Abstract
This review discusses how temperature signals are integrated into the Arabidopsis circadian clock and proposes Temperature-Dependent Alternative Splicing (TDAS) of core clock genes as an additional mechanism to adapt the circadian system to temperature changes. We present examples of TDAS in a range of organisms, pointing towards a conserved mechanism that enables temperature adaptation.
Similar content being viewed by others
Temperature affects every biological process
Global warming is threatening ecosystems across the world and reshaping the balance of life. These impacts are far-reaching, ranging from declining marine biodiversity and rising animal extinction risks to reduced agricultural productivity1,2. Among the most vulnerable species are plants, whose sessile nature leaves them especially exposed to shifting climates. Increasing temperatures not only disrupt plant growth and development but also reduce exposure to low temperatures necessary for reproductive processes3. The impacts of climate change are evident at the molecular level, where temperature influences all aspects of protein biosynthesis. For example, temperature has been shown to cause differential protein expression in plants4,5,6 and to affect protein stability by disrupting protein folding and ligand binding7,8. These environmental changes make it imperative that we understand how plants perceive and respond to temperature, especially with regard to the circadian system that provides context to these environmental cues.
Circadian systems are a ubiquitous concept found in a wide range of organisms
The Earth’s daily rotation has ensured that the majority of species possess an endogenous timekeeper which controls physical and behavioural changes in a repeating 24-h cycle9,10. This circadian system (or clock) is an intrinsic, molecular time keeping mechanism that coordinates cellular activity with daily environmental patterns, facilitating adaptation to changing environments11. Circadian rhythms were first observed in plants through leaf movements under constant light and were later linked to specific genes in Drosophila melanogaster, with their mammalian homologues revealing core clock components12,13,14. Although circadian systems differ between kingdoms and phyla, the genetic aspect of the circadian system has converged over evolutionary time to operate via interlinked Transcription-Translation Feedback Loops (TTFL) that drive rhythmic gene expression and circadian gating of biological events12,13,14,15. Metabolic circadian mechanisms have also been described; in the cyanobacterium Synechococcus, a post-translational circadian clock coordinates gene expression for photosynthesis during the day and nitrogen fixation at night via rhythmic phosphorylation of Kai proteins16. In eukaryotes, non-transcriptional oscillation in the form of circadian-driven redox cycles has been found in elements of the antioxidant system; these cycles are entrainable, offering an alternative or additional mechanism alongside the TTFL circadian system17,18,19.
The resetting of the circadian system to environmental cues ensures entrainment with dawn and dusk throughout the year while also allowing temperature compensation to increase the reliability of the biological timer as temperatures vary across daily and seasonal timescales. Reliable circadian timing enables emergent properties, including the external coincidence control of flowering and circadian gating, whereby molecular responses are modulated dependent upon time-of-day9. In this review, we assess how the Arabidopsis circadian system adapts to persistent temperature change and consider the consequences of global warming to plant circadian timing.
The Arabidopsis circadian clock can be divided into three notional sub-groups
Building upon the foundational observations of de Mairan, studies have described the botanic molecular oscillator to fit the classic TTFL model, exemplified in the model plant Arabidopsis thaliana (Arabidopsis, Fig. 1)14,20,21,22. Conceptually, the Arabidopsis circadian clock can be split into three temporally distinct groups: the Evening Complex (EC), the PSEUDO-RESPONSE REGULATORS (PRRs), and MYB-like transcription factors. The EC was the first identified circadian complex23,24: EARLY FLOWERING3 (ELF3) acts as a scaffold, interacting with ELF4 via its central domain and with LUX ARRHYTHMO (LUX) via its C-terminus23,24,25. ELF3 also binds phytochrome B (phyB), linking the circadian clock to light signalling25. Chromatin immunoprecipitation (ChIP) studies show that ELF3 and ELF4 bind more frequently to the promoters of genes like PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) and PIF5 in the presence of LUX23,26. The interactions between ELF3, ELF4, and LUX are essential for the ECs' regulatory function27.
The plant circadian clock comprises at least three families of transcription factors that reciprocally regulate promoter activity, ultimately generating a biological timer with an approximate 24-h period. CIRCADIAN CLOCK ASSOCIATED1 (CCA1) acts with LATE ELONGATED HYPOCOTYL (LHY) to repress circadian gene expression immediately after dawn, whereas PSEUDO-RESPONSE REGULATORS (PRRs) repress circadian gene expression during the day. The Evening Complex, comprising LUX ARRHYTHMO, EARLY FLOWERING3 (ELF3), and ELF4 represses gene expression overnight to complete the sequential repression of circadian gene expression throughout the 24 h cycle. REVEILLE8 (RVE8) works as part of a complex to activate circadian gene expression in the morning.
