Abstract
Background
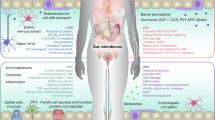
The gut microbiota influences breast cancer development through the estrobolome, a collection of bacterial genes involved in estrogen metabolism. While estrogen and the gut microbiota mutually affect each other, the long-term effects of oral endocrine therapy (ET) on the gut microbiota remain unclear. Furthermore, the relationship between gut microbiota profiles and breast cancer recurrence is not well understood. This study aims to investigate the long-term impact of oral ET on gut microbiota composition in disease-free and recurrent breast cancer patients.
Methods
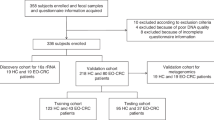
We enrolled 48 participants divided into four groups: tamoxifen only (Tam), letrozole only (Let), chemotherapy plus letrozole without recurrence (CLet), and chemotherapy plus letrozole with recurrence (Recu). Fecal samples were collected for 16S rRNA sequencing. Blood samples for cell-free DNA (cfDNA) analysis and tissue samples for EndoPredict (EPclin) scoring.
Results
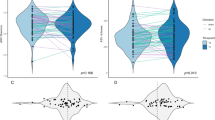
Here we show that long-term ET administration does not significantly alter overall gut microbial composition. However, patients with recurrence display lower α-diversity and higher abundances of Sutterella and Ruminococcus compared with non-recurrent patients. cfDNA profiles do not differ significantly between groups. Notably, high EPclin scores predict chemotherapy benefit, but recurrence still occurs in some cases. In such patients, gut microbial markers effectively distinguish recurrence and are associated with poorer progression-free survival, particularly in those with larger tumors.
Conclusions
This study provides the first human evidence with long-term ET administration to reveal that, besides genetic profiles, the gut microbiota is another critical factor that we should consider in the influence and prediction of breast cancer recurrence in the future.
Plain Language Summary
Breast cancer treatments often include long-term hormone therapy, but little is known about how these drugs affect the bacteria living in our gut. In this study, we followed women receiving different endocrine therapies and analyzed their gut microbiota before and after one year of treatment. We discovered that women whose cancer later returned had less diverse gut bacteria and higher amounts of certain species, including Sutterella and Ruminococcus. These bacterial patterns were linked to poorer treatment outcomes, especially in patients with larger tumors. Our findings suggest that gut bacteria may influence how breast cancer responds to therapy and could serve as an additional factor to help predict disease recurrence in the future.
Similar content being viewed by others
Data availability
Due to contractual obligations with the funding foundation and ethical restrictions defined in the informed consent, the datasets generated and analyzed in this study are not publicly available. Data may be made available from the corresponding author upon reasonable request and subject to approval by the funding organization and the institutional review board.
References
Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74, 229–263 (2024).
Jordan, V. C. Tamoxifen: a most unlikely pioneering medicine. Nat. Rev. Drug Discov. 2, 205–213 (2003).
Saphner, T., Tormey, D. C. & Gray, R. Annual hazard rates of recurrence for breast cancer after primary therapy. J. Clin. Oncol. 14, 2738–2746 (1996).
Pan, H. et al. 20-year risks of breast-cancer recurrence after stopping endocrine therapy at 5 years. N. Engl. J. Med. 377, 1836–1846 (2017).
Eslami, S. Z., Majidzadeh, A. K., Halvaei, S., Babapirali, F. & Esmaeili, R. Microbiome and breast cancer: new role for an ancient population. Front Oncol. 10, 120 (2020).
Di Modica, M., Arlotta, V., Sfondrini, L., Tagliabue, E. & Triulzi, T. The link between the microbiota and HER2+ breast cancer: the new challenge of precision medicine. Front. Oncol. 12, 947188 (2022).
Parida, S. et al. A procarcinogenic colon microbe promotes breast tumorigenesis and metastatic progression and concomitantly activates notch and beta-catenin axes. Cancer Discov. 11, 1138–1157 (2021).
Kwa, M., Plottel, C. S., Blaser, M. J. & Adams, S. The intestinal microbiome and estrogen receptor-positive female breast cancer. J. Natl Cancer Inst. 108, djw029 (2016).
Thirunavukkarasan, M. et al. Short-chain fatty acid receptors inhibit invasive phenotypes in breast cancer cells. PLoS One 12, e0186334 (2017).
Lam, K. C. et al. Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell 184, 5338–5356 e5321 (2021).
