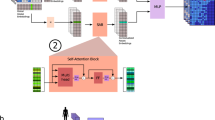
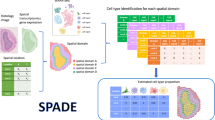
Abstract
Spatial epigenomics (SE) technologies profile epigenomic landscapes within intact tissues, preserving spatial context and enabling the study of gene regulatory mechanisms in situ. However, current SE datasets typically suffer from low signal detection, substantial noise and extremely sparse peak matrices, which pose considerable challenges for downstream analysis. Here we introduce SPEED (spatial epigenomic data denoising), a deep matrix factorization framework that leverages atlas-level single-cell epigenomic data and spatial context to impute and denoise SE data. In comprehensive benchmarks on both simulated data and real SE tissue datasets, SPEED outperformed five state-of-the-art methods across diverse tissues and technologies. Moreover, SPEED’s denoised outputs facilitated downstream analyses such as differential chromatin accessibility analysis, epigenomic spatial domain identification and gene activity inference. Collectively, our results indicate that SPEED is a generalizable tool for improving data quality and biological insights in SE.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All SE, single-cell RNA-seq and scATAC-seq datasets used in this study can be downloaded from public websites or databases: E11.5, E12.5 and E13.5 mouse embryo scATAC-seq data from 16 tissues at https://ngdc.cncb.ac.cn/gsa/browse/CRA003910 (ref. 15). E12.5, E13.5 and E14.5 embryonic mouse cerebellum snATAC-seq data are available in the GEO database under accession GSE178546 (ref. 28). E17.5 embryonic mouse heart scATAC-seq data are available in the GEO database under accession GSE190977 (ref. 27). E12.5, E13.5 and E15.5 mouse embryo snATAC-seq data are available in the GEO database under accession GSE214991 (ref. 12). Three samples of E18 mouse embryo brain snATAC-seq data are available at https://www.10xgenomics.com/datasets (ref. 16). Mouse embryo scRNA-seq data are available in the GEO database under accession GSE119945 (ref. 48). Human brain scATAC-seq data are available in the GEO database under accession GSE147672 (ref. 57). Adult mouse brain scATAC-seq data are available in the GEO database under accession GSE246791 (ref. 22). E13 mouse embryo spatial-ATAC-RNA-seq data are available in the GEO database under accession GSE205055 (ref. 8). E11–E18.5 mouse embryo MISAR-seq data are available at https://www.biosino.org/node/project/detail/OEP003285 (ref. 13). P22 mouse brain Spatial-CUT&Tag-RNA-seq data are available in the GEO database under accession GSE205055 (ref. 8). Human hippocampus spatial-ATAC-RNA-seq data are available in the GEO database under accession GSE205055 (ref. 8). P22 mouse brain spatial-ATAC-RNA-seq data are available in the GEO database under accession GSE205055 (ref. 8). EAE mouse brain spatial-Mux-seq data are available in the GEO database under accession GSE263333 (ref. 11). E13.5 mouse embryonic forebrain, hindbrain, midbrain and limb bulk ATAC-seq data from ENCODE are available at https://www.encodeproject.org (ref. 29). Chromatin state annotations for the E13.5 mouse embryonic forebrain and hindbrain are available at https://genome.ucsc.edu/cgi-bin/hgTrackUi?hgsid=2471038369_O34GqlYujAEy04rHeqMnjX560AHY&g=encode3RenChromHmm (refs. 30,31). Source data are provided with this paper.
Code availability
The open-source package of SPEED is available via GitHub at https://github.com/QuKunLab/SPEED. All codes and scripts used for the analyses and figure plotting in this study are available via Zenodo at https://doi.org/10.5281/zenodo.14948507 (ref. 58).
References
Bergmann, S. et al. Spatial profiling of early primate gastrulation in utero. Nature 609, 136–143 (2022).
Chen, A. et al. Single-cell spatial transcriptome reveals cell-type organization in the macaque cortex. Cell 186, 3726–3743 (2023).
Chen, A. et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell 185, 1777–1792 (2022).
Deng, Y. et al. Spatial-CUT&Tag: spatially resolved chromatin modification profiling at the cellular level. Science 375, 681–686 (2022).
Unterauer, E. M. et al. Spatial proteomics in neurons at single-protein resolution. Cell 187, 1785–1800 (2024).
Liu, Y. et al. High-plex protein and whole transcriptome co-mapping at cellular resolution with spatial CITE-seq. Nat. Biotechnol. 41, 1405–1409 (2023).
Lu, T., Ang, C. E. & Zhuang, X. Spatially resolved epigenomic profiling of single cells in complex tissues. Cell 185, 4448–4464 (2022).
