Abstract
The persistent evolution of SARS-CoV-2 highlights the crucial role of genomic surveillance in tracking emerging variants and guiding public health interventions. We performed whole-genome sequencing on 235 SARS-CoV-2-positive samples collected across Morocco between 2021 and 2024 to characterize viral evolution and variant dynamics. Our analysis revealed a temporal shift in variant prevalence that paralleled global trends: initial co-circulation of Alpha and Delta variants, followed by complete replacement by Omicron and its sub-lineages in 2022. Phylogenetic reconstruction identified key mutations associated with enhanced immune evasion and transmissibility, demonstrating the virus’s adaptive capacity. These findings underscore the critical importance of sustained genomic surveillance for monitoring viral evolution, informing vaccine and therapeutic strategies, and guiding public health responses. Our results advocate for expanded collaborative surveillance networks to proactively address threats from SARS-CoV-2 and other betacoronaviruses.
Similar content being viewed by others
Introduction
The Severe Acute Respiratory Syndrome Coronavirus 2 (SARS‑CoV‑2), first identified in December 2019, has caused an unprecedented global pandemic characterized by the emergence of Variants of Concern (VOCs) with enhanced transmissibility and immune evasion1,2. Variants such as Alpha (B.1.1.7), Delta (B.1.617.2), and Omicron (B.1.1.529) have significantly impacted pandemic dynamics, vaccine efficacy, and public health strategies3. Genomic surveillance has thus become indispensable for tracking viral evolution, detecting emerging lineages, and guiding timely interventions4.
Situated at the crossroads of Africa and Europe, Morocco—a nation of over 36 million people—serves as a critical hub for international travel and trade. The country reported its first confirmed SARS-CoV-2 case on March 2, 2020, in a Moroccan citizen returning from Italy5, followed by its first COVID-19-related death on March 10, 20206. Subsequent epidemic waves in Morocco mirrored global trends, each driven by the dominance of distinct VOCs7.
SARS-CoV-2, a positive-sense RNA betacoronavirus, possesses a high mutation rate due to its error-prone RNA-dependent RNA polymerase joint to a widespread human-to-human transmission capacity8. Its ~29.9 kb genome encodes four structural proteins (spike [S], envelope [E], membrane [M], and nucleocapsid [N]), non-structural proteins involved in replication and immune evasion, and accessory proteins (e.g., Orf3a, Orf6, Orf7a, Orf8) critical for pathogenesis9. The high mutation rate affecting SARS-CoV-2 genome has fueled the emergence of variants with altered virulence, transmissibility, and resistance to therapeutics10.
Between 2021 and 2024, Morocco’s SARS-CoV-2 genomic landscape evolved rapidly, transitioning from Alpha and Delta to Omicron and its sub-lineages, reflecting global phylogenetic patterns11. By early 2024, the emergence of the JN.1 Omicron sub-lineage further underscored the virus’s continuous adaptation12. This dynamic evolution highlights the necessity of sustained genomic surveillance to monitor mutations, predict variant trajectories, and inform public health policies13. International collaboration is essential to rapidly identify novel variants and assess their epidemiological implications14.
To date, all published studies on SARS-CoV-2 evolution in Morocco have relied exclusively on publicly shared sequences from the GISAID database. In contrast, this study presents the first real-time genomic surveillance of SARS-CoV-2 in Morocco, tracking the introduction, establishment, and transmission dynamics of circulating lineages throughout successive epidemic waves (2021–2024). By analyzing variant distribution and evolutionary trends from prospectively sequenced samples, we demonstrate how real-time genomic data can guide public health responses, optimize vaccine strategies, and mitigate emerging threats. Our findings establish a model for integrating rapid genomic surveillance into national outbreak preparedness frameworks.
Results
Demographic and clinical characteristics
In our cohort of 235 SARS-CoV-2-positive patients, a female predominance was observed, with 56.6% (n = 133) females and 43.4% (n = 102) males. The age distribution showed that 48.5% (n = 114) of patients were between 20–40 years old, 31.5% (n = 74) were aged 41–60 years, 14.5% (n = 34) were over 60 years, and 5.5% (n = 13) were under 20 years. The majority of cases were symptomatic (79.1%, n = 186), while 14.5% (n = 34) were asymptomatic; symptom status was unavailable for 6.4% (n = 15) of patients (Table 1).
