Abstract
While the rise of antibiotic resistance poses a global health challenge, the development of new antibiotics has slowed down over the past decades. This turned the attention of researchers towards the rational design of drug combination therapies to combat antibiotic resistance. In this review we discuss how drug combinations can exploit the deleterious pleiotropic effects of antibiotic resistance and conclude that each drug interaction has its prospective therapeutic application.
Similar content being viewed by others
Introduction
Antibiotics are one of the most commonly used medicines in modern times, saving innumerable amounts of lives every day since their discovery in the early 20th century. Hence, the rise and spread of bacterial resistance to antibiotics has become one of the leading causes of worry in healthcare as well as in animal husbandry1. Globally, 1.27 million deaths were directly attributable to bacterial antimicrobial resistance in 2019 - meaning that drug-resistant infections killed more people than AIDS or malaria1. While antibiotic resistance is ancient and predates clinical use of antibiotics, the constant selective pressure imposed by prolonged exposure in clinical and agricultural settings has led to the spread of resistance conferring mutations and mobile genetic elements2,3,4. Even though these resistance determinants emerged against commonly used antibiotics, due to shared resistance mechanisms and similar chemical structures, they provide resistance against antibiotics currently undergoing clinical trials5.
The clinical testing of antibiotic resistance is based on long-standing global standards defined by the Clinical and Laboratory Standards Institute (CLSI) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST). These standards are based on minimum inhibitory concentration measurements using broth microdilution and disk diffusion tests on agar plates6,7. Similarly, the conventional way of determining antibiotic interactions is by measuring growth inhibition in the presence of various concentrations of two different antibiotics, in checkerboard assays8. However, neither of these growth inhibition based measurements give information on antibiotic efficacy as defined by bacterial cell death and thus has two major limitations. First, these methods might overlook the dormant, persistent pathogens represented by a small subpopulation of slow or non-growing bacterial cells with an enhanced ability to escape the bactericidal effects of antibiotics9,10. Second, the reduction of antimicrobial killing rates by tolerance-conferring effects of the bacterial populations - such as energy depletion or activation of stress pathways - mediated by the population’s pre-existing genetic repertoire is also not captured by conventional bacterial growth assays10,11,12,13,14. Measuring bacterial survival after prolonged exposures to antimicrobials is inherently difficult as it requires the quantification of low densities of surviving cells, and the use of the tedious conventional agar-plating techniques limit the number of conditions that can be tested15,16. Despite the fact that the success in antibiotic therapy is defined by bacterial clearance, the ease of rapidly and automatically measuring growth inhibition by optical density metres commonly makes this metric outshine its limitations in clinical settings.
The search for novel antibiotics is still ongoing – in this calendar year (2024), a new class of antibiotics has been discovered: ethered macrocyclic peptides are able to block the transport of bacterial lipopolysaccharides in carbapenem-resistant Acinetobacter baumannii17. While this revelation is significant and groundbreaking, the discovery of novel antibiotic mechanisms of action has slowed down considerably over the years. Due to the shortage of new antibiotics, the attention of researchers turned towards the repurposing of existing antibiotics by combinational therapies. Certain antibiotic combinations are already applied efficiently to combat infections, and combination treatment is now the standard for specific bacterial infections18,19. In the case of Helicobacter pylori infections, quadruple therapies of proton pump inhibitors and metronidazole combined with either amoxicillin or bismuth and clarithromycin or tetracycline has been dubbed the Toronto Consensus18. Similarly, the guidelines recommended against tuberculosis by various centres in the United States such as the Centre of Disease Control start with a quadruple therapy of ethambutol, isoniazid, pyrazinamide, and rifampin, followed by a dual therapy of isoniazid and rifampin19. The therapeutic application of antibiotic combinations is often justified by one of the following three reasons i) to broaden the spectrum of activity ii) to increase treatment efficacy or iii) to prevent or delay the emergence of resistance.
Routine single-antibiotic treatment practices may have two conflicting effects: the desired, immediate effect of pathogen inhibition, and the undesired, long-term effect of promoting the evolution of antibiotic resistance20. Such an undesired effect, when concentrated in environments where infected patients congregate such as hospitals, can lead to the development of resistant pathogens, resulting in an infection that is significantly harder to treat. Combinations of two or more antibiotics within the same patient produce complex adaptive landscapes different from its component treatments due to physiological and evolutionary drug interactions, influencing the balance of pathogen clearance and resistance evolution. This adaptive landscape is formed by interactions of multiple resistance mutations in producing a phenotype (epistasis) and the effect of a single mutation on multiple phenotypes (pleiotropy). Hence, understanding the complex phenotypic effects of genetic mutations could help in charting the landscape of and constraining antibiotic resistance evolution. In this review, our aim is to highlight the pros and cons of drug combinations in the perspective of antimicrobial resistance evolution, and explore the possibilities of their potential therapeutic applications.
Collateral sensitivity
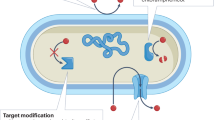
Different antibiotic resistance mutations may increase the resistance of pathogens to multiple antibiotics – a phenomenon called cross-resistance21. However, certain antibiotic resistance mutations may also simultaneously enhance susceptibility to other antibiotics from distinct structural classes, a phenomenon called collateral sensitivity. Collateral sensitivity and its therapeutic potential to hinder antibiotic resistance has been extensively studied, revealing complex networks of evolutionary trade-offs (Fig. 1; specific examples with known mechanisms included in Supplementary Table 1)22,23,24,25,26,27,28,29,30. Interestingly, collateral sensitivity responses can be linked to not just interactions between antibiotics and/or nonantibiotics, as the lysis of certain bacteriophages depend on cell membrane proteins and other surface structures also involved in antibacterial resistance mechanisms. An example is the phage U136B, which relies on the efflux pump protein TolC and the structural barrier molecule lipopolysaccharide, thereby selecting for the loss or disruption of these resistance mechanisms, sensitising bacterial populations to antibiotics31.