The MYB-like family in Arabidopsis includes core clock components such as CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), as well as the less-characterised REVEILLE (RVE) transcription factors (RVE1-8), all sharing a conserved MYB-like DNA-binding motif (Fig. 128,29,30,31). CCA1 and LHY exhibit peak expression at dawn and ensure that the clock is synchronised with dawn32. Although disruption of RVE1 function does not alter circadian timing, the other RVE genes have each been described to regulate timekeeping31,33,34,35,36. Like the EC, RVEs interact with various proteins to expand their molecular functions. For example, RVE8 associates with other RVEs (RVE4, 5, 6), TOLERANT TO CHILLING AND FREEZING1 (TCF1), a cold response regulator, and REGULATOR OF CHROMOSOME CONDENSATION1-LIKE (RCC1L)37. In the morning, RVE8 forms a complex with LIGHT NIGHT-INDUCIBLE AND CLOCK-REGULATED 1 (LNK1) and LNK2, transcriptional coactivators that influence gene expression (Fig. 134,37). In the evening, COLD-REGULATED 27 (COR27) and COR28 join this complex to suppress RVE8 activity by regulating its abundance. Both CCA1 and RVE8 modulate TOC1 expression via histone H3 modifications, with RVE8 promoting acetylation (associated with activation) and CCA1 inducing deacetylation (repression)38,39.
The PRR family is comprised of PRR5, PRR7, PRR9, and TIMING OF CAB EXPRESSION1 (TOC1), which function as a series of repressors expressed sequentially from mid-morning to dusk, following the peak of CCA1 and LHY at dawn (Fig. 140,41,42,43). They play a key role in negative feedback within the clock, as TOC1 indirectly promotes CCA1 by reducing its repressor, whereas CCA1 and LHY inhibit TOC1 expression to form a feedback loop44. Like the other key clock families, PRRs form protein complexes to regulate gene expression. For instance, PRR5, PRR7, and PRR9 bind the promoters of CCA1 and LHY via their C-terminal CONSTANTS, CONSTANS-LIKE, and TOC1 (CCT) domains, recruiting TOPLESS (TPL) proteins that help histone deacetylases repress CCA1 and TOC1 transcription41. This highlights the role of PRRs in maintaining clock stability and coordinating interactions between distinct components of the clock network.
Temperature integration within the circadian system
Understanding how plants perceive temperature signals is critical for manipulating plant growth within our evolving environment. While a consensus exists regarding the broad aspects of light integration45, the identification of thermosensors is more complex as temperature changes generally influence all chemical reactions and many subcellular components. In alignment with photoreceptor definitions, we consider a biological thermosensor must directly translate the temperature stimulus into a biological signal via alteration of its own structure, activity, or interaction with other molecular components, triggering a downstream response46. In this context, ELF3, PHYTOCHROME INTERACTION FACTOR 7 (PIF7) mRNA, and THERMO-WITH ABA-RESPONSE (TWA1), have been identified as thermosensors47,48,49. Additionally, photoreceptors, including phytochromes and phototropins have temperature-dependent activity that enables integration of light and temperature signals50,51,52.
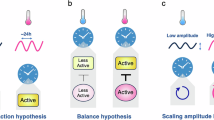
Despite decades of research within circadian biology, the field continues to describe mechanisms by which light and temperature signals are integrated with the endogenous timing system (Fig. 2). Recent evidence highlights the role of the EC to integrate light and temperature signals. ELF3 undergoes conformational changes in response to temperature47. Increasing temperatures promote the accumulation of ELF3 into nuclear bodies via phase separation, preventing ELF3 from interacting with signalling partners47. The sequestration of ELF3 could enable integration of temperature information into the clock since the association of ELF3 with target gene promoters is temperature-dependent53,54. A temperature-dependent role for the EC is supported by the rescue of the lux arhythmic circadian phenotype at lower temperatures55. Alongside temperature signalling, phyB biological activity is also affected by temperature, with the half-life of the Pr form of phytochrome reduced as temperatures increase, allowing temperature to influence photoreceptor activity as part of the Evening Complex25,50,52,56.
The plant circadian clock comprises at least three families of transcription factors that reciprocally regulate promoter activity, ultimately generating a biological timer with an approximate 24-h period. Distinct mechanisms of temperature-modulated regulation have been characterised for certain genes within the circadian clock, although these are not mutually exclusive, and doubtless additional regulations will be elucidated over time.