Zhang, D. et al. Spatial epigenome-transcriptome co-profiling of mammalian tissues. Nature 616, 113–122 (2023).
Deng, Y. et al. Spatial profiling of chromatin accessibility in mouse and human tissues. Nature 609, 375–383 (2022).
Russell, A. J. C. et al. Slide-tags enables single-nucleus barcoding for multimodal spatial genomics. Nature 625, 101–109 (2024).
Guo, P. F. et al. Multiplexed spatial mapping of chromatin features, transcriptome and proteins in tissues. Nat. Methods 22, 520–529 (2025).
Llorens-Bobadilla, E. et al. Solid-phase capture and profiling of open chromatin by spatial ATAC. Nat. Biotechnol. 41, 1085–1088 (2023).
Jiang, F. et al. Simultaneous profiling of spatial gene expression and chromatin accessibility during mouse brain development. Nat. Methods 20, 1048–1057 (2023).
Kong, D. et al. Spatial profiling of chromatin accessibility reveals alteration of glial cells in Alzheimer’s disease mouse brain. Preprint at bioRxiv https://doi.org/10.1101/2025.05.01.651759 (2025).
Jiang, S. et al. Single-cell chromatin accessibility and transcriptome atlas of mouse embryos. Cell Rep. 42, 112210 (2023).
10x Genomics Datasets. 10X Genomics https://www.10xgenomics.com/resources/datasets (2019).
Yuan, H. & Kelley, D. R. scBasset: sequence-based modeling of single-cell ATAC-seq using convolutional neural networks. Nat. Methods 19, 1088–1096 (2022).
Xiong, L. et al. SCALE method for single-cell ATAC-seq analysis via latent feature extraction. Nat. Commun. 10, 4576 (2019).
Li, Z. et al. Chromatin-accessibility estimation from single-cell ATAC-seq data with scOpen. Nat. Commun. 12, 6386 (2021).
Bravo Gonzalez-Blas, C. et al. cisTopic: cis-regulatory topic modeling on single-cell ATAC-seq data. Nat. Methods 16, 397–400 (2019).
Tian, T., Zhang, J., Lin, X., Wei, Z. & Hakonarson, H. Dependency-aware deep generative models for multitasking analysis of spatial omics data. Nat. Methods 21, 1501–1513 (2024).
Zu, S. et al. Single-cell analysis of chromatin accessibility in the adult mouse brain. Nature 624, 378–389 (2023).
Li, Y. E. et al. A comparative atlas of single-cell chromatin accessibility in the human brain. Science 382, eadf7044 (2023).
Zhang, K. et al. A single-cell atlas of chromatin accessibility in the human genome. Cell 184, 5985–6001 (2021).
Xue, H. J., Dai, X. Y., Zhang, J. B., Huang, S. J. & Chen, J. J. Deep matrix factorization models for recommender systems. In Proc. 26th International Joint Conference on Artificial Intelligence (ed. Sierra, C.) 3203–3209 (IJCAI, 2017).
Yi, B. L. et al. Deep matrix factorization with implicit feedback embedding for recommendation system. IEEE Trans. Ind. Inf. 15, 4591–4601 (2019).
Yamada, S. et al. TEAD1 trapping by the Q353R-Lamin A/C causes dilated cardiomyopathy. Sci. Adv. 9, eade7047 (2023).
Khouri-Farah, N., Guo, Q., Morgan, K., Shin, J. & Li, J. Y. H. Integrated single-cell transcriptomic and epigenetic study of cell state transition and lineage commitment in embryonic mouse cerebellum. Sci. Adv. 8, eabl9156 (2022).
Encode Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Gorkin, D. U. et al. An atlas of dynamic chromatin landscapes in mouse fetal development. Nature 583, 744–751 (2020).
Perez, G. et al. The UCSC Genome Browser database: 2025 update. Nucleic Acids Res. 53, D1243–D1249 (2025).
Xu, H. et al. SPACEL: deep learning-based characterization of spatial transcriptome architectures. Nat. Commun. 14, 7603 (2023).
Harris, J. A. et al. Hierarchical organization of cortical and thalamic connectivity. Nature 575, 195–202 (2019).
Bartosovic, M. & Castelo-Branco, G. Multimodal chromatin profiling using nanobody-based single-cell CUT&Tag. Nat. Biotechnol. 41, 794–805 (2023).
Bartosovic, M., Kabbe, M. & Castelo-Branco, G. Single-cell CUT&Tag profiles histone modifications and transcription factors in complex tissues. Nat. Biotechnol. 39, 825–835 (2021).