This schematic figure represents the growth inhibition-based standard broth dilution assay for defining the minimal inhibitory concentration (left part) of an antibiotic resistant (orange) and sensitive (grey) bacterial strain and their survival rate (right part). Bacterial survival is measured at a high, clinically relevant dose of the antibiotics after a predetermined amount of treatment time. In the presence of the green antibiotic, the effect on the resistant strain’s growth is not different compared to its sensitive counterpart (marked by the decrease of intensity in the orange and grey circles, respectively). However, the similar results in growth inhibition still leave two possibilities open: collateral sensitivity, when the resistant population (orange) is cleared orders of magnitude more efficiently than its sensitive counterpart (grey) or no interaction where both variants are equally affected.
However, collateral sensitivity is not necessarily robust - bacteria can acquire resistance to an antibiotic in diverse ways, and the phenotypic effects of each viable resistance mutation may or may not overlap (Fig. 2)22,24,30,32,33,34. In fact, systematic screens showed the emergence of both cross-resistance and collateral sensitivity patterns in parallel evolved populations from the same genetic background in the presence of the same antibiotic35,36,37. Therefore, the application of non-robust collateral sensitivity pairs where cross-resistance is another possible evolutionary trajectory could be disadvantageous in terms of applicability. Since even a single, rare cross-resistant mutant would be able to outcompete the collateral sensitive variants and could inevitably result in the selection of resistant subpopulations to both antibiotics37,38.
This hypothetical antibiotic treatment (orange pill) of a heterogeneous population (leftmost flask, represented by different shades of grey bacteria) shows four possible outcomes. A During treatment the initially antibiotic sensitive bacterial population (grey bacteria) will evolve resistance through different mutations (represented by a range of colours). Hence, when the treatment is switched to the blue antibiotic, only those fractions of the resistant population will be eliminated that had specific resistance mutations causing collateral sensitivity (orange bacteria). B If a collateral sensitivity pattern is robust and conserved, the majority of the heterogeneous population will evolve resistance mutations to the orange antibiotic that cause collateral sensitivity to the blue antibiotic. Hence, the second treatment eliminates the population. C However, if this collaterally sensitive resistance mechanism is transiently induced with a specific drug (white tablet) throughout the whole sensitive population, the resistant population can be eliminated. D A similar principle can apply for rapidly spreading resistance determinants on plasmids available in the environment: Here, antibiotic treatment causes the rapid uptake of this resistance plasmid (circular DNA) – this makes the population robustly hypersusceptible to a second antibiotic, clearing the population with great efficacy.
Another weakness arises when comparing collateral sensitivity phenotypes originating from diverse genetic backgrounds. Isolates with different genetic backgrounds contain various pre-existing mutations that can modify the phenotypic effects of a mutation by genetic epistasis. Therefore, different bacterial strains could have differing collateral sensitivity phenotypes, even when the acquired resistance mutation is the same. For example, comparing the phenotypic effect of specific antibiotic resistance mutations in two closely related bacterial species revealed that collateral sensitivity interactions can differ completely39. In addition, a retrospective analysis of around 450,000 antimicrobial susceptibility tests, spanning a 4-year period at different hospitals, showed that collateral sensitivity was rarely conserved at the species level, with only 0.7% of 875 antibiotic pairs showing evidence of collateral sensitivity40. This implies that treatment strategies exploiting collateral sensitivity might require strain-level pathogen identification and be limited to a few antibiotic pairings39,40. Therefore, efforts to identify the few cases in which collateral sensitivity phenotypes are robust and conserved in different genomic backgrounds are crucial. The exploration and exploitation of collateral sensitivity is further impeded by the fact that resistance mechanisms against antibiotics can evolve from adaptation to non-antibiotic drugs a patient might be administered41. This suggests that collateral sensitivity treatments should also take into account medications prescribed for preexisting conditions of the patient.
Another limitation of collateral sensitivity is the general lack of clearance efficacy studies as the usual method is to examine growth inhibition, potentially limiting the available antibiotic pairs showing collateral sensitivity that could also be used in clinical settings (Fig. 1). It has been shown that treatment effects can change when studying bacterial clearance efficacy or measuring growth inhibition42. Tolerance and persistence mechanisms seem to be more conserved between different species, regulated by essential genes (e.g. RpoS) or general stress response pathways (e.g. (pp)pGpp system, ROS and SOS responses). The conservation of tolerance and persistence mechanisms could mean that a more robust or conserved collateral sensitivity network might only be revealed by deliberately and extensively studying antibiotic clearance efficacy43,44. Furthermore, collateral sensitivity studies measuring growth inhibition often identify small changes of the minimal inhibitory concentration between the antibiotic sensitive wild type and the resistant variant29. Consequently, such small changes might result in a very limited clinical concentration range of the collateral sensitive antibiotic in which the resistant mutants are under counter-selection.
Despite the fact that collateral sensitivity studies often identify only small changes in inhibitory concentration, even these subtle collateral sensitivity responses can still reduce the mutant prevention concentration, constraining resistance development29. In addition, cycling or combining robust bidirectional collateral sensitive partners could further delay the evolution of antibiotic resistance by constraining the set of available mutational trajectories36,38,45,46,47,48,49. An evolutionary study on Staphylococcus aureus measured the efficacy of alternating therapies of neomycin (NEO), trimethoprim and ciprofloxacin (CIP), finding that in the CIP-NEO alternating treatment, CIP resistance was gained significantly slower46.
Collateral sensitive cyclic treatments could also help target prevalent hypertransmissible pathogens in chronic infections. It has been shown that cystic fibrosis patients carry hypertransmissible Pseudomonas aeruginosa strains, already prevalent in geographically disparate regions50. Furthermore, recent revelations regarding the phenotypic convergence of Pseudomonas aeruginosa revealed that even highly diverse clinical isolates or isogenic resistant mutants evolve rapid and robust collateral sensitivity patterns that seem to predict the success of such treatment26,34,48,51,52. Since previous treatments could lead to robust collateral sensitivity development, the high clonal presence of a bacterial strain in chronic infections could be exploited by taking the patient’s treatment history into account34,53,54.