A comparative analysis of SARS-CoV-2 variants based on demographic characteristics and disease presentation was performed (Fig. 1). No significant differences in age distribution were observed across the variants (Fig. 1a; overall p = 0.903): Alpha vs. Delta (p = 0.697), Alpha vs. Omicron (p = 0.935), and Delta vs. Omicron (p = 0.786). Notably, Delta cases were significantly more symptomatic than Alpha (p = 0.0001) or Omicron (p = 0.007) cases, whereas the difference in symptomatology between Alpha and Omicron was not significant (p = 0.220) (Fig. 1b). Regarding sex distribution, no significant differences were observed between Alpha and Delta (p = 0.060) or Delta and Omicron (p > 0.999); however, a significant difference was found between Alpha and Omicron (p = 0.043), and the overall comparison among all three variants was significant (p = 0.044) (Fig. 1c).
Variant distribution and temporal dynamics
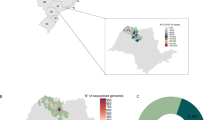
Sequencing data revealed distinct shifts in variant prevalence from 2021 to 2024 (Table 2). In 2021, Alpha (B.1.1.7, 37.5%, n = 42/112) and Delta (B.1.617.2, 39.2%, n = 44/112) dominated, with minor circulation of Eta (B.1.525, 1.8%, n = 2/112) and an already notable presence of early Omicron (B.1.1.529, 21.4%. n = 24/112). Omicron and its sub-lineages fully replaced all other variants, accounting for 100% of cases in 2022, 2023, and 2024, and maintained complete dominance throughout this period.
Figure 2 illustrates this transition: 2021 exhibited high variant diversity, BA.1.17 (Alpha variant), AY.33 (Delta sub-lineage) and BA.1 (Omicron sub-lineage), while 2022 saw consolidation into BA.2, BA.5.1, and BA.5.2 Omicron sub-lineages. In 2023, a single Omicron sub-lineage (JN.1.1) predominated, followed by the emergence of JN.1.45 (Omicron sub-lineage) in 2024, which rapidly displaced earlier variants.
Mutation accumulation and immune evasion
Delta (AY.112, AY.122) and Alpha variants in 2021 carried modest genetic changes (34–38 amino acid substitutions, 4–4 deletions). Omicron BA.1 (2022) showed marked complexity (44 substitutions, 16 deletions), the lineages derived from BA.2 and BA.5 showed a high mutational load, with BA.2.9.3 exhibiting 54 amino acid substitutions and BA.5.1 15 deletions, underlining their significant genetic divergence within Omicron. By 2024, JN.1.1 and JN.1.45 dominated, exhibiting unprecedented mutational loads (88–89 substitutions, 17 deletions) (Table 3), correlating with enhanced immune evasion and transmissibility.
Phylogeographic and evolutionary analysis
Phylogenetic reconstruction revealed distinct evolutionary patterns across the study period (Fig. 3a, b). In 2021, the Alpha (20I) and Delta (21A/J) variants showed limited global connectivity, with short phylogenetic branches indicative of predominantly localized transmission within Morocco. The emergence of Omicron in late 2021 and its sustained circulation through 2022–2023 marked a significant shift. Sub-lineages BA.1, BA.2, and BA.5 exhibited long branch lengths and high divergence in phylogenetic analyses, reflecting both rapid viral evolution and extensive international spread across multiple continents, including Europe, Sub-Saharan Africa, and Asia. By 2024, the JN.1.45 sub-lineage had formed a distinct phylogenetic clade, demonstrating adaptive radiation likely driven by immune selection pressures.
Migration pattern analysis (Fig. 3b) revealed multiple bidirectional viral exchanges between Morocco and other international regions, particularly during the Omicron wave.
Discussion
Our four-year genomic surveillance effort (2021–2024) has provided crucial insights into SARS-CoV-2’s evolutionary trajectory in Morocco, revealing three key phases of viral adaptation that mirror global patterns while highlighting regional specificities. The initial co-circulation of Alpha and Delta variants gradually gave way to complete Omicron dominance by 2022, a transition driven by the latter’s superior immune evasion capabilities2,15,16. This shift was particularly evident in the third phase characterized by the emergence of highly mutated JN.1.1 and JN.1.45 sub-lineages (2023–2024), which accumulated 88–89 amino acid substitutions and 17 deletions- including critical immune-escape mutations (E484K, N501Y) previously associated with reduced vaccine efficacy in Brazil and India17,18. However, as Japanese studies caution, this mutational burden may compromise viral fitness, suggesting an evolutionary trade-off between immune evasion and replicative efficiency13.
