Abstract
Background
It is difficult for clinicians to make predictions for cancer progression or outcomes based on AJCC staging for individual patients. Models individualising risk prediction for clinical outcomes are developed using patient level data, advanced statistical techniques, and artificial intelligence.
Methods
A systematic search identified cutaneous melanoma prognostic prediction tools published between January 1985–March 2023. Population comparisons of key clinico-pathological variables, external prediction of receiver operating characteristics and calibration analysis are applied to an unselected group of patients undergoing sentinel lymph node biopsy in a UK University hospital setting (n = 1564).
Results
Twenty-nine models were identified which predicted survival, disease recurrence or sentinel lymph node positivity (Internal validation n = 19 and external validation n = 14). 3 out of 7 tools for sentinel node positivity were contemporaneous with available characteristics for external validation. External validation of models by Lo et al. Friedman et al. & Bertolli et al. highlighted good discriminative performance (AUC 68.1% (64.5–71.8%), 77.1% (66.8–85.7%) & 68.6% (63.3–74.1%) respectively) but were sub-optimally calibrated for the UK patient cohort (Calibration intercept & slope Friedman: −4.01 & 32.92, Lo: −1.17 & 0.44, Bertolli: −2.75 & 4.88).
Conclusions
This work highlights the complexity of predictive modelling and the rigorous validation process necessary to ensure accurate predictions. Our search highlights a tendency to focus on discriminative performance over calibration, and the possibility for inconsistent predictions when tools are applied to populations with differing characteristics.
Similar content being viewed by others
Background
Melanoma is the 5th most common cancer in the UK, accounting for approximately 5% of all new cancer cases [1]. Age-standardised incidence rates of melanoma have increased by 140% in the UK since the early 1990s. Whilst prognosis for early-stage melanoma (AJCC stages I/II) is good, many more deaths occur overall in patients diagnosed in these stages than those with more advanced disease (AJCC stages III/IV) [2]. This demonstrates the difficult challenge for clinicians in making accurate predictions about survival at initial diagnosis and suggests that using AJCC staging alone in early-stage cutaneous melanoma is not sufficient.
Cancer staging systems are designed to combine disease variables, with established prognostic associations, to provide estimations of expected outcomes of relevance such as survival or disease progression. They are widely used to group together patients with a similar expected survival to guide decision making regarding further investigations and treatments. The American Joint Committee on Cancer (AJCC) staging systems are commonly used for numerous cancer types and are frequently updated to improve accuracy and reflect current survival trends [3].
With an increasing interest in individualised medicine, prognostic prediction tools have been developed for a variety of cancer types. These tools take additional variables into consideration, combining them with disease specific information to provide survival predictions as well as other interim outcomes of interest to patients. Such models are specifically defined in guidelines designed to standardise the reporting of new tools as: “a mathematical equation that relates multiple predictors for a particular individual to the probability of risk for the presence (diagnosis) or future occurrence (prognosis) of a particular outcome” [4].
The TRIPOD reporting guidelines [4] have been designed to increase the transparency of newly developed models. The aim is to enable potential users to understand model development, the populations in which the model has been developed and how well it performs in that population. This enables users to assess how useful the model might be to patient groups of interest, especially in different patient populations. The development of such tools are based on a variety of statistical techniques and are increasingly utilising artificial intelligence and machine learning based methods. Making the decision about whether to use a particular tool is not a straightforward one.
The popularity of some prognostic prediction tools is impressively large, suggesting both clinicians and patients find them a useful adjunct to current standards of care including endorsement by professional bodies such as the American Joint Committee on Cancer (AJCC) and the National Institute for Health and Care Excellence (NICE) in the UK [5, 6].
To highlight the complexities of selecting a prognostic prediction tool to use, this review systematically examines prognostic tools currently available for use in patients diagnosed with primary cutaneous melanoma. It examines the methodological basis of the tools and validates a subset of those recently published on a dataset derived from an unselected group of patients from a University Hospital in the UK [7].
Input variables for prognostic prediction tools in melanoma
Existing tools utilise clinicopathological variables to predict outcomes. These data are easily attainable and require little further processing of tumour tissue or patient data than would already be done routinely. Those clinicopathological variables with established relationships with disease severity are naturally the most frequent input variables included in such models. Variables such as: histological subtype, Breslow thickness, ulceration, mitotic rate, sex & age are well supported in the literature as being useful prognostic indicators. Any variables utilised in addition to these may be included based on particular data being available to an institution and/or noted to significantly improve model performance.