Another way to exploit collateral sensitivity is to target highly conserved, horizontally spreading resistance mechanisms (Fig. 2). It has been shown that adaptation to β-thujaplicin, a natural product extract that preferentially selects against strains carrying the tetA-tetR efflux pump operon, leads to the loss of this resistance mechanism, reverting the resistant strain back to sensitivity55. Similarly, the expression of mobile beta-lactamases - the most common resistance mechanism in Enterobacterales - produces robust collateral sensitivity to colistin and azithromycin in multiple, phylogenetically unrelated Escherichia coli strains56,57. These results pave the way for exploiting collateral sensitive combination treatments not only against spontaneous chromosomal mutations, but already prevalent, plasmid-mediated mobile resistance mechanisms (Fig. 2).
Collateral sensitivity can also be transiently induced by administering a sensitising agent, thereby evading the need for the stable evolution of antibiotic resistance to exploit its pleiotropy (Fig. 2). For example, it is possible in Pseudomonas aeruginosa isolates to overexpress the MexCD-OprJ efflux pump by using dequalinium chloride, inducing the same effect as downregulator loss-of-function mutants. Such an overexpression results in ciprofloxacin resistance, which is robustly coupled with tobramycin hypersensitivity, without interference by the diversity of possible resistance mutations32.
Finally, recent advancements in collateral sensitivity research developed a metric to infer collateral sensitivity and cross resistance interactions in an Escherichia coli strain using available chemical genetics data35. Chemical genetic screens are based on measuring the antimicrobial activity of a range of drugs against a bacterial knockout library, which is then used to predict novel evolutionary interactions. By performing evolutionary experiments to verify these inferred results, it was confirmed that chemical genetics data can pinpoint robust collateral sensitivity and cross resistance mechanisms that emerge and get selected during experimental adaptation33,35. Combining a prediction metric with the recently established high-throughput chemogenomic approach for drug activity screening on a barcoded knockout library could reveal a vast network of collateral sensitivity even between antibiotics and non-antibiotics35,41.
Suppression
Suppression occurs when two antibiotics interact in a seemingly undesired way, where one of the administered antibiotics reduces the effect of the other, hence the combined effect of the mixed antibiotics is weaker compared to the effect of one of the antibiotics alone (Fig. 3).
The barplot compares the survival of a bacterial strain resistant to the orange antibiotic (orange) and a strain sensitive to it (grey) under single-antibiotic and combination treatments in cases of different antibiotic interactions. In single-antibiotic treatments, the sensitive strain’s survival is reduced by both antibiotics, at varying rates. In suppressive combination treatments, the sensitive strain is less affected compared to the blue antibiotic’s effects, but the orange resistant strain suffers the unimpeded effects of the blue antibiotic, providing the sensitive bacteria a competitive advantage. In synergistic combination treatments, the sensitive strain’s survival is highly reduced compared to the effects of the single antibiotics, but the resistant strain to the orange antibiotic only suffers the effect of the blue antibiotic, allowing the mono-resistant strain to potentially outcompete the sensitive strain.
At first glance, suppressive combinations pose a great disadvantage, since the purpose of antimicrobial treatments is eradicating the infection as soon as possible. Since suppression reduces the efficacy of the antibiotics administered, suppressive combinations could be less potent against infections sensitive to the drugs used, subsequently prolonging treatment time58. This disadvantage of suppressive treatments might explain the fact that these combinations are statistically underrepresented in scientific works59. When the patient’s condition is severe or the infection needs to be swiftly removed for any reason, suppressive treatments could be disadvantageous. To explore the efficacy of combinational treatments, libraries of approved drugs (both antibiotic and non-antibiotic) have been screened to explore the impact of approved drugs in combination against a given pathogen60,61,62,63,64,65. An interesting example in the case of Staphylococcus aureus is that when measuring growth inhibition, protein-synthesis inhibitors suppress the efficacy of trimethoprim - however, this interaction is not present when measuring killing efficacy63.
Second, since suppressive combination treatments lessen the efficacy of an antibiotic, higher doses might be necessary to reach a desired effect - and such increased dosages could carry toxicological risks to the patient itself. This can be a limiting factor, especially when considering medications which have severe side effects and could prove to be highly toxic when administered in increased dosages66. Higher administered dosage could also be unfeasible when treating patients with preexisting conditions limiting the metabolism of the administered drugs.
The advantage of suppressive treatments is to limit bacterial resistance evolution by exploiting the pleiotropy of mono-resistance mutations. Since suppression reduces the efficacy of one antibiotic when taken with another, a strain resistant to the suppressor antibiotic suffers a greater hit from the suppressed antibiotic compared to the antibiotic sensitive variant when co-administered (Fig. 3)59,63,67,68. Suppression is common, especially between bacteriostatic and metabolism-dependent bactericidal antibiotics61,69,70. In spite of the fact that a drug’s effect depends on the species treated, oxygen depletion, and the concentration used, these categorizations might be used as preliminary indicative factors when searching for suppressive combinations against resistant infections60,71,72,73,74. While suppression is typically non-reciprocal - one antibiotic suppresses the effect of the other, but not vice-versa - interactions can also be reciprocal. Reciprocal suppression - where both antibiotics in a pairwise combination suppress the effects of one another - can be even more beneficial by increasing long-term clearance of pathogens resistant to any one antibiotic of a reciprocally suppressive drug pair63.
Suppression is not only revealed in pairwise combinations, but in three-way interactions as well75. Utilising three drugs significantly increases the number of available interactions, and so, increases the chance of discovering suppressive ones as well. Overwhelmingly, though, the triple combinations found were not suppressive towards single-drug treatments, but towards pairwise combinations of the antibiotics used (named “hidden” suppressors)75. This might prove useful when suppressive treatments are considered against strains resistant to two of the drugs in a three-way combination. Drug interactions exceeding three-way combinations mainly use mathematical modelling to predict treatment efficacy, since the amount of combinations increases quadratically when incrementing the number of antibiotics in a given combination76,77,78,79.