Phylogenetic reconstruction revealed important spatiotemporal patterns in variant spread. The 2022 diversification event, characterized by the simultaneous circulation of multiple Omicron sub-lineages (BA.2, BA.5), eventually resolved into dominant strains through competitive exclusion, a process well-documented in Danish populations, where natural and vaccine-induced immunity influenced the dynamics of BA.2 and BA.5 infections2,19. Notably, transmission mapping identified Morocco as both a recipient and exporter of variants, with JN.1 following an establishment pattern within regional clusters before international dissemination, a dynamic similar to that observed for Omicron sub-lineages BA.4 and BA.5 in West Africa, where these variants emerged and spread locally prior to global dissemination20.
Migration pattern analysis underscores Morocco’s role as a key regional transmission hub during the Omicron waves, characterized by significant bidirectional viral exchanges with neighboring regions. This connectivity pattern highlights Omicron’s distinct capacity for rapid transmission and global dispersal, emphasizing the critical need for enhanced regional surveillance systems and coordinated, cross-border public health strategies.
These findings carry significant implications for pandemic response strategies. First, regarding vaccine development, Israel’s demonstration of bivalent vaccines’ superior efficacy against Omicron sub-lineages21 strongly supports the need for regularly updated, antigenically matched boosters. Second, in surveillance systems, South Korea’s successful integration of Artificial Intelligence driven analytics with real-time sequencing22 provides a model for predictive outbreak monitoring that could be adapted to the Moroccan context. Third, for global health security, proven frameworks exist for coordinated variant containment, including Australia’s targeted border controls23 and South Africa’s regional surveillance network approach24. Moving forward, we propose three key recommendations: (1) establishing sentinel surveillance sites at major travel hubs (airports, harbors, bus terminals) to monitor variant importation patterns, (2) investing in adaptable platform vaccine technologies that can be rapidly modified for emerging variants, and (3) strengthening North African genomic data-sharing networks through World Health Organization-supported initiatives. This study underscores the dual value of national genomic surveillance programs - both for understanding local transmission dynamics and for contributing to global pandemic preparedness. The operational insights gained from tracking SARS-CoV-2 evolution in Morocco will undoubtedly inform more effective responses to future respiratory pathogen threats.
This study has some limitations. First, while all 235 genomes were successfully sequenced, the study was primarily descriptive in scope and therefore did not incorporate advanced phylogenetic modeling or formal phylogeographic inference. Second, mutation profiling focused solely on frequency and did not assess functional or structural implications. These limitations were explicitly considered during the interpretation of the results and do not diminish the significant surveillance value of the dataset.
This four-year genomic surveillance study (2021–2024) provides compelling evidence for the indispensable role of viral sequencing in pandemic management, documenting SARS-CoV-2’s remarkable evolutionary capacity and its profound public health consequences. Our findings reveal three critical insights: (1) the predictable yet rapid succession of variants from Alpha through Omicron sub-lineages, (2) region-specific patterns of variant establishment and spread that reflect Morocco’s position as a North African transmission hub, and (3) the growing mutational complexity of circulating strains, particularly the immune-evasive JN.1 lineage. When contextualized with global data, these observations underscore Omicron’s unprecedented adaptive advantage and the urgent need for coordinated international surveillance. We must emphasize a crucial point that justifies genomic surveillance of SARS-CoV-2. While the virulence of SARS-CoV-2 has tended to decrease during the pandemic, particularly due to a predilection of the latest variants for the upper levels of the respiratory tree, we have observed in parallel an increase in the transmissibility of the virus. However, there will always remain in each general population subset of people who are susceptible or predisposed to developing a severe form of SARS-CoV-2 infection due to their age, comorbidities, or lack of previous immunity. The high transmissibility of the virus means that these groups can be more easily and therefore more widely affected than with the initial variants. In these circumstances, a good public health policy must proceed, according to the means available, to regular surveillance of viral forms in circulation2,3,25. Moving forward, we identify three priority investments: sustained funding for genomic infrastructure, development of next-generation vaccine platforms, and establishment of regional data-sharing partnerships. These measures will be essential not only for managing SARS-CoV-2’s continuing evolution but also for building resilience against future pandemic threats.
Methods
Sample collection and SARS-CoV-2 testing
Between 2021 and 2024, 235 nasopharyngeal swabs were collected from SARS-CoV-2-positive patients across multiple healthcare facilities in Casablanca, including Ibn Rochd University Hospital Center and private clinics. All samples were confirmed as SARS-CoV-2-positive using the Speedy RT-PCR kit (PCL Inc., Republic of Korea) at Morizgo Laboratory, Casablanca. RNA extraction and sequencing were performed at the Viral Hepatitis Laboratory, Institut Pasteur of Morocco.