Plottel, C. S. & Blaser, M. J. Microbiome and malignancy. Cell Host Microbe 10, 324–335 (2011).
Ervin, S. M. et al. Gut microbial beta-glucuronidases reactivate estrogens as components of the estrobolome that reactivate estrogens. J. Biol. Chem. 294, 18586–18599 (2019).
Hu, S. et al. Gut microbial beta-glucuronidase: a vital regulator in female estrogen metabolism. Gut Microbes 15, 2236749 (2023).
Gloux, K. et al. A metagenomic beta-glucuronidase uncovers a core adaptive function of the human intestinal microbiome. Proc. Natl. Acad. Sci. USA 108, 4539–4546 (2011).
McIntosh, F. M. et al. Phylogenetic distribution of genes encoding beta-glucuronidase activity in human colonic bacteria and the impact of diet on faecal glycosidase activities. Environ. Microbiol 14, 1876–1887 (2012).
Ghimire, S. et al. Dietary isoflavones alter gut microbiota and lipopolysaccharide biosynthesis to reduce inflammation. Gut Microbes 14, 2127446 (2022).
Huang, L., Zheng, T., Hui, H. & Xie, G. Soybean isoflavones modulate gut microbiota to benefit the health weight and metabolism. Front. Cell Infect. Microbiol. 12, 1004765 (2022).
Chen, P. et al. The bioavailability of soy isoflavones in vitro and their effects on gut microbiota in the simulator of the human intestinal microbial ecosystem. Food Res. Int. 152, 110868 (2022).
Toi, M. et al. Probiotic beverage with soy isoflavone consumption for breast cancer prevention: a case-control study. Curr. Nutr. Food Sci. 9, 194–200 (2013).
Meng, Q. et al. The gut microbiota during the progression of atherosclerosis in the perimenopausal period shows specific compositional changes and significant correlations with circulating lipid metabolites. Gut Microbes 13, 1–27 (2021).
Chen, K. L. A. et al. Long-term administration of conjugated estrogen and bazedoxifene decreased murine fecal beta-glucuronidase activity without impacting overall microbiome community. Sci. Rep. 8, 8166 (2018).
Song, C. H. et al. 17beta-Estradiol supplementation changes gut microbiota diversity in intact and colorectal cancer-induced ICR male mice. Sci. Rep. 10, 12283 (2020).
Mazo, C., Aura, C., Rahman, A., Gallagher, W. M. & Mooney, C. Application of artificial intelligence techniques to predict risk of recurrence of breast cancer: a systematic review. J. Pers. Med. 12, 1496 (2022).
Peters, B. A. et al. The microbiome in lung cancer tissue and recurrence-free survival. Cancer Epidemiol. Biomark. Prev. 28, 731–740 (2019).
Huo, R. X. et al. Gut mucosal microbiota profiles linked to colorectal cancer recurrence. World J. Gastroenterol. 28, 1946–1964 (2022).
Hou, M. F. et al. Comprehensive profiles and diagnostic value of menopausal-specific gut microbiota in premenopausal breast cancer. Exp. Mol. Med. 53, 1636–1646 (2021).
Lu, Y. et al. MicrobiomeAnalyst 2.0: comprehensive statistical, functional and integrative analysis of microbiome data. Nucleic Acids Res. 51, W310–W318 (2023).
Filipits, M. et al. A new molecular predictor of distant recurrence in ER-positive, HER2-negative breast cancer adds independent information to conventional clinical risk factors. Clin. Cancer Res. 17, 6012–6020 (2011).
Constantinidou, A. et al. Clinical validation of EndoPredict in pre-menopausal women with ER-positive, HER2-negative primary breast cancer. Clin. Cancer Res. 28, 4435–4443 (2022).
Shih, C. T., Yeh, Y. T., Lin, C. C., Yang, L. Y. & Chiang, C. P. Akkermansia muciniphila is negatively correlated with hemoglobin A1c in refractory diabetes. Microorganisms 8, 1360 (2020).
Sabui, S. et al. Tamoxifen-induced, intestinal-specific deletion of Slc5a6 in adult mice leads to spontaneous inflammation: involvement of NF-kappaB, NLRP3, and gut microbiota. Am. J. Physiol. Gastrointest. Liver Physiol. 317, G518–G530 (2019).
Li, H. et al. Potential risk of tamoxifen: gut microbiota and inflammation in mice with breast cancer. Front. Oncol. 13, 1121471 (2023).