The antibiotic sensitive gut microbiota is an essential part of the gastrointestinal system, and suppressive treatments could also prove to be helpful in preserving its functionality and integrity60. A healthy gut microbiota performs several vital functions by aiding host digestion, stimulating and regulating the immune system, and preventing the growth of virulent pathogens. Both antibiotic treatments and non-antibacterial treatments can disturb gut commensals, reducing the overall diversity of species and increasing susceptibility to colonisation with infectious pathogens60,68,80,81,82,83,84,85,86,87,88. However, it has been shown that the collateral damage of certain broad-spectrum antibiotic treatments on gut commensals could be mitigated by drugs such as tolfenamic acid that specifically suppressed the antibiotic treatment’s effects on Bacteroides species, but not on pathogenic bacteria60.
Suppressive treatments could also help avoid the spread of existing but rare mechanisms of resistance to newly developed or critically important antimicrobial drugs by co-application with a suppressive partner in treatments to preserve the rarity of resistance availability. Similarly, suppressive treatments could preserve widely-used antibiotics which are invaluable in the treatment of severe diseases - evidence of this is already present in the protocols of tuberculosis treatments. Combinational treatments with rifampicin are advocated for by the World Health Organization and the International Union against Tuberculosis and Lung Disease to preserve the efficacy of the drug and counteract resistance mutations19,89,90.
Synergy
Synergy occurs in a drug combination when one or more of the administered antibiotics strengthen the effect of the other, resulting in the drug combination being amplified compared to the expected, combined effect (such as 2001’s best-selling drug Augmentin, a combination of the beta-lactam amoxicillin and the beta-lactamase inhibitor clavulanic acid targeting beta-lactamase resistant strains; Fig. 3, Fig. 4). This interaction is the one that seems most beneficial when taken at face value - however, it is not free from complications and disadvantages.
The typical checkerboard assay consists of gradients of two antibiotics, which increase in concentration on two different axes. Here, antibiotics blue and orange (blue pill and orange pill), are presented on a checkerboard assay, where antibiotic blue increases in concentration along the x axis, and antibiotic orange increases in concentration on the y axis. As the concentrations increase, bacterial growth is reduced, marked by a decrease in the darkness of the grey circles. This creates a front of inhibition, where bacteria stop growing. The shape of this inhibition front is hyperbolic, as shown by the red dashed line, suggesting a synergistic combination compared to the expected additive effect (grey dashed line). However, inspection of inhibited wells for clearance efficacy after a predetermined amount of treatment time (barplot on the upper right), the survival of bacteria when receiving the maximal dose of the antibiotics and their combinations betrays a suppressive interaction, the exact opposite of the effect expected by growth inhibition data.
Since the synergistic effect relies on a drug amplifying the effect of another, if a strain becomes resistant to any single drug of the combination, the efficacy of the administered treatment is reduced91,92,93 (Fig. 3). Even though the evolution of resistance to antibiotic combinations is generally slower than mono-therapies, the rate at which bacteria adapt to multidrug treatments is correlated with the degree of synergy between the constituent antibiotics38,92. Since synergism relies on the drugs’ effects enhancing one another, even modest single-drug resistant mutants can show large losses of sensitivity to synergistic treatments, resulting in greater fitness benefits that can further promote the emergence of resistance to both antibiotics. The setback caused by synergism’s nullification by mono-resistance evolution might also provide a populational risk as well by increasing the prevalence of multidrug resistance in the environment. Furthermore, synergistic interactions, thanks to their enhanced effects on antibiotic-susceptible bacteria, can impact the gut microbiota, worsening the adverse side effects of antimicrobial combinations compared to single-drug treatments94.
Most preclinical screenings aim to find synergistic combinations - however, the majority of studies have been performed by measuring growth inhibition instead of clearance efficacy61,64,70,78,95,96. As the effects of combinational treatments on growth inhibition and clearance efficacy do not necessarily overlap, synergistic combinations based on growth inhibition might not be synergistic in practice (Fig. 4)63,69. In fact, they might even prove to be suppressive, meaning the effectiveness of treatments - and, consequently, the selection advantages of resistant and sensitive bacterial mutants - might be reversed compared to the expected outcomes.
Despite the aforementioned limitations, synergistic combinations could be used to reduce the required doses of antibiotics when designing a treatment, which is especially important in the case of antibiotics that are toxic to a patient even in minute dosages - such as ciprofloxacin co-administered with colistin66,69. This could prove to be important by widening the possible range of antibiotics used in treatments for patients who might experience increased toxicity due to preexisting conditions or intolerances towards certain antibiotics.
Additionally, synergistic combinations must not be overlooked either when considering the proper treatment for an infection. Synergistic combinations, when used with care, could be an invaluable asset in clearing severe infections that must be dealt with swiftly. In such cases, synergistic combinations reduce treatment time and speed up recovery in tuberculosis patients89,97. Furthermore, there is evidence that synergistic drug combinations can effectively eradicate biofilm-forming infections98,99,100,101,102. In fact, in the case of Staphylococcus aureus and Staphylococcus epidermidis, it has been found that anprocide, a steroidal amine, was effective against biofilms of these species, and was synergistically active with bacitracin and oxacillin as well100. In the case of Pseudomonas aeruginosa, the combined treatments of isepamicin and fosfomycin was successful against the biofilm-forming infection, even in rat models, while the components alone were not sufficient to alleviate the infection in the in vivo model99.
A mathematical model has been described to find balance between the advent of resistance evolution and treatment efficacy, aimed at finding the optimal spot between drug interactions and the emergence of resistance93. This model also describes a synergistic ceiling - a limit where additional interactions no longer provide any benefits for treatment, but still increase the prevalence of drug resistance. Mathematical models like the one described could be an asset when finding the optimal synergistic treatment regimen for patients.