Additional examples of the integration of light and temperature signalling arise from the regulation of PIF proteins. Phytochrome thermal reversion ensures PIF accumulation is temperature dependent50,52,57. PIF3 interacts with C-REPEAT BINDING FACTOR3 (CBF3) to stabilise phyB in response to cold temperatures, ultimately leading to increased freezing tolerance58. PIF4 and PIF5 have roles in heat stress responses, with PIF4 promoting growth and transition to flowering but decreasing immunity59,60,61. Circadian gating has also been linked to PIF4, suggesting that PIF4 thermosensitivity could contribute to the adaptation of the circadian system to temperature62. Furthermore, PIF7 translation is temperature sensitive, emphasising the pervasive effects of temperature upon biology49,63.
Beyond these proteins, numerous other temperature-dependent effects have been noted within the circadian system. Temperature-dependent phosphorylation of CCA1 by CASEIN KINASE2 alters circadian timing64, and temperature-dependent accumulation of circadian transcription factors and their interacting proteins has been reported. For example, RVE5 and RVE7 both accumulate at higher temperatures to alter gene expression65,66, while PRR7 and PRR9 contribute to temperature compensation67. Since TWA1 interacts with TOPLESS in a temperature-dependent manner, it will be interesting to determine whether TWA1 also contributes to temperature-dependent circadian gene expression via interactions with PRR proteins41,48. However, it remains unclear how temperature affects protein accumulation and activity in these cases.
Alternative Splicing as a global thermometer
Alternative Splicing (AS), the process whereby a single gene generates multiple transcript isoforms through diverse combinations of exons and introns, contributes to proteomic diversity68,69. Isoforms exhibit diverse functionalities, with some contributing to distinct cellular processes while others may result in non-functional proteins due to the presence of premature termination codons, leading to RNA degradation via nonsense mediated decay (NMD)68,69. The presence of various isoforms alters the transcriptome and proteome, leading to a specific temperature-dependent response and enabling global changes in AS to provide thermosensitivity6,70,71. Temperature-dependent AS has been reported in commercially important crops such as tomato, rice, sugarcane, and barley, as well as Arabidopsis71,72,73,74,75,76,77. Indeed, this phenomenon is not restricted to plants but can also be found in yeast, fish, flies, and even mammals, pointing towards an evolutionary benefit of AS transmission of temperature signals70,78,79,80,81,82.
AS is facilitated by the spliceosome, a complex macromolecule composed of small nuclear ribonucleoproteins (snRNPs) designated U1-2, U4-6, and multiple ancillary proteins and RNA molecules83. Splicing factor recruitment by cis-regulatory sequences situated in exons and introns governs the selection of splice sites, leading to either activation or inhibition of splicing83. Since RNA structure is dependent on transcription rate, it appears plausible that interactions between spliceosome and ribosome activities enable temperature-dependent AS49,84,85,86,87. Alternatively, RNA riboswitches have been reported, with RNA structure changing in a temperature-dependent manner87,88. In either case, temperature-induced conformational changes within the spliceosome or RNA can modify the accessibility of splice sites, changing splicing outcomes89.
Temperature-dependent AS of clock components
Several core components of the circadian clock, including CCA1, LHY, PRR7, TOC1, and ELF3, exhibit temperature-dependent AS, although the functional consequences of these alternative isoforms are mostly uncharacterised71,73,90,91. CCA1 accumulates primarily in two isoforms: the full-length CCA1α and the truncated CCA1β, which are observed in comparable proportions at ambient temperature; however, a shift to CCA1β has been noted at elevated temperatures92,93. Whereas both isoforms compete for binding partners, only CCA1α encodes a DNA-binding protein, as it contains the MYB-like domain94. The presence of low temperatures affects AS of CCA1, promoting the accumulation of CCA1α71,94. Similarly, increases in canonical LHY transcript accumulation are observed at 12 °C compared to 20 °C, although this aligns with an increase in total LHY transcript levels. LHY transcript isoforms, including a 5′ UTR intronic retention, are elevated at 12 °C, although the biological function of this isoform compared to other isoforms remains to be determined95.
Temperature-dependent AS has not only been shown for CCA1 and LHY, but also for RVE2. RVE2 predominantly accumulates as an intron-retaining isoform at 20 °C, lacking translation of the functional MYB domains due to a premature stop codon71. Upon chilling, the canonical isoform becomes the most abundant. RVE2 has been demonstrated to influence the periodicity of the circadian clock and cold responses through the CBF signalling pathway71. Interestingly, RVE2 contributes to flowering time, although the temperature-dependent role has not been appreciated until recently71,96. It is possible that temperature-dependent AS of RVE2 provides a signalling mechanism enabling ambient temperature to ‘re-program’ crucial developmental decisions.
Future work prospects and conclusions
Recent discoveries are finally revealing specific mechanisms that define how plants respond to temperature and how these environmental cues are integrated into regulatory systems such as the circadian system. Temperature cues modulate the activity of circadian transcription factors to provide time-of-day context to environmental responses. However, the contribution of AS to circadian regulation has been limited by the lack of robust data detailing transcript isoform accumulation. Our review highlights the conserved mechanism of AS, which transmits temperature information and reprograms the transcriptome accordingly to the environmental changes, and theorises on the underlying causes that could promote AS in response to temperature. Enhanced comprehension of plant temperature signal perception will enable the manipulation of plant responses to the evolving environmental conditions driven by climate change.
Data availability
No datasets were generated or analysed during the current study.
References
Root, T. L. et al. Fingerprints of global warming on wild animals and plants. Nature 421, 57–60 (2003).
Ortiz-Bobea, A., Ault, T. R., Carrillo, C. M., Chambers, R. G. & Lobell, D. B. Anthropogenic climate change has slowed global agricultural productivity growth. Nat. Clim. Change 11, 306–312 (2021).
Penfield, S., Warner, S. & Wilkinson, L. Molecular responses to chilling in a warming climate and their impacts on plant reproductive development and yield. J. Exp. Bot. 19, erab375 (2021).
Provart, N. J. et al. Gene expression phenotypes of Arabidopsis associated with sensitivity to low temperatures. Plant Physiol. 132, 893–906 (2003).
Song, J., Liu, Q., Hu, B. & Wu, W. Comparative transcriptome profiling of Arabidopsis Col-0 in responses to heat stress under different light conditions. Plant Growth Regul. 79, 209–218 (2016).
Calixto, C. P. G. et al. Rapid and dynamic alternative splicing impacts the arabidopsis cold response transcriptome. Plant Cell Online 30, 1424–1444 (2018).
Hamborg, L. et al. Global analysis of protein stability by temperature and chemical denaturation. Anal. Biochem. 605, 113863 (2020).
Somero, G. N. Proteins and temperature. Annu. Rev. Physiol. 57, 43–68 (1995).
Xu, X. et al. Circadian clock in plants: linking timing to fitness. J. Integr. Plant Biol. 64, 792–811 (2022).
Folta, K. M. & Kaufman, L. S. Phototropin 1 is required for high-fluence blue-light-mediated mRNA destabilization. Plant Mol. Biol. 51, 609–618 (2003).
Millar, A. J. The intracellular dynamics of circadian clocks reach for the light of ecology and evolution. Annu. Rev. Plant Biol. 67, 595–618 (2016).
Dubowy, C. & Sehgal, A. Circadian rhythms and sleep in drosophila melanogaster. Genetics 205, 1373–1397 (2017).
Emery, P., Klarsfeld, A., Stanewsky, R. & Shafer, O. T. Sensitive timing: a reappraisal of chronobiology’s foundational texts. J. Biol. Rhythms 38, 245–258 (2023).
Harmer, S. L. The time machine: feedback loops, post-transcriptional regulation, and environmental integration in the plant circadian oscillator. Plant J. 122, e70275 (2025).
Paajanen, P., Kimmey, J. M. & Dodd, A. N. Circadian gating: concepts, processes, and opportunities. Philos. Trans. R. Soc. Lond. B Biol. Sci. 380, 20230346 (2025).
Golden, S. S. Principles of rhythmicity emerging from cyanobacteria. Eur. J. Neurosci. 51, 13–18 (2020).
Edgar, R. S. et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature 485, 459–464 (2012).
O’Neill, J. S. et al. Circadian rhythms persist without transcription in a eukaryote. Nature 469, 554–558 (2011).
Wulund, L. & Reddy, A. B. A brief history of circadian time: the emergence of redox oscillations as a novel component of biological rhythms. Perspect. Sci. 6, 27–37 (2015).
Kennis, J. T. et al. Primary reactions of the LOV2 domain of phototropin, a plant blue-light photoreceptor. Biochemistry 42, 3385–3392 (2003).
McClung, C. R. Plant circadian rhythms. Plