S. & Zavadlav, J. Learning neural network potentials from experimental data via differentiable trajectory reweighting. Nat. Commun. 12, 6884 (2021).
Zhang, Shi-Xin, Wan, Zhou-Quan & Yao, H. Automatic differentiable Monte Carlo: theory and application. Phys. Rev. Res. 5, 033041 (2023).
González-Foutel, NicolásS. et al. Conformational buffering underlies functional selection in intrinsically disordered protein regions. Nat. Struct. Mol. Biol. 29, 781–790 (2022).
Lin, Yi-Hsuan & Chan, HueSun Phase separation and single-chain compactness of charged disordered proteins are strongly correlated. Biophys. J. 112, 2043–2046 (2017).
G. Greener, J. Differentiable simulation to develop molecular dynamics force fields for disordered proteins. Chem. Sci. 15, 4897–4909 (2024).
Mugnai, M. L. et al. Sizes, conformational fluctuations, and SAXS profiles for intrinsically disordered proteins. Protein Sci. 34, e70067 (2025).
Riback, J. A. et al. Commonly used FRET fluorophores promote collapse of an otherwise disordered protein. Proc. Natl Acad. Sci. USA 116, 8889–8894 (2019).
Krueger, R. K. & Ward, M. JAX-RNAfold: scalable differentiable folding. Bioinformatics 41, btaf203 (2025).
Emenecker, R. J., Griffith, D. & Holehouse, A. S. Metapredict V2: an update to metapredict, a fast, accurate, and easy-to-use predictor of consensus disorder and structure. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/2022.06.06.494887v1 (2022).
Das, R. K. & Pappu, R. V. Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc. Natl Acad. Sci. USA 110, 13392–13397 (2013).
Zhang, M. et al. The intrinsically disordered region from PP2C phosphatases functions as a conserved CO2 sensor. Nat. Cell Biol. 24, 1029–1037 (2022).
Dignon, G. L., Zheng, W., Kim, Y. C. & Mittal, J. Temperature-controlled liquid–liquid phase separation of disordered proteins. ACS Central Sci. 5, 821–830 (2019).
Borgia, A. et al. Extreme disorder in an ultrahigh-affinity protein complex. Nature 555, 61–66 (2018).
Schuler, B. et al. Binding without folding—the biomolecular function of disordered polyelectrolyte complexes. Curr. Opin. Struc. Biol. 60, 66–76 (2020).
Adachi, K. & Kawaguchi, K. Predicting heteropolymer interactions: demixing and hypermixing of disordered protein sequences. Phys. Rev. X 14, 031011 (2024).
Ginell, G. M. et al. Sequence-based prediction of intermolecular interactions driven by disordered regions. Science 388, eadq8381 (2025).
Portz, B., Lee, Bo. Lim & Shorter, J. FUS and TDP-43 phases in health and disease. Trends Biochem. Sci. 46, 550–563 (2021).
Wang, J. et al. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell 174, 688–699.e16 (2018).
Roden, C. & Gladfelter, A. S. RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol. 22, 183–195 (2021).
Rauh, A. S., Hedemark, G. S., Tesei, G. & Lindorff-Larsen, K. A coarse-grained model for simulations of phosphorylated disordered proteins. Biophys J. https://doi.org/10.1016/j.bpj.2025.07.001 (2025).
Kilgore, H. R. et al. Distinct chemical environments in biomolecular condensates. Nat. Chem. Biol. 20, 291–301 (2024).
Choi, Jeong-Mo & Pappu, R. V. Improvements to the ABSINTH force field for proteins based on experimentally derived amino acid specific backbone conformational statistics. J. Chem. Theory Comput. 15, 1367–1382 (2019).
Wessén, J., Das, S., Pal, T. & Chan, HueSun Analytical formulation and field-theoretic simulation of sequence-specific phase separation of protein-like heteropolymers with short- and long-spatial-range interactions. J. Phys. Chem. B 126, 9222–9245 (2022).
Shrinivas, K. & Brenner, M. P. Phase separation in fluids with many interacting components. Proc. Natl Acad. Sci. USA 118, e2108551118 (2021).
Frank, C. et al. Scalable protein design using optimization in a relaxed sequence space. Science 386, 439–445 (2024).