RNA extraction and library preparation
Viral RNA was extracted from nasopharyngeal samples using the QIAamp Viral RNA Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions. For library preparation, we employed the Illumina COVIDSeq Assay kit (Illumina Inc., USA) following the standardized protocol. The process began with reverse transcription of viral RNA into complementary DNA (cDNA). The SARS-CoV-2 genome was then amplified through PCR using ARTIC v4 primer pools 1 and 2, which are designed to generate overlapping amplicons covering the complete viral genome. The resulting amplicons underwent fragmentation and were subsequently tagged with dual-index adapters (IDT for Illumina UD Set 1–4) using Enrichment BLT (EBLTS) chemistry. Finally, the prepared libraries were quantified using a Qubit 3.0 fluorometer (Invitrogen Inc.) and normalized to a concentration of 4 nM to ensure equal representation of all samples in the pooled library prior to sequencing.
Whole genome SARS-CoV-2 sequencing
Libraries were sequenced on an Illumina NextSeq 2000 platform using a P2 300-cycle kit (2× 150 bp paired-end reads). A loading concentration of 1000 pM ensured >1000× coverage per sample.
Consensus sequence creation and phylogenetic analysis
The bioinformatic pipeline began with consensus sequence generation, where raw sequencing reads were aligned to the SARS-CoV-2 reference genome (NC_045512) using the Illumina DRAGEN COVID Lineage pipeline (version 4) on the BaseSpace platform. After quality control and consensus sequence generation, a total of 235 high-quality genomes were retained for downstream analysis. These sequences have been submitted to the GISAID database, with accession numbers listed in Supplementary Table 1.
For lineage classification, the resulting consensus sequences were subsequently analyzed using the NextClade web tool (https://clades.nextstrain.org/), which assigned Pango lineages and clades based on characteristic mutations. To investigate evolutionary relationships, we performed phylogenetic analysis using the Nextstrain command-line interface (CLI). This analysis incorporated both the newly generated sequences from our Moroccan cohort and a globally representative dataset obtained from Nextstrain’s Africa-focused repository (https://nextstrain.org/ncov/gisaid/africa/all-time), enabling comparative assessment of viral evolution within both regional and global contexts. Among the 235 sequences, only 27 met the strict quality criteria defined by the Nextstrain workflow (e.g., genome completeness, low proportion of ambiguous bases, and high alignment quality) and were thus retained in the final phylogenetic tree.
Statistical analysis
Statistical analyses were performed using GraphPad Prism v.9.0. Age, treated as a continuous variable, was compared between SARS-CoV-2 variant groups (Alpha, Delta, Omicron) using one-way ANOVA. Categorical variables, such as sex and symptom status (symptomatic/asymptomatic), were analyzed using the chi-square test. For variables with ordinal characteristics or to assess trends across ordered variant groups, the Chi-square test for trend was applied.
A p-value < 0.05 was considered statistically significant.
Data availability
All data presented in this study are available upon reasonable request from the corresponding author. Additionally, the 235 SARS-CoV-2 genome sequences generated have been deposited in the GISAID database, with the full list of accession numbers provided in Supplementary Table 1.
Code availability
The custom scripts used to perform sequence alignment and variant analysis in this study were run using NextClade. The tool is publicly available at https://clades.nextstrain.org. no additional custom code was used.
References
Huang, C. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020).
Viana, R. et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 603, 679–686 (2022).
Carabelli, A. M. et al. SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat. Rev. Microbiol. 21, 162–177 (2023).
Tosta, S. et al. Global SARS-CoV-2 genomic surveillance: what we have learned (so far). Infect. Genet. Evol. 108, 105405 (2023).
Kahkahi, R. E., Moustaine, M., Hafidi, M., Zouhair, R. & Errakhi, R. Coronavirus disease (COVID-19) in Morocco: situation update and proposed remedial measures. Germs 10, 129–131 (2020).
Senhaji, M. Le Maroc annonce le premier cas de décès du nouveau coronavirus, https://lematin.ma/express/2020/maroc-annonce-premier-cas-deces-nouveau-coronavirus/333247.html (2020).
Djorwé, S. et al. Epidemiology, clinical characteristics and risk factors of coronavirus disease 2019 (COVID-19) in Casablanca. Access Microbiol. 5, acmi000400 (2023).
Naqvi, A. A. T. et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta Mol. Basis Dis. 1866, 165878 (2020).