Data sources to develop prediction tools
Tools are generated by analysing patient data for those who have been diagnosed and undergone treatment for melanoma. A variety of methods can then be used to create a model of the data by determining the relationship between combinations of variables with the outcomes of interest, such as survival or sentinel lymph node positivity.
Regardless of the methods used, the data used to determine the relationships is crucial to the success of the model. For instance, a dataset that does not include any patients over the age of 60 is unlikely to produce a model that performs well when making predictions in an older patient group. This also extends to the diversity of the individuals in the dataset, and its similarity between the populations where the model is developed to where it is applied. Datasets collected entirely from tertiary centres are at risk of containing a disproportionate number of patients with advanced disease or that which requires specialist treatment, including enrolment in trials. The development of models is specifically focused on patients with a particular stage or type of disease and the user must be aware of these criteria before attempting to apply any resulting tool in a wider patient group.
Validation & performance of prediction tools
Validation is the process of assessing the performance of a predictive model. This is performed on the data used to derive the model (internal validation) and should also be performed on separate data not involved in model derivation (external validation). Of these, it is external validation that is most of interest since our primary focus is in the model making predictions in unseen data.
Performance is assessed by measuring the calibration and discrimination of the model. Calibration is a measure of the agreement between estimated risks of an outcome and the observed outcome frequencies. Discrimination is the ability of the model to differentiate between individuals that experience and outcome and those that remain event free. In models that predict survival it can be thought of as the ability of the model to correctly rank individuals by their risk.
It is important that both these aspects of model performance are assessed. Good models are both well calibrated and can effectively discriminate, excellent performance in one domain is not sufficient to account poor performance in the other.
Methods
Systematic search strategy
A database search strategy was developed searching for relevant manuscripts & online tools from January 1985–March 2023 following PRISMA principles [8]. An example search term used is provided in Supplementary Methods. The search was only conducted for articles published in English, with full text availability and focusing specifically on cutaneous melanoma.
A prognostic tool was defined as any equation, nomogram, risk classification system, electronic calculator, or other tool format that had a foundation in a statistical model or algorithm, developed with the purpose of predicting survival in clinical practice [9]. All references in identified articles were scrutinised for additional relevant work meeting the search criteria.
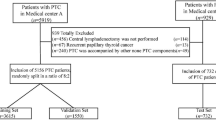
This search initially yielded 196 results, and an additional study was identified during anonymous manuscript expert review (Fig. 1). Following removal of duplicates (102), articles relating to melanoma of sites other than the skin [2], those articles not related to the development of a prognostic prediction tool for clinical use [10], not published in English [1], specifically relating to prediction for individual subtypes of melanoma or metastatic disease in one anatomical location [4] and using genetic or other non-clinicopathological predictors [8], 29 studies remained for inclusion in the review.
Articles were assessed using the CHARMS [11] and TRIPOD [4] guidelines designed to aid systematic review of prognostic prediction tools, model development and validation. The criteria set out by the AJCC for individualised risk prediction models was also utilised as a reference for model assessment [12].
External validation of existing tools for predicting a positive sentinel lymph node biopsy result
Sentinel lymph node biopsy has become part of standardised care pathways for melanoma. In the tools identified by our search, seven [13,14,15,16,17,18,19] are designed to provide predictions for the probability of receiving a positive result from undertaking this procedure. Such a prediction has potential for use in clinical settings to determine the risk more accurately for an individual and better assess the balance of risks and benefits of conducting this procedure.
A dataset containing patient data for individuals who underwent sentinel lymph node biopsy for cutaneous melanoma between 2008–2023 (n = 1564) was curated from a tertiary university melanoma centre (Addenbrooke’s Hospital, Cambridge, UK) [7]. This dataset was utilised to externally validate selected prediction tools identified in the literature search. The results provide an indication of the suitability of these models in the UK population. This study was reviewed by the Cambridge University Hospitals EHR Research and Innovation (ERIN) Database Access Committee (Reference A096904). We did not have data for patients not undergoing SLNB to validate survival or recurrence models.
Given the changing guidance regarding eligibility for SLNB it was felt that assessment of the most recently published prediction tools would be most appropriate. Those models utilising the AJCC 8th edition staging criteria were identified and included models published by Tripathi et al. (2023) [19], Bertolli et al. (2021) [18], Lo et al. (2020) [17] and Friedman et al. (2019) [16].