Conclusion
In this review, we outlined the influence that the pleiotropy of resistance can have on the effectiveness of multidrug therapies. Our current knowledge suggests that antibiotic interactions - and thus, combination treatments - should be viewed as a versatile toolbox to tackle bacterial infections, where depending on the therapeutic aim, each interaction has its rational field of application. In the complex landscape of drug interactions, synergistic and suppressive combinations could easily overlap with collateral sensitivity or cross-resistance. This can cause multi-layer interactions, where collateral sensitivity responses overlapping with synergy might be able to cause unexpected advantages for the synergistic pairs by limiting resistance evolution49. Similarly, certain suppressive interactions combined with a high chance of cross-resistance evolution could lose all possible benefits of suppressive pairs, making such treatments unsuitable for clinical use. Therefore, in the space of drug interactions, it is equally important to explore the synergy - suppression axis and the cross resistance - collateral sensitivity axis before drawing any definitive conclusion about a drug combination. Moreover, there is evidence that synergistic and suppressive treatments can also show different interactions in different species, further incrementing the dimensionality of the drug interaction space61.
It should also be noted that our current strategies for developing and applying antibiotic combination treatments are hampered by the ignorance of bacterial strategies to overcome the bactericidal effects of drugs such as biofilm formation and bacterial persistence. Microbial biofilms are becoming increasingly difficult to treat in medical settings due to their intrinsic resistance to antibiotics103. To combat this, several biofilm dispersal agents in combination with conventional antibiotics are currently being developed as treatments for biofilm infections104. Certain drug combinations can increase the rate of bacterial persistence, which have the potential to re-establish infections after treatment63. Recently, a number of robust and scalable bacterial viability measures have been developed, paving the way to advance bacterial clearance measures into drug combination design and the emergence of tolerance and persistence-tailored treatments63,105,106,107.
Although the use of combinations of antimicrobial agents is common practice in medicine, clinical experience and in vitro research hasn’t yet been fully connected. The emergence of pre-clinical research in combinational therapies paves the way to new sustainable approaches of antimicrobial treatment, therefore, these strategies warrant further exploration by prospective studies of the clinical treatment of various infectious diseases40. The retrospective exploration of empirically applied treatments in hospital settings could also further shed light on the underlying drug interactions, allowing us to extrapolate and find more candidates68.
References
Murray, C. J. L. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet 399, 629–655 (2022).
D’Costa, V. M. et al. Antibiotic resistance is ancient. Nature 477, 457–461 (2011).
Levy Stuart, B., FitzGerald George, B. & Macone Ann, B. Changes in Intestinal Flora of Farm Personnel after Introduction of a Tetracycline-Supplemented Feed on a Farm. N. Engl. J. Med. 295, 583–588 (1976).
Humeniuk, C. et al. β-Lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob. Agents Chemother. 46, 3045–3049 (2002).
Daruka, L. et al. Antibiotics of the future are prone to resistance in Gram-negative pathogens. 2023.07.23.550022 Preprint at https://doi.org/10.1101/2023.07.23.550022 (2023).
Kohlmann, R. & Gatermann, S. G. Analysis and presentation of cumulative antimicrobial susceptibility test data – the influence of different parameters in a routine clinical microbiology laboratory. PLoS ONE 11, e0147965 (2016).
Balouiri, M., Sadiki, M. & Ibnsouda, S. K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 6, 71–79 (2016).
Garcia. Synergism Testing: Broth Microdilution Checkerboard and Broth Macrodilution Methods. 140–162 (American Society of Microbiology, 2010). https://doi.org/10.1128/9781555817435.ch5.12.
Ayrapetyan, M., Williams, T. C., Baxter, R. & Oliver, J. D. Viable but nonculturable and persister cells coexist stochastically and are induced by human serum. Infect. Immun. 83, 4194–4203 (2015).
Brauner, A., Fridman, O., Gefen, O. & Balaban, N. Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 14, 320–330 (2016).
Sáez-López, E., Millán-Placer, A. C., Lucía, A. & Ramón-García, S. Amoxicillin/clavulanate in combination with rifampicin/clarithromycin is bactericidal against Mycobacterium ulcerans. PLoS Negl. Trop. Dis. 18, e0011867 (2024).
Meylan, S., Andrews, I. W. & Collins, J. J. Targeting antibiotic tolerance, pathogen by pathogen. Cell 172, 1228–1238 (2018).
Levin-Reisman, I., Brauner, A., Ronin, I. & Balaban, N. Q. Epistasis between antibiotic tolerance, persistence, and resistance mutations. Proc. Natl. Acad. Sci. 116, 14734–14739 (2019).
Brauner, A., Shoresh, N., Fridman, O. & Balaban, N. Q. An experimental framework for quantifying bacterial tolerance. Biophys. J. 112, 2664–2671 (2017).
Kwak, N. et al. M ycobacterium abscessus pulmonary disease: individual patient data meta-analysis. Eur. Respir. J. 54, 1801991 (2019).
Lee, S. et al. Comparative outcomes of cefazolin versus nafcillin for methicillin-susceptible Staphylococcus aureus bacteraemia: a prospective multicentre cohort study in Korea. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 24, 152–158 (2018).
Pahil, K. S. et al. A new antibiotic traps lipopolysaccharide in its intermembrane transporter. Nature 625, 572–577 (2024).
Fallone, C. A. et al. The Toronto consensus for the treatment of helicobacter pylori infection in adults. Gastroenterology 151, 51–69.e14 (2016).
Nahid, P. et al. Executive Summary: Official American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America Clinical Practice Guidelines: Treatment of Drug-Susceptible Tuberculosis. Clin. Infect. Dis. 63, 853–867 (2016).
Levy, S. B. Factors impacting on the problem of antibiotic resistance. J. Antimicrob. Chemother. 49, 25–30 (2002).
Szybalski, W. & Bryson, V. Genetic studies on microbial cross resistance to toxic agents. I. Cross resistance of Escherichia coli to fifteen antibiotics. J. Bacteriol. 64, 489–499 (1952).
Maltas, J. & Wood, K. B. Pervasive and diverse collateral sensitivity profiles inform optimal strategies to limit antibiotic resistance. PLoS Biol 17, e3000515 (2019).
Lázár, V. et al. Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides. Nat. Microbiol. 3, 718–731 (2018).
Lázár, V. et al. Bacterial evolution of antibiotic hypersensitivity. Mol. Syst. Biol. 9, 700 (2013).