Alam, Y. et al. Variation in human gut microbiota impacts tamoxifen pharmacokinetics. mBio, 16, e0167924 (2024).
Zheng, C. et al. Gut microbiome as a biomarker for predicting early recurrence of HBV-related hepatocellular carcinoma. Cancer Sci. 114, 4717–4731 (2023).
Okubo, R. et al. Impact of chemotherapy on the association between fear of cancer recurrence and the gut microbiota in breast cancer survivors. Brain Behav. Immun. 85, 186–191 (2020).
Mosca, A., Leclerc, M. & Hugot, J. P. Gut microbiota diversity and human diseases: should we reintroduce key predators in our ecosystem?. Front. Microbiol. 7, 455 (2016).
Kriss, M., Hazleton, K. Z., Nusbacher, N. M., Martin, C. G. & Lozupone, C. A. Low diversity gut microbiota dysbiosis: drivers, functional implications and recovery. Curr. Opin. Microbiol. 44, 34–40 (2018).
Chang, J. Y. et al. Decreased diversity of the fecal Microbiome in recurrent Clostridium difficile-associated diarrhea. J. Infect. Dis. 197, 435–438 (2008).
Wang, L. et al. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol. Autism 4, 42 (2013).
Lim, M. Y. et al. The effect of heritability and host genetics on the gut microbiota and metabolic syndrome. Gut 66, 1031–1038 (2017).
Kaakoush, N. O. Sutterella species, IgA-degrading bacteria in ulcerative colitis. Trends Microbiol 28, 519–522 (2020).
Mori, G. et al. Shifts of faecal microbiota during sporadic colorectal carcinogenesis. Sci. Rep. 8, 10329 (2018).
Su, Q. et al. Association between gut microbiota and gastrointestinal cancer: a two-sample bi-directional Mendelian randomization study. Front. Microbiol. 14, 1181328 (2023).
Katayama, Y. et al. The role of the gut microbiome on the efficacy of immune checkpoint inhibitors in Japanese responder patients with advanced non-small cell lung cancer. Transl. Lung Cancer Res. 8, 847–853 (2019).
Deng, X. et al. Comparison of microbiota in patients treated by surgery or chemotherapy by 16S rRNA sequencing reveals potential biomarkers for colorectal cancer therapy. Front. Microbiol. 9, 1607 (2018).
Goldbaum, A. A. et al. The role of diet and the gut microbiota in the obesity-colorectal cancer link. Nutr. Cancer 77, 626–639 (2025).
Zaman, S., Chen, G., Segarra, D., Patel, A. & Protiva, P. S208 increased primary tumor sutterella bacterial RNAseq signatures are associated with increased M2 macrophage abundance and worse overall survival in rectal adenocarcinoma. J. Am. Coll. Gastroenterol. ACG 117, e150–e151 (2022).
Zhai, L. et al. Ruminococcus gnavus plays a pathogenic role in diarrhea-predominant irritable bowel syndrome by increasing serotonin biosynthesis. Cell Host Microbe 31, 33–44 e35 (2023).
Crost, E. H., Coletto, E., Bell, A. & Juge, N. Ruminococcus gnavus: friend or foe for human health. FEMS Microbiol. Rev. 47, fuad014 (2023).
Alexander, J. L. et al. Pathobionts in the tumour microbiota predict survival following resection for colorectal cancer. Microbiome 11, 100 (2023).
Shively, C. A. et al. Consumption of Mediterranean versus western diet leads to distinct mammary gland microbiome populations. Cell Rep. 25, 47–56 e43 (2018).
Soto-Pantoja, D. R. et al. Diet alters entero-mammary signaling to regulate the breast microbiome and tumorigenesis. Cancer Res. 81, 3890–3904 (2021).
Pernigoni, N. et al. Commensal bacteria promote endocrine resistance in prostate cancer through androgen biosynthesis. Science 374, 216–224 (2021).
Dejea, C. M. et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 359, 592–597 (2018).
Benzo, Y. et al. Acyl-CoA synthetase 4 modulates mitochondrial function in breast cancer cells. Heliyon 10, e30639 (2024).
Orlando, U. D. et al. Acyl-CoA synthetase-4 is implicated in drug resistance in breast cancer cell lines involving the regulation of energy-dependent transporter expression. Biochem. Pharm. 159, 52–63 (2019).