Sufficient information on the statistical models derived was available in the articles published by Lo et al. and Bertolli et al. Sufficient detail was provided by authors of the Friedman et al. paper on request. Model details from Tripathi et al. were not received to enable validation of the model they describe in our dataset.
Variables from our dataset were recoded according to the requirements of each model. The Friedman et al. model is specifically designed to make predictions for individuals diagnosed with thin melanomas (Breslow thickness 0.5–1.00 mm) [16]. The model does not permit the input of unknown or missing values for any variable and hence only individuals with thin melanomas and recorded values for all required variables were included (n = 215). Whilst the Lo et al. calculator does allow unknown values for several variables, only those with complete data for all required variables were used for analysis (n = 1348) [17]. To validate the Bertolli et al. model only those patients with complete data for all variables required by the model were included (n = 714) [18]. We compared the patient populations used to derive each model by variable. All provided population data was categorical and compared to the UK population using either the chi-squared test or Fisher’s exact test with a significance threshold set at 0.05.
Model discrimination was assessed by plotting the receiver operating characteristics for each models and calculating the area under the curve utilising the R package pROC [20]. 95% confidence intervals for the area were computed using this package with 2000 stratified bootstrap replicates.
Model calibration was assessed by plotting the observed frequency of the outcome and predicted probabilities produced by models and comparing the slope and intercept for each. It was additionally assessed by regressing the outcome onto the probability prediction produced by the model.
Results
Identification of current prognostic prediction tools
The literature search identified 29 clinical prognostic tools for use in cutaneous melanoma [14,15,16,17,18,19, 21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42]. Twelve of these tools were available to use with an online interface to calculate and display patient risk [13, 14, 17, 19, 21,22,23,24,25,26,27,28], with one of these also available as an Android app [24]. The remainder were available as a publication only. A detailed summary of identified tools can be seen in Fig. 2.
AJCC American Joint Committee on Cancer, TILs tumour infiltrating lymphocytes, Cox Reg Cox Regression Model (proportional hazards or logistic), LR Logistic Regression, KM Kaplan Meier, ML machine learning, Survival model predicting patient survival (disease specific or overall), Recurrence model predicting disease recurrence, SLNB result model predicting the result of a sentinel lymph node biopsy procedure, SEER Surveillance, Epidemiology, and End Results, EORTC European Organisation for Research And Treatment of Cancer).
Fifty-five input variables were used in the models identified. Breslow thickness was the most used pathological variable and age the most common patient factor variable, appearing in 24 and 23 models, respectively. The top 10 variables are illustrated in Fig. 2 with a complete list of input variables included in Supplementary Table 1.
All articles exclude children, with the exception of Bertolli et al. [18]. Whilst the age cut off for this varies between tools, no tool includes data on any individual diagnosed with cutaneous melanoma at less than fifteen years of age, except for Bertolli et al. which has an age range of 5–89.
From the models identified, the data used to develop them came from twenty different sources. Six population level datasets; The Surveillance, Epidemiology, and End Results (SEER) database (USA) [19, 27, 29, 30], Dutch Pathology Registry (PALGA, Netherlands) [26], Queensland Cancer Registry (Australia) [31], Swedish melanoma registry (Sweden [16, 19]) [32], Veneto Cancer Registry (Italy) [28], National Cancer Database, USA [16, 19]. Nine multi-centre datasets: Pigmented Lesion Group, University of Pennsylvania (USA) [33, 34], AJCC Melanoma Database (USA) [23], Cancer Genome Atlas (USA) [25], Scottish Melanoma Group Database (Scotland) [35], Sunbelt Melanoma Trial [36] Data [24], European Organisation for Research and Treatment of Cancer (EORTC) Melanoma Group Centres [37], six European melanoma centres [38] and combined datasets from 5 melanoma centres in the UK [15]. Eleven tools made use of single centre datasets from; The John Wayne Cancer Institute (USA) [39], The Melanoma Institute Australia [17], Memorial Sloan Kettering (USA) [13], Edmonton, Alberta (Canada) [40], Mayo Clinic, Rochester (USA) [41], Princess Alexandra Hospital, Queensland, Australia [42], Mass General Brigham, Dana-Farber Cancer Institute, Boston, USA [43, 44], Massachusetts General Hospital, USA [14, 21, 22] and the A.C. Camargo Cancer Centre, Brazil [18].
The sample sizes used for initial model creation ranged from 68 patients to 156,154 (Median 2647, IQR (979–25,930). One tool provided details of a sample size calculation and verification performed to ensure that a large enough sample was used for the chosen methodology [43].