Barbosa, C., Römhild, R., Rosenstiel, P. & Schulenburg, H. Evolutionary stability of collateral sensitivity to antibiotics in the model pathogen Pseudomonas aeruginosa. eLife 8, e51481 (2019).
Hernando-Amado, S., Sanz-García, F. & Martínez, J. L. Rapid and robust evolution of collateral sensitivity in Pseudomonas aeruginosa antibiotic-resistant mutants. Sci. Adv. 6, eaba5493 (2020).
Kavanaugh, L. G., Flanagan, J. N. & Steck, T. R. Reciprocal antibiotic collateral sensitivity in Burkholderia multivorans. Int. J. Antimicrob. Agents 56, 105994 (2020).
Oz, T. et al. Strength of selection pressure is an important parameter contributing to the complexity of antibiotic resistance evolution. Mol. Biol. Evol. 31, 2387–2401 (2014).
Podnecky, N. L. et al. Conserved collateral antibiotic susceptibility networks in diverse clinical strains of Escherichia coli. Nat. Commun. 9, 3673 (2018).
Barbosa, C. et al. Alternative evolutionary paths to bacterial antibiotic resistance cause distinct collateral effects. Mol. Biol. Evol. 34, 2229–2244 (2017).
Burmeister, A. R. et al. Pleiotropy complicates a trade-off between phage resistance and antibiotic resistance. Proc. Natl. Acad. Sci. USA. 117, 11207–11216 (2020).
Hernando-Amado, S., Laborda, P. & Martínez, J. L. Tackling antibiotic resistance by inducing transient and robust collateral sensitivity. Nat. Commun. 14, 1723 (2023).
Lázár, V. et al. Genome-wide analysis captures the determinants of the antibiotic cross-resistance interaction network. Nat. Commun. 5, 4352 (2014).
Laborda, P., Martínez, J. L. & Hernando‐Amado, S. Convergent phenotypic evolution towards fosfomycin collateral sensitivity of Pseudomonas aeruginosa antibiotic‐resistant mutants. Microb. Biotechnol. 15, 613–629 (2021).
Sakenova, N. et al. Systematic mapping of antibiotic cross-resistance and collateral sensitivity with chemical genetics. 2024.01.25.576750 Preprint at https://doi.org/10.1101/2024.01.25.576750 (2024).
Rodriguez de Evgrafov, M., Gumpert, H., Munck, C., Thomsen, T. T. & Sommer, M. O. A. Collateral resistance and sensitivity modulate evolution of high-level resistance to drug combination treatment in Staphylococcus aureus. Mol. Biol. Evol. 32, 1175–1185 (2015).
Munck, C., Gumpert, H. K., Wallin, A. I. N., Wang, H. H. & Sommer, M. O. A. Prediction of resistance development against drug combinations by collateral responses to component drugs. Sci. Transl. Med. 6, 262ra156 (2014).
Jahn, L. J. et al. Compatibility of evolutionary responses to constituent antibiotics drive resistance evolution to drug pairs. Mol. Biol. Evol. 38, 2057–2069 (2021).
Apjok, G. et al. Limited evolutionary conservation of the phenotypic effects of antibiotic resistance mutations. Mol. Biol. Evol. 36, 1601–1611 (2019).
Beckley, A. M. & Wright, E. S. Identification of antibiotic pairs that evade concurrent resistance via a retrospective analysis of antimicrobial susceptibility test results. Lancet Microbe 2, e545–e554 (2021).
Noto Guillen, M., Li, C., Rosener, B. & Mitchell, A. Antibacterial activity of nonantibiotics is orthogonal to standard antibiotics. Science 384, 93–100 (2024).
Balaban, N. Q. et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 17, 441–448 (2019).
Trastoy, R. et al. Mechanisms of Bacterial Tolerance and Persistence in the Gastrointestinal and Respiratory Environments. Clin. Microbiol. Rev. 31, e00023–18 (2018).
Irving, S. E., Choudhury, N. R. & Corrigan, R. M. The stringent response and physiological roles of (pp)pGpp in bacteria. Nat. Rev. Microbiol. 19, 256–271 (2021).
Fuentes-Hernandez, A. et al. Using a sequential regimen to eliminate bacteria at sublethal antibiotic dosages. PLoS Biol 13, e1002104 (2015).
Kim, S., Lieberman, T. D. & Kishony, R. Alternating antibiotic treatments constrain evolutionary paths to multidrug resistance. Proc. Natl. Acad. Sci. USA. 111, 14494–14499 (2014).
Imamovic, L. & Sommer, M. O. A. Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci. Transl. Med. 5, 204ra132 (2013).
Hernando-Amado, S. et al. Rapid Phenotypic Convergence towards Collateral Sensitivity in Clinical Isolates of Pseudomonas aeruginosa Presenting Different Genomic Backgrounds. Microbiol. Spectr. 11, e02276–22 (2022).
Barbosa, C., Beardmore, R., Schulenburg, H. & Jansen, G. Antibiotic combination efficacy (ACE) networks for a Pseudomonas aeruginosa model. PLoS Biol 16, e2004356 (2018).
Armstrong, D. et al. Evidence for Spread of a Clonal Strain of Pseudomonas aeruginosa among Cystic Fibrosis Clinics. J. Clin. Microbiol. 41, 2266–2267 (2003).
Hernando-Amado, S., Laborda, P., Valverde, J. R. & Martínez, J. L. Mutational background influences P. aeruginosa ciprofloxacin resistance evolution but preserves collateral sensitivity robustness. Proc. Natl. Acad. Sci. USA. 119, e2109370119 (2022).
Diaz Caballero, J. et al. Mixed strain pathogen populations accelerate the evolution of antibiotic resistance in patients. Nat. Commun. 14, 4083 (2023).
Imamovic, L. et al. Drug-driven phenotypic convergence supports rational treatment strategies of chronic infections. Cell 172, 121–134.e14 (2018).
Stracy, M. et al. Minimizing treatment-induced emergence of antibiotic resistance in bacterial infections. Science 375, 889–894 (2022).
Stone, L. K. et al. Compounds that select against the tetracycline resistance efflux pump. Nat. Chem. Biol. 12, 902–904 (2016).
Herencias, C. et al. Collateral sensitivity associated with antibiotic resistance plasmids. eLife 10, e65130 (2021).
β-lactamase expression induces collateral sensitivity in Escherichia coli | bioRxiv. https://www.biorxiv.org/content/10.1101/2023.11.22.568265v1
Lepper, M. H. & Dowling, H. F. Treatment of pneumococcic meningitis with penicillin compared with penicillin plus aureomycin; studies including observations on an apparent antagonism between penicillin and aureomycin. AMA Arch. Intern. Med. 88, 489–494 (1951).
Singh, N. & Yeh, P. J. Suppressive drug combinations and their potential to combat antibiotic resistance. J. Antibiot. (Tokyo) 70, 1033–1042 (2017).
Maier, L. et al. Unravelling the collateral damage of antibiotics on gut bacteria. Nature 599, 120–124 (2021).
Brochado, A. R. Species-specific activity of antibacterial drug combinations. Nature 559, 259–263 (2018).
Hind, C. K. et al. Evaluation of a library of FDA-approved drugs for their ability to potentiate antibiotics against multidrug-resistant Gram-negative pathogens. Antimicrob. Agents Chemother. 63, e00769–19 (2019).
Lázár, V., Snitser, O., Barkan, D. & Kishony, R. Antibiotic combinations reduce Staphylococcus aureus clearance. Nature 610, 540–546 (2022).
Cacace, E. et al. Systematic analysis of drug combinations against Gram-positive bacteria. Nat. Microbiol. 8, 2196–2212 (2023).
Ejim, L. et al. Combinations of antibiotics and nonantibiotic drugs enhance antimicrobial efficacy. Nat. Chem. Biol. 7, 348–350 (2011).
Sorlí, L. et al. Trough colistin plasma level is an independent risk factor for nephrotoxicity: a prospective observational cohort study. BMC Infect. Dis. 13, 380 (2013).
Baym, M., Stone, L. K. & Kishony, R. Multidrug evolutionary strategies to reverse antibiotic resistance. Science 351, aad3292 (2016).
Liu, J., Gefen, O., Ronin, I., Bar-Meir, M. & Balaban, N. Q. Effect of tolerance on the evolution of antibiotic resistance under drug combinations. Science 367, 200–204 (2020).
Zheng, E. J., Stokes, J. M. & Collins, J. J. Eradicating bacterial persisters with combinations of strongly and weakly metabolism-dependent antibiotics. Cell Chem. Biol. 27, 1544–1552.e3 (2020).
Ocampo, P. S. et al. Antagonism between bacteriostatic and bactericidal antibiotics is prevalent. Antimicrob. Agents Chemother. 58, 4573–4582 (2014).
Pankey, G. A. & Sabath, L. D. Clinical relevance of bacteriostatic versus bactericidal mechanisms of action in the treatment of Gram-positive bacterial infections. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 38, 864–870 (2004).
Klepser, M. E., Nicolau, D. P., Quintiliani, R. & Nightingale, C. H. Bactericidal activity of low-dose clindamycin administered at 8- and 12-hour intervals against Staphylococcus aureus, Streptococcus pneumoniae, and Bacteroides fragilis. Antimicrob. Agents Chemother. 41, 630–635 (1997).
Zahedi Bialvaei, A., Rahbar, M., Yousefi, M., Asgharzadeh, M. & Samadi Kafil, H. Linezolid: a promising option in the treatment of Gram-positives. J. Antimicrob. Chemother. 72, 354–364 (2017).
Léger, L. β-Lactam Exposure Triggers Reactive Oxygen Species Formation in Enterococcus faecalis via the Respiratory Chain Component DMK. Cell Rep. 29, 2184–2191 3 (2019).
Beppler, C. et al. When more is less: Emergent suppressive interactions in three-drug combinations. BMC Microbiol 17, 107 (2017).
Katzir, I., Cokol, M., Aldridge, B. B. & Alon, U. Prediction of ultra-high-order antibiotic combinations based on pairwise interactions. PLoS Comput. Biol. 15, e1006774 (2019).
Wood, K., Nishida, S., Sontag, E. D. & Cluzel, P. Mechanism-independent method for predicting response to multidrug combinations in bacteria. Proc. Natl. Acad. Sci. 109, 12254–12259 (2012).
Zimmer, A., Katzir, I., Dekel, E., Mayo, A. E. & Alon, U. Prediction of multidimensional drug dose responses based on measurements of drug pairs. Proc. Natl. Acad. Sci. USA. 113, 10442–10447 (2016).
Zimmer, A., Tendler, A., Katzir, I., Mayo, A. & Alon, U. Prediction of drug cocktail effects when the number of measurements is limited. PLOS Biol 15, e2002518 (2017).
Maier, L. et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555, 623–628 (2018).
Bahl, D. et al. In vitro activities of ciprofloxacin and rifampin alone and in combination against growing and nongrowing strains of methicillin-susceptible and methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 41, 1293–1297 (1997).
Bollenbach, T., Quan, S., Chait, R. & Kishony, R. Nonoptimal Microbial Response to Antibiotics Underlies Suppressive Drug Interactions. Cell 139, 707–718 (2009).
Brown, T. H. & Alford, R. H. Antagonism by chloramphenicol of broad-spectrum beta-lactam antibiotics against Klebsiella pneumoniae. Antimicrob. Agents Chemother. 25, 405–407 (1984).
Jawetz, E., Gunnison, J. B., Speck, R. S. & Coleman, V. R. Studies on antibiotic synergism and antagonism; the interference of chloramphenicol with the action of penicillin. AMA Arch. Intern. Med. 87, 349–359 (1951).
Johansen, H. K., Jensen, T. G., Dessau, R. B., Lundgren, B. & Frimodt-Moller, N. Antagonism between penicillin and erythromycin against Streptococcus pneumoniae in vitro and in vivo. J. Antimicrob. Chemother. 46, 973–980 (2000).