Schwarzenbach, H., Hoon, D. S. & Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 11, 426–437 (2011).
Canzoniero, J. V. & Park, B. H. Use of cell free DNA in breast oncology. Biochim. Biophys. Acta 1865, 266–274 (2016).
Gao, Q. et al. Circulating cell-free DNA for cancer early detection. Innovation 3, 100259 (2022).
Bettegowda, C. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6, 224ra224 (2014).
Liebs, S. et al. Detection of mutations in circulating cell-free DNA in relation to disease stage in colorectal cancer. Cancer Med. 8, 3761–3769 (2019).
Zhang, Y. et al. Pan-cancer circulating tumor DNA detection in over 10,000 Chinese patients. Nat. Commun. 12, 11 (2021).
Killock, D. Diagnosis: CancerSEEK and destroy - a blood test for early cancer detection. Nat. Rev. Clin. Oncol. 15, 133 (2018).
Teo, Y. V. et al. Cell-free DNA as a biomarker of aging. Aging Cell 18, e12890 (2019).
Liang, D. H. et al. Cell-free DNA as a molecular tool for monitoring disease progression and response to therapy in breast cancer patients. Breast Cancer Res. Treat. 155, 139–149 (2016).
Jongbloed, E. M. et al. A systematic review of the use of circulating cell-free DNA dynamics to monitor response to treatment in metastatic breast cancer patients. Cancers 13, 1811 (2021).
Xu, J. et al. Circulating tumor DNA: from discovery to clinical application in breast cancer. Front. Immunol. 15, 1355887 (2024).
Filipits, M. et al. Prediction of distant recurrence using EndoPredict among women with ER(+), HER2(-) node-positive and node-negative breast cancer treated with endocrine therapy only. Clin. Cancer Res. 25, 3865–3872 (2019).
Buus, R. et al. Comparison of EndoPredict and EPclin with oncotype DX recurrence score for prediction of risk of distant recurrence after endocrine therapy. J. Natl Cancer Inst. 108, djw149 (2016).
Sestak, I. et al. Prediction of chemotherapy benefit by EndoPredict in patients with breast cancer who received adjuvant endocrine therapy plus chemotherapy or endocrine therapy alone. Breast Cancer Res. Treat. 176, 377–386 (2019).
Sestak, I. et al. Comparison of the performance of 6 prognostic signatures for estrogen receptor-positive breast cancer: a secondary analysis of a randomized clinical trial. JAMA Oncol. 4, 545–553 (2018).
Terrisse, S., Zitvogel, L. & Kroemer, G. Impact of microbiota on breast cancer hormone therapy. Cell Stress 7, 12–19 (2023).
Alexander, J. L. et al. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroenterol. Hepatol. 14, 356–365 (2017).
Ma, W. et al. Gut microbiota shapes the efficiency of cancer therapy. Front. Microbiol. 10, 1050 (2019).
Helmink, B. A., Khan, M. A. W., Hermann, A., Gopalakrishnan, V. & Wargo, J. A. The microbiome, cancer, and cancer therapy. Nat. Med. 25, 377–388 (2019).
Acknowledgements
The authors would like to thank the YongLin Healthcare Foundation for their sponsorship and Germark Biotechnology Co., Ltd for performing the bioinformatics analyses. This study was supported by the YongLin Healthcare Foundation under study protocol No. QCR18001 and supported by grants 109-2314-B-037-142-MY2, 111-2314-B-037-092-, 112-2314-B-037-007-, 114-2314-B-040-038- from the National Science and Technology Council and the grants KMUH110-0M38, KMUH110-0R39, KMUH110-0R40, KMUH111-1R33, KMTTH-111-R013, KMTTH-112-R006, and KMGH-113G002 from the Kaohsiung Medical University Hospital and Kaohsiung Medical University Gangshan Hospital.
Author information
Authors and Affiliations
Contributions
M.-F.H. and C.-L.L. performed the experiments; S.-H.M. performed the cfDNA and statistical analysis; F.-M.C., J.-Y.C., S.-L.S., J.-Y.K., S.-F.Y. carried out the patient recruitment and specimen collection; C.-P.C. designed, analyzed the data and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Medicine thanks Maya Dadiani and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hou, MF., Li, CL., Moi, SH. et al. Longitudinal multiomics analysis of endocrine therapy effects and gut microbiota in breast cancer recurrence. Commun Med (2026). https://doi.org/10.1038/s43856-026-01384-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43856-026-01384-1