A range of statistical tools were utilised to generate the prediction tools reviewed. These are outlined in Supplementary Table 2. ‘Classical’ statistical methods such as Cox and Logistic regression were the most frequently utilised. Techniques based on Cox regression were most common, utilised in sixteen of the tools reviewed. Machine learning techniques were less frequently used and as expected, appeared in work published much more recently.
Nineteen tools are provided with details of internal validation methodology, however only sixteen provide statistical results for this. The most common statistic provided was a concordance statistic or equivalent (e.g., Harrel C statistic for models utilising censored data).
Fourteen tools provided details of an external validation process with nine providing details of external validation statistics within the publication describing the tool. It is noted that some tools have subsequently been externally validated in separate publications and sometimes by separate authors, these are not included in this review. Concordance statistics were again the most frequently presented. Datasets used for external validation purposes were sourced from the same source as training data in two tools, but at different time points providing temporal validation. The other nine tools used data from an external source, including from a dataset originating from another country.
External validation of tools predicting positive sentinel lymph node biopsy results
Population comparisons
Populations utilised by Friedman et al., Lo et al. and Bertolli et al. in development of their respective tools were compared to the patient population used to externally validate them [7]. Tables 1–3 display comparisons between populations utilised by each model and our own dataset used to assess their performance. In the case of all model development populations there is a significant difference in disease variables when compared with our own dataset.
The comparison with the thin melanoma cohort from Friedman et al. showed more variables with adverse outcomes present in our cohort [16]. For example, there was a higher proportion of thinner melanomas (32.2% vs. 17.2% in the 0.5–0.8 mm category, p = >0.005), fewer patients with absent mitotic figures (15.4% vs 32.4%, p < 0.005), and fewer patients with dermal regression (7.0% vs. 15.2%, p < 0.005) in the Cambridge cohort [Table 1]. This may explain why the proportion of positive sentinel lymph node biopsies was lower in the Cambridge cohort compared to Friedman et al. (4.0% vs 7.3%, p = <0.005). This is likely reflective of higher thresholds of SLNB use in the UK population for pT1b melanomas.
In the case of the Lo et al. the population comparison highlighted that the Australian cohort had a significantly greater proportion of patients with adverse features [17] [Table 2]. A greater proportion of patients had thicker melanomas (≥2mm) (46.7% vs 40.1%, p = <0.005), ulcerated tumours (29.6% vs 21.9%, p = < 0.005), more mitoses (54.5% vs 35.7% in ≥4 category, p = < 0.005) & evidence of lymphovascular invasion (5.8% vs 2.7%, p = <0.005). These tie in with the significantly greater proportion of positive SLNB observed (21.0% vs 20.1%, p = <0.005). This is likely reflective of more adverse melanomas presenting in the Australian population.
Comparison of the Bertolli population with our Cambridge database population notably demonstrates some key differences [18] [Table 3]. Median melanoma Breslow thickness is higher in the Brazilian cohort (2.26 vs 1.4) with a greater proportion presenting with ulcerated tumours 25.8% vs 16.1%. A greater proportion of tumours were of the acral subtype than in the Cambridge group, associated with a significantly worse prognosis (8.9% vs 2.1%).
Model performance comparisons
The Friedman et al. model had an AUC of 77.1% (95% CI: 66.8% to 85.7%) (Fig. 3a), a significantly better result to that reported by the original paper of 67% (95% CI: 65–70%). Figure 3b, c displays the calibration plot for the model, demonstrating consistent underestimation of risk by the model that appears to worsen with increasing frequency of observed events. It is worth reiterating that this model is only designed to make predictions for those individuals with thin melanoma (0.5–1.0 mm) and hence has only been assessed on such patients from our dataset.
a–c Comparisons with the Friedman et al. model [16] with (a) Receiver Operating Characteristics area under the curve 77.1% (95% CI 66.8–85.7%). b Calibration Plot with slope = 32.92 and intercept = −4.01. c Differences between predicted and observed probabilities in the Cambridge University dataset. d–f Comparisons with the Lo et al. model [17] with (d) Receiver Operating Characteristics area under the curve 68.1% (95% CI 64.5–71.8%). e Calibration Plot slope = 0.44 and intercept = −1.17. f Differences between predicted and observed probabilities in the Cambridge University dataset. g–i Comparisons with the Bertolli et al. model [18] with (g) Receiver Operating Characteristics area under the curve 68.6% (95% CI 63.3–74.1%). h Calibration Plot with slope = 4.88 and intercept = −2.75. i Differences between predicted and observed probabilities in the Cambridge University dataset.