Matthies, M. C., Krueger, R., Torda, A. E. & Ward, M. Differentiable partition function calculation for RNA. Nucleic Acids Res. 52, e14 (2024).
Krueger, R. K., Engel, M. C., Hausen, R. & Brenner, M. P. Fitting coarse-grained models to macroscopic experimental data via automatic differentiation. Preprint at https://arxiv.org/abs/2411.09216 (2025).
Janson, G. & Feig, M. Transferable deep generative modeling of intrinsically disordered protein conformations. PLoS Comput. Biol. 20, e1012144 (2024).
Liu, C. et al. Diffusing protein binders to intrinsically disordered proteins. Nature 644, 809–817 (2025)
Pesce, F. et al. Design of intrinsically disordered protein variants with diverse structural properties. Sci. Adv. 10, eadm9926 (2024).
Sanchez-Burgos, I., Espinosa, J. R., Joseph, J. A. & Collepardo-Guevara, R. RNA length has a non-trivial effect in the stability of biomolecular condensates formed by RNA-binding proteins. PLoS Comput. Biol. 18, e1009810 (2022).
Zhu, H. et al. The chromatin regulator HMGA1a undergoes phase separation in the nucleus. ChemBioChem 24, e202200450 (2023).
Alston, J. J., Soranno, A. & Holehouse, A. S. Conserved molecular recognition by an intrinsically disordered region in the absence of sequence conservation. Biophys. J. 123, 26a (2024).
Wessén, J., Das, S., Pal, T. & Chan, HueSun Analytical formulation and field-theoretic simulation of sequence-specific phase separation of protein-like heteropolymers with short-and long-spatial-range interactions. J. Phys. Chem. B 126, 9222–9245 (2022).
Garaizar, A. et al. Aging can transform single-component protein condensates into multiphase architectures. Proc. Natl Acad. Sci. USA 119, e2119800119 (2022).
Taneja, I. & Lasker, K. Machine learning based methods to generate conformational ensembles of disordered proteins. Biophys. J. 123, 101–113 (2023).
Wang, X., Ramírez-Hinestrosa, Simón, Dobnikar, J. & Frenkel, D. The Lennard–Jones potential: when (not) to use it. Phys. Chem. Chem. Phys. 22, 10624–10633 (2020).
Krueger, R. K. & Shrinivas, K. rkruegs123/idp-design: file format change for figures. Zenodo https://doi.org/10.5281/zenodo.15311353 (2025).
Acknowledgements
We thank M. Ward for his collaboration on sequence design via overparameterization in the context of differentiable RNA folding, which inspired this work, J. Smith for helpful discussions relating to stochastic gradient estimators, W. Snead for helpful discussions on IDP design, and J. Boodry, N. Tyagi and members of the Shrinivas laboratory, for discussions and experimentation with the codebase. We acknowledge support from the Simons Foundation through the Simons Foundation Investigator award (R.K.K., M.P.B. and K.S). We acknowledge support from the NSF AI Institute of Dynamic Systems (grant no. 2112085), the Office of Naval Research (grant no. N00014-17-1-3029) and the Harvard Materials Research Science and Engineering Center (DMR no. 20-11754 to R.K.K and M.P.B.). We acknowledge support from NSF-Simons Center for Mathematical and Statistical Analysis of Biology at Harvard (grant no. 1764269) and Northwestern University for startup funding (K.S.). The computations in this paper were in part run on the FASRC Cannon cluster supported by the FAS Division of Science Research Computing Group at Harvard University. This research was supported in part through the computational resources and staff contributions provided for the Quest high performance computing facility at Northwestern University.
Author information
Authors and Affiliations
Contributions
R.K.K., M.P.B. and K.S. designed the study. R.K.K. and K.S. performed the research. All authors contributed to the writing and revision of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Computational Science thanks Mikael Lund and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Kaitlin McCardle, in collaboration with the Nature Computational Science team. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8, Tables 1–4 and Sections 1–9.
Supplementary Data 1
All optimized sequences from Figs. 1–6.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Krueger, R.K., Brenner, M.P. & Shrinivas, K. Generalized design of sequence–ensemble–function relationships for intrinsically disordered proteins. Nat Comput Sci (2025). https://doi.org/10.1038/s43588-025-00881-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s43588-025-00881-y