He, K. M., Zhang, X. Y., Ren, S. Q. & Sun, J. Deep residual learning for image recognition. In Proc. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 770–778 (IEEE, 2016).
Chen, R. J. et al. Towards a general-purpose foundation model for computational pathology. Nat. Med. 30, 850–862 (2024).
Xu, H. et al. A whole-slide foundation model for digital pathology from real-world data. Nature 630, 181–188 (2024).
Rumelhart, D. E., Hinton, G. E. & Williams, R. J. Learning representations by back-propagating errors. Nature 323, 533–536 (1986).
Wightman, R. PyTorch image models. GitHub https://github.com/rwightman/pytorch-image-models (2019).
Kingma, D. P. & Ba, J. Adam: a method for stochastic optimization. Proceedings of the 3rd International Conference on Learning Representations https://arxiv.org/abs/1412.6980 (2015).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Granja, J. M. et al. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet. 53, 403–411 (2021).
Stuart, T., Srivastava, A., Madad, S., Lareau, C. A. & Satija, R. Single-cell chromatin state analysis with Signac. Nat. Methods 18, 1333–1341 (2021).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Bredikhin, D., Kats, I. & Stegle, O. MUON: multimodal omics analysis framework. Genome Biol. 23, 42 (2022).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Luecken, M. D. et al. Benchmarking atlas-level data integration in single-cell genomics. Nat. Methods 19, 41–50 (2022).
Kaufman, M. H. The Atlas of Mouse Development (Academic Press, 1992).
van Dijk, D. et al. Recovering gene interactions from single-cell data using data diffusion. Cell 174, 716–729 (2018).
spVAE. GitHub https://github.com/ttgump/spaVAE/blob/main/src/spaPeakVAE/run_spaPeakVAE.py (2024).
scBasset. GitHub https://github.com/calico/scBasset/blob/main/README.md (2022).
Using pycisTopic on human cerebellum single-cell multiome data. pycisTopic https://pycistopic.readthedocs.io/en/latest/notebooks/human_cerebellum.html (2022).
scopen. GitHub https://github.com/CostaLab/scopen/blob/master/README.md (2021).
SCALE. GitHub https://github.com/jsxlei/SCALE/blob/master/README.md (2019).
Corces, M. R. et al. Single-cell epigenomic analyses implicate candidate causal variants at inherited risk loci for Alzheimer’s and Parkinson’s diseases. Nat. Genet. 52, 1158–1168 (2020).
Wang, S. Scripts and data for paper titled “Denoising spatial epigenomic data via deep matrix factorization”. Zenodo https://doi.org/10.5281/zenodo.14948507 (2025).
Acknowledgements
This work was supported by the National Natural Science Foundation of China grants (grant nos T2125012 and 92574202 to K.Q.; 323B2014 to H.X.), the National Key R&D Program of China (grant no. 2022YFA1303200 to K.Q.), Strategic Priority Research Program of Chinese Academy of Sciences (grant no. XDB0940301 to K.Q.) and USTC Research Funds of the Double First-Class Initiative (grant nos YD9100002026 and YD9100002032 to K.Q.). We thank the USTC supercomputing center and the School of Life Science Bioinformatics Center for providing computing resources for this project.
Author information
Authors and Affiliations
Contributions
K.Q. conceived the project. S.W. and H.X. designed the framework and performed data analysis with help from J.W., Y.X., S.D., J.L., R.C. and X.C. K.Q., S.W. and H.X. wrote the paper with input from all authors. K.Q. supervised the entire project. All authors read and approved the final paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Computational Science thanks Chaoyong Yang and Zexian Zeng for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editors: Michelle Badri and Ananya Rastogi, in collaboration with the Nature Computational Science team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–17.
Supplementary Data 1
Detailed information on five single-cell ATAC-seq mouse embryo datasets.
Supplementary Data 2
TSCAS identified from single-cell and bulk E13.5 mouse embryo data.
Supplementary Data 3
Annotation details for spatial transcriptomic and spatial epigenomic datasets.
Supplementary Data 4
Runtime, memory usage, and CPU and GPU requirements of SPEED across datasets.
Supplementary Data 5
Source data for Supplementary Figs. 1, 3 and 6–17.
Source data
Source Data Fig. 2
Statistical source data for Fig. 2.
Source Data Fig. 3
Statistical source data for Fig. 3.
Source Data Fig. 4
Statistical source data for Fig. 4.
Source Data Fig. 5
Statistical source data for Fig. 5.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, S., Xu, H., Wang, J. et al. Denoising spatial epigenomic data via deep matrix factorization. Nat Comput Sci (2026). https://doi.org/10.1038/s43588-025-00941-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s43588-025-00941-3