The Lo et al. model had an AUC of 68.1% (95% CI: 64.5–71.8%) demonstrating a reasonable discriminative performance (Fig. 3d). This compares with the 74.1% (95% CI: 72.1% to 76.0%) result from the internal validation reported by the original paper and 75.0% (95% CI: 73.2 to 76.7%) from external validation using data from MD Anderson Cancer Centre [17]. Fig. 3e, f displays the calibration plot for the model and demonstrates a tendency for the model to overestimate the risk of a positive SLNB result, particularly in the group of patients with highest clinicopathological risk.
The Bertolli et al. model had an AUC of 68.6% (95% CI: 63.3–74.1%) demonstrating a reasonable discriminative performance (Fig. 3g). The original paper describing this model reports a value of 75.1% (no CI provided) from internal validation. Figure 3h, i display the calibration of the model for our dataset, they demonstrate overestimation of risk for positive SLNB result with most accurate predictions at extremes of patient observed risk.
Discussion
This systematic literature search has identified published prognostic prediction tools designed for use in patients diagnosed with cutaneous melanoma. They aim to make a variety of predictions, but most commonly focus on melanoma specific survival, recurrence, and probability of a positive sentinel lymph node result. These models have been developed on a range of datasets that range from single centre to national cohorts of patients. The techniques used range from classical statistical techniques to newer machine learning derived methods. The data utilised and techniques employed to create the models are of interest to the end user since they can materially impact the suitability of the model for use in other patient groups.
Validation of such prediction models are essential and whilst such analysis is reported alongside models, it is not done so in all cases. There is also a focus on presenting validation statistics that relate only to the discriminative performance of models with the calibration component either not performed or not specifically reported upon. Good discriminative performance cannot make up for poor calibration and indeed can result in inaccurate predictions [45].
The three models that underwent external validation on our own dataset demonstrate poor calibration in our patient group. Two tended to overestimate risk and the other to underestimate, whilst demonstrating reasonable discriminative performance. These results suggest that they should be used in a UK population with caution and an awareness of these specific tendencies. In comparing the populations used in the development of these models with our own patient dataset, we have identified significant differences in the distribution of disease specific variables such as Breslow thickness, ulceration, and mitotic count. This further suggests that these models can be improved for use in a UK population.
In the case of the Friedman model, our dataset used for external validation is small, with a low number of positive SLNBs. This is secondary to UK melanoma guidelines for performing SLNB in patients with thin melanomas. For T1b melanomas, there are variations internationally in SLNB uptake (18.2% Sweden versus 28.1% Australia) [46]. Our criterion for offering SLNB has evolved over time in line with changes to AJCC 7th edition staging where a single mitotic figure classified as thin melanomas <1mm Breslow thickness as pT1b, and current NICE guidelines where SLNB can be considered if thin melanomas have a mitotic rate ≥3. Variations in practice as indications for SLNB change both over time, and in different countries should be considered when developing and applying different risk prediction models.
Technical developments in the fields of genetics and genomics have enabled the development of tools based on additional molecular features such as: gene expression profiles, ctDNA and individual biomarkers. These have deliberately been excluded from this review of clinically validated prediction tools. These tools are the consequences of developments in technology and our understanding of the genetic basis of disease. They may present opportunities for improvement in prognostic prediction and treatments, but there are several issues with using them in clinical practice at present. A review of utilising GEPs in cutaneous melanoma [47] highlights that they are not endorsed by either the American Academy of Dermatology [48] or National Comprehensive Cancer Network [49]. No guidance exists to specify interventions based on GEP test results, although data on case use scenarios continue to develop.
Current GEP tests largely assign an individual’s tumour to a prognostic class (high vs low risk, or class 1 vs 2), rather than calculating specific survival. This can lead to grouping of patients to either high or low risk, despite significant differences in those individuals clinicopathological factors, that would normally be associated with a different expected survival [50]. Available tools development and validation appear to be based on small case numbers. The studies forming the basis of the two largest commercially available GEP based tools report using between 217 [10]-260 [51] patient samples (DecisionDX-Melanoma) and 245 [52] patient samples (Melagenix). Some authors have also expressed concerns regarding the minimal overlap among gene panels across various studies. The discussion of the review suggests that “there is insufficient data to support routine use of the currently available GEP tests” [47].
We suggest that utilising a large national dataset is warranted to develop a prognostic prediction tool for patient with cutaneous melanoma in the UK. This would serve as a valuable resource for patient and clinicians to enable better communication about risk and the decision-making process for further investigations and treatment. It would also serve to contrast and understand differences between melanoma patient cohorts internationally.
Data availability
The patient dataset used for validation purposes is not publicly available. Reasonable requests for anonymised information can be made directly to the corresponding author.
References
Cancer Research UK. Melanoma skin cancer statistics [Internet]. 2024 [cited 2024 Sep 8]. https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/melanoma-skin-cancer
Whiteman DC, Baade PD, Olsen CM. More people die from thin melanomas (≤1 mm) than from thick melanomas (>4 mm) in Queensland, Australia. J Investig Dermatol. 2015;135:1190–3.
Amin M, Edge S, Greene F, Byrd D, Brookland R, Washington M, et al. editors. AJCC Cancer Staging Manual 8th Edition. 8th ed. Springer; 2017.
Moons KGM, Altman DG, Reitsma JB, Ioannidis JPA, Macaskill P, Steyerberg EW, et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med. 2015;162:W1–73.
PREDICT Breast Cancer [Internet]. [cited 2020 Oct 31]. https://breast.predict.nhs.uk/
PREDICT Prostate Cancer [Internet]. [cited 2020 Oct 31]. https://prostate.predict.nhs.uk/
Pasha T, Arain Z, Buscombe J, Aloj L, Durrani A, Patel A, et al. Association of complex lymphatic drainage in head and neck cutaneous melanoma with sentinel lymph node biopsy outcomes. JAMA Otolaryngol–Head Neck Surg. 2023;149:416.
Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71.
Steyerberg EW. Clinical Prediction Models. 1st ed. New York: Springer; 2008.
Gerami P, Cook RW, Russell MC, Wilkinson J, Amaria RN, Gonzalez R, et al. Gene expression profiling for molecular staging of cutaneous melanoma in patients undergoing sentinel lymph node biopsy. J Am Acad Dermatol. 2015;72:780–785.e3.
Moons KGM, de Groot JAH, Bouwmeester W, Vergouwe Y, Mallett S, Altman DG, et al. Critical appraisal and data extraction for systematic reviews of prediction modelling studies: the CHARMS checklist. PLoS Med. 2014;11:e1001744.
Kattan MW, Hess KR, Amin MB, Lu Y, Moons KGM, Gershenwald JE, et al. American Joint Committee on Cancer acceptance criteria for inclusion of risk models for individualized prognosis in the practice of precision medicine. CA Cancer J Clin. 2016;66:370–4.
Wong SL, Kattan MW, McMasters KM, Coit DG. A nomogram that predicts the presence of sentinel node metastasis in melanoma with better discrimination than the American Joint Committee on CancerStaging System. Ann Surg Oncol. 2005;12:282–8.
Michaelson JS. Melanoma nodal status calculator LifeMath.net [Internet]. 2007 [cited 2024 Sep 29]. http://lifemath.net/cancer/
Mitra A, Conway C, Walker C, Cook M, Powell B, Lobo S, et al. Melanoma sentinel node biopsy and prediction models for relapse and overall survival. Br J Cancer. 2010;103:1229–36.
Friedman C, Lyon M, Torphy RJ, Thieu D, Hosokawa P, Gonzalez R, et al. A nomogram to predict node positivity in patients with thin melanomas helps inform shared patient decision making. J Surg Oncol. 2019;120:1276–83.
Lo SN, Ma J, Scolyer RA, Haydu LE, Stretch JR, Saw RPM, et al. Improved risk prediction calculator for sentinel node positivity in patients with melanoma: the melanoma institute Australia nomogram. J Clin Oncol. 2020;38:2719–27.
Bertolli E, Calsavara VF, de Macedo MP, Pinto CAL, Duprat Neto JP. Development and validation of a Brazilian nomogram to assess sentinel node biopsy positivity in melanoma. Tumor J. 2021;107:440–5.
Tripathi R, Larson K, Fowler G, Han D, Vetto JT, Bordeaux JS, et al. A clinical decision tool to calculate pretest probability of sentinel lymph node metastasis in primary cutaneous melanoma. Ann Surg Oncol. 2023;30:4321–8.
Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinforma. 2011;12:77.