Brant, A. C., Tian, W., Majerciak, V., Yang, W. & Zheng, Z. M. SARS-CoV-2: from its discovery to genome structure, transcription, and replication. Cell Biosci. 11, 136 (2021).
El Mazouri, S. et al. Genetic diversity and evolutionary dynamics of the Omicron variant of SARS-CoV-2 in Morocco. Pathog. Glob. Health 118, 241–252 (2024).
Bouddahab, O. et al. Report of SARS-CoV-2 JN.1 variant in Morocco. Microbiol. Resour. Announc. 13, e0055924 (2024).
Ghammaz, H. et al. Genomic evolution of SARS-CoV-2 in Morocco: insights from whole genome sequences collected from 2020 to 2024. Virus Res. 353, 199530 (2025).
Liu, W. et al. Evolution of the SARS-CoV-2 Omicron variants: genetic impact on viral fitness. Viruses 16, https://doi.org/10.3390/v16020184 (2024).
Reuschl, A. K. et al. Evolution of enhanced innate immune suppression by SARS-CoV-2 Omicron subvariants. Nat. Microbiol. 9, 451–463 (2024).
Galasso, V. et al. Gender differences in COVID-19 attitudes and behavior: panel evidence from eight countries. Proc. Natl. Acad. Sci. USA 117, 27285–27291 (2020).
Andrews, N. et al. Covid-19 vaccine effectiveness against the Omicron (B.1.1.529) variant. N. Engl. J. Med. 386, 1532–1546 (2022).
Yadav, P. D. et al. Neutralization of Beta and Delta variant with sera of COVID-19 recovered cases and vaccinees of inactivated COVID-19 vaccine BBV152/Covaxin. J. Travel Med. 28, https://doi.org/10.1093/jtm/taab104 (2021).
Adamoski, D. et al. SARS-CoV-2 Delta and Omicron variants surge in Curitiba, Southern Brazil, and its impact on overall COVID-19 lethality. Viruses 14, https://doi.org/10.3390/v14040809 (2022).
Hansen, C. H. et al. Risk of reinfection, vaccine protection, and severity of infection with the BA.5 omicron subvariant: a nation-wide population-based study in Denmark. Lancet Infect. Dis. 23, 167–176 (2023).
Sow, M. S. et al. Genomic characterization of SARS-CoV-2 in Guinea, West Africa. PLoS One. 19, e0299082 (2024).
Feikin, D. R. et al. Assessing COVID-19 vaccine effectiveness against Omicron subvariants: report from a meeting of the World Health Organization. Vaccine 41, 2329–2338 (2023).
Kim, I. H. et al. Genomic epidemiology of SARS-CoV-2 variants in South Korea between January 2020 and February 2023. Virology 587, 109869 (2023).
Hanly, M. J. et al. The impact of re-opening the international border on COVID-19 hospitalisations in Australia: a modelling study. Med. J. Aust. 216, 39–42 (2022).
Tegally, H. et al. The evolving SARS-CoV-2 epidemic in Africa: insights from rapidly expanding genomic surveillance. Science 378, eabq5358 (2022).
Vo, A. D. et al. Factors associated with severe COVID-19 among vaccinated adults treated in US Veterans Affairs Hospitals. JAMA Netw. Open 5, e2240037 (2022).
Acknowledgements
This study was supported by a ACIP (Action Concertées Interpasteuriennes) grant (ACIP n°505) from Institut Pasteur (Paris) and Africa Centres for Disease Control and Prevention (Africa CDC).
Author information
Authors and Affiliations
Contributions
S.E., M.S., A.B., M.L., and A.M. conceived and designed the study. O.B., S.A., R.N., A.E., O.L., A.A., H.B., A.O. participated in the sample collection and methodology. H.C. performed the bioinformatical analysis. O.B. drafted the manuscript. S.E., M.S., A.B., A.E., M.L., H.C. and P.P. reviewed and edited the manuscript. S.E. acquired the funding. M.L. and S.E. participated in the supervision. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
This study was conducted in accordance with the Declaration of Helsinki. The study was approved by the Ethics Committee of Ibn Rochd Hospital in Casablanca (2020–2022) and the Ethics Committee of the Faculty of Medicine of Rabat (Approval Number: S/22). Informed consent was obtained from all participants involved in the study.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Bouddahab, O., Aqillouch, S., Charoute, H. et al. Genomic surveillance in Morocco tracks SARS-CoV-2 variant shift from Alpha to Omicron sublineage JN1. npj Viruses 3, 63 (2025). https://doi.org/10.1038/s44298-025-00145-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s44298-025-00145-6