Lange, K., Buerger, M., Stallmach, A. & Bruns, T. Effects of Antibiotics on Gut Microbiota. Dig. Dis. 34, 260–268 (2016).
Modi, S. R., Collins, J. J. & Relman, D. A. Antibiotics and the gut microbiota. J. Clin. Invest. 124, 4212–4218 (2014).
Zinner, S. H., Provonchee, R. B., Elias, K. S. & Peter, G. Effect of clindamycin on the in vitro activity of amikacin and gentamicin against gram-negative bacilli. Antimicrob. Agents Chemother. 9, 661–664 (1976).
Chaisson, R. E. Treatment of Chronic Infections with Rifamycins: Is Resistance Likely To Follow? Antimicrob. Agents Chemother. 47, 3037–3039 (2003).
Rieder, H. L. Interventions for Tuberculosis Control and Elimination. (International Union against Tuberculosis and Lung Disease, Paris, 2002).
Pena-Miller, R. et al. When the Most Potent Combination of Antibiotics Selects for the Greatest Bacterial Load: The Smile-Frown Transition. PLoS Biol 11, e1001540 (2013).
Hegreness, M., Shoresh, N., Damian, D., Hartl, D. & Kishony, R. Accelerated evolution of resistance in multidrug environments. Proc. Natl. Acad. Sci. USA. 105, 13977–13981 (2008).
Torella, J. P., Chait, R. & Kishony, R. Optimal Drug Synergy in Antimicrobial Treatments. PLOS Comput. Biol. 6, e1000796 (2010).
Zhang, Y., Limaye, P. B., Renaud, H. J. & Klaassen, C. D. Effect of various antibiotics on modulation of intestinal microbiota and bile acid profile in mice. Toxicol. Appl. Pharmacol. 277, 138–145 (2014).
Russ, D. & Kishony, R. Additivity of inhibitory effects in multidrug combinations. Nat. Microbiol. 3, 1339–1345 (2018).
Yeh, P., Tschumi, A. I. & Kishony, R. Functional classification of drugs by properties of their pairwise interactions. Nat. Genet. 38, 489–494 (2006).
Zhang, Y., Shi, W., Zhang, W. & Mitchison, D. Mechanisms of pyrazinamide action and resistance. Microbiol. Spectr. 2, 1–12 (2013).
Zhang, K., Li, X., Yu, C. & Wang, Y. Promising therapeutic strategies against microbial biofilm challenges. Front. Cell. Infect. Microbiol. 10, 359 (2020).
Cai, Y., Fan, Y., Wang, R., An, M.-M. & Liang, B.-B. Synergistic effects of aminoglycosides and fosfomycin on Pseudomonas aeruginosa in vitro and biofilm infections in a rat model. J. Antimicrob. Chemother. 64, 563–566 (2009).
Pettit, R. K. et al. In vivo activity of anprocide alone, and in vitro activity in combination with conventional antibiotics against Staphylococcus aureus and Staphylococcus epidermidis biofilms. J. Med. Microbiol. 58, 1203–1206 (2009).
Cernohorská, L. & Votava, M. Antibiotic synergy against biofilm-forming Pseudomonas aeruginosa. Folia Microbiol. (Praha) 53, 57–60 (2008).
Rose, W. E. & Poppens, P. T. Impact of biofilm on the in vitro activity of vancomycin alone and in combination with tigecycline and rifampicin against Staphylococcus aureus. J. Antimicrob. Chemother. 63, 485–488 (2009).
Vestby, L. K., Grønseth, T., Simm, R. & Nesse, L. L. Bacterial Biofilm and its Role in the Pathogenesis of Disease. Antibiot. Basel Switz. 9, 59 (2020).
Hawas, S., Verderosa, A. D. & Totsika, M. Combination Therapies for Biofilm Inhibition and Eradication: A Comparative Review of Laboratory and Preclinical Studies. Front. Cell. Infect. Microbiol. 12, 850030 (2022).
Gefen, O., Chekol, B., Strahilevitz, J. & Balaban, N. Q. TDtest: easy detection of bacterial tolerance and persistence in clinical isolates by a modified disk-diffusion assay. Sci. Rep. 7, 41284 (2017).
Zheng, E. J. et al. Discovery of antibiotics that selectively kill metabolically dormant bacteria. Cell Chem. Biol. 31, 712–728.e9 (2024).
Meyer, C. T. et al. A high-throughput and low-waste viability assay for microbes. Nat. Microbiol. 8, 2304–2314 (2023).
Acknowledgements
We would like to thank the graphical design of the figures to Enikő Koliger. V.L. is supported by the Lendulet “Momentum” program of the Hungarian Academy of Sciences [grant agreement LP2022-12/2022] and the EMBO Installation Grant [grant number 5709_2024]; R.S. is supported by the The Janos Bolyai Research Fellowship of the Hungarian Academy of Sciences [grant number BO/608/21].
Author information
Authors and Affiliations
Contributions
BB and VL wrote the manuscript. RS edited and reviewed the manuscript. VL contributed to reviewing and editing the manuscript, and supervised the process. BB, RS and VL conceptualised the manuscript and the figures.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Bognár, B., Spohn, R. & Lázár, V. Drug combinations targeting antibiotic resistance. npj Antimicrob Resist 2, 29 (2024). https://doi.org/10.1038/s44259-024-00047-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s44259-024-00047-2
This article is cited by
-
Iterative discovery of potent polymeric antibiotics via multi-stage and multi-task learning against antimicrobial resistance
Nature Communications (2026)
-
Combining Bile Salt with Gentamicin: A Promising Strategy Against Vibrio Parahaemolyticus and Escherichia Coli
Current Microbiology (2026)
-
Evolutionary drivers of divergent collateral sensitivity responses during antibiotic therapy
Nature Ecology & Evolution (2025)
-
A Novel Synergized Nanoformulation Revives the Activity of Ineffective Ceftibuten Antibiotic: Synthesis, Characterization, In Vitro Validation and Molecular Interactions Study
BioNanoScience (2025)