Michaelson JS. Melanoma conditional outcome calculator LifeMath.net [Internet]. 2007 [cited 2024 Sep 30]. http://lifemath.net/cancer/melanoma/condsurv/index.php
Michaelson JS. Melanoma outcome calculator LifeMath.net [Internet]. 2007 [cited 2024 Sep 30]. http://lifemath.net/cancer/melanoma/outcome/index.php
Soong SJ, Ding S, Coit D, Balch CM, Gershenwald JE, Thompson JF, et al. Predicting survival outcome of localized melanoma: an electronic prediction tool based on the AJCC melanoma database. Ann Surg Oncol. 2010;17:2006–14.
Callender GG, Gershenwald JE, Egger ME, Scoggins CR, Martin RCG, Schacherer CW, et al. A novel and accurate computer model of melanoma prognosis for patients staged by sentinel lymph node biopsy: comparison with the American Joint Committee on cancer model. J Am Coll Surg. 2012;214:608–17.
Arora C, Kaur D, Lathwal A, Raghava GPS. Risk prediction in cutaneous melanoma patients from their clinico-pathological features: superiority of clinical data over gene expression data. Heliyon 2020;6:e04811.
El Sharouni MA, Ahmed T, Varey AHR, Elias SG, Witkamp AJ, Sigurdsson V, et al. Development and validation of nomograms to predict local, regional, and distant recurrence in patients with thin (T1) melanomas. J Clin Oncol. 2021;39:1243–52.
Liu W, Zhu Y, Lin C, Liu L, Li G. An online prognostic application for melanoma based on machine learning and statistics. J Plast Reconst Aesthet Surg. 2022;75:3853–8.
Cozzolino C, Buja A, Rugge M, Miatton A, Zorzi M, Vecchiato A, et al. Machine learning to predict overall short-term mortality in cutaneous melanoma. Discov Oncol. 2023;14:13.
Gimotty PA, Guerry DP, Ming ME, Elenitsas R, Xu X, Czerniecki B, et al. Thin primary cutaneous malignant melanoma: a prognostic tree for 10-year metastasis is more accurate than American Joint Committee on cancer staging. J Clin Oncol. 2004;22:3668–76.
Yang J, Pan Z, Zhou Q, Liu Q, Zhao F, Feng X, et al. Nomogram for predicting the survival of patients with malignant melanoma: a population analysis. Oncol Lett. 2019;18:3591–8.
Baade PD, Royston P, Youl PH, Weinstock MA, Geller A, Aitken JF. Prognostic survival model for people diagnosed with invasive cutaneous melanoma. BMC Cancer. 2015;15:27.
Lyth J, Hansson J, Ingvar C, Månsson-Brahme E, Naredi P, Stierner U, et al. Prognostic subclassifications of T1 cutaneous melanomas based on ulceration, tumour thickness and Clark’s level of invasion: results of a population-based study from the Swedish Melanoma Register. Br J Dermatol. 2013;168:779–86.
Clark WH, Elder DE, Guerry D, Braitman LE, Trock BJ, Schultz D, et al. Model predicting survival in stage I melanoma based on tumor progression. J Natl Cancer Inst. 1989;81:1893–904.
Schuchter L, Schultz D, Synnestvedt M, Trock B, Guerry D, Elder D, et al. A prognostic model for predicting 10-year survival in patients with primary melanoma. Ann Intern Med. 1996;125:369.
Aitchison TC, Sirel JM, Watt DC, MacKie RM, Group f. t. SM. Prognostic trees to aid prognosis in patients with cutaneous malignant melanoma. BMJ. 1995;311:1536–9.
Egger ME, Scoggins CR, McMasters KM. The Sunbelt melanoma trial. Ann Surg Oncol. 2020;27:28–34.
Verver D, van Klaveren D, Franke V, van Akkooi ACJ, Rutkowski P, Keilholz U, et al. Development and validation of a nomogram to predict recurrence and melanoma-specific mortality in patients with negative sentinel lymph nodes. Br J Surg. 2019;106:217–25.
Maurichi A, Miceli R, Camerini T, Mariani L, Patuzzo R, Ruggeri R, et al. Prediction of survival in patients with thin melanoma: results from a multi-institution study. J Clin Oncol. 2014;32:2479–85.
COCHRAN A, ELASHOFF D, MORTON D, ELASHOFF R. Individualized prognosis for melanoma patients. Hum Pathol. 2000;31:327–31.
Cadili A, Dabbs K, Scolyer RA, Brown PT, Thompson JF. Re-evaluation of a scoring system to predict nonsentinel-node metastasis and prognosis in melanoma patients. J Am Coll Surg. 2010;211:522–5.
Thomé SD, Loprinzi CL, Heldebrant MP. Determination of potential adjuvant systemic therapy benefits for patients with resected cutaneous melanomas. Mayo Clin Proc. 2002;77:913–7.
Khosrotehrani K, van der Ploeg APT, Siskind V, Hughes MC, Wright A, Thomas J, et al. Nomograms to predict recurrence and survival in stage IIIB and IIIC melanoma after therapeutic lymphadenectomy. Eur J Cancer. 2014;50:1301–9.
Wan G, Nguyen N, Liu F, DeSimone MS, Leung BW, Rajeh A, et al. Prediction of early-stage melanoma recurrence using clinical and histopathologic features. NPJ Precis Oncol 2022;6:79.
Gimotty PA, Elder DE, Fraker DL, Botbyl J, Sellers K, Elenitsas R, et al. Identification of high-risk patients among those diagnosed with thin cutaneous melanomas. J Clin Oncol. 2007;25:1129–34.
Van Calster B, McLernon DJ, van Smeden M, Wynants L, Steyerberg EW. Calibration: the Achilles heel of predictive analytics. BMC Med. 2019;17:230.
Isaksson K, Nielsen K, Mikiver R, Nieweg OE, Scolyer RA, Thompson JF, et al. Sentinel lymph node biopsy in patients with thin melanomas: Frequency and predictors of metastasis based on analysis of two large international cohorts. J Surg Oncol. 2018;118:599–605.
Grossman D, Okwundu N, Bartlett EK, Marchetti MA, Othus M, Coit DG, et al. Prognostic gene expression profiling in cutaneous melanoma: identifying the knowledge gaps and assessing the clinical benefit. JAMA Dermatol. 2020;156:1004–11.
Swetter SM, Tsao H, Bichakjian CK, Curiel-Lewandrowski C, Elder DE, Gershenwald JE, et al. Guidelines of care for the management of primary cutaneous melanoma. J Am Acad Dermatol. 2019;80:208–50.
National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology Version 2.2024 Melanoma: Cutaneous [Internet]. Pennsylvania; 2024 Apr [cited 2024 Sep 8]. https://www.nccn.org/professionals/physician_gls/pdf/cutaneous_melanoma.pdf
Chan WH, Tsao H. Consensus, controversy, and conversations about gene expression profiling in melanoma. JAMA Dermatol. 2020;156:949–51.
Gerami P, Cook RW, Wilkinson J, Russell MC, Dhillon N, Amaria RN, et al. Development of a prognostic genetic signature to predict the metastatic risk associated with cutaneous melanoma. Clin Cancer Res. 2015;21:175–83.
Amaral TMS, Hoffmann MC, Sinnberg T, Niessner H, Sülberg H, Eigentler TK, et al. Clinical validation of a prognostic 11-gene expression profiling score in prospectively collected FFPE tissue of patients with AJCC v8 stage II cutaneous melanoma. Eur J Cancer. 2020;125:38–45.
Acknowledgements
This work has been presented in part at the 20th European Association of Dermato-oncology (EADO) Congress, Paris, April 2024.
Funding
RNM is supported by a Royal College of Surgeons of England Research Fellowship & Freemasons Royal Arch Research Fellowship. AR is a Clinician Scientist supported by the Royal College of Surgeons of England and Cancer Research UK [C64667/A27958].
Author information
Authors and Affiliations
Contributions
RNM performed systematic search, initial review of all abstracts and performed all analysis, validation of selected tools and interpretation of results. AR conceived the idea for this manuscript and conducted secondary review of all abstracts. All authors have contributed to the manuscript writing and review.
Corresponding author
Ethics declarations
Competing interests
AR has received honoraria from the Alliance for Cancer Early Detection, and the British Association for Plastic, Reconstructive and Aesthetic Surgery. RNM has no conflict of interest to declare. AR is an Associate Editor for BJC Reports, he was not involved in any aspect of handling of this manuscript or any editorial decisions.
Ethics approval and consent to participate
This study was reviewed by the Cambridge University Hospitals NHS Trust EHR Research and Innovation (ERIN) Database Data Access Committee (Reference A096904). This study was performed in accordance with the Declaration of Helsinki.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Manton, R.N., Roshan, A. Systematic review of risk prediction tools for primary cutaneous melanoma outcomes and validation of sentinel lymph node positivity prediction in a UK tertiary cohort. BJC Rep 2, 86 (2024). https://doi.org/10.1038/s44276-024-00110-5
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s44276-024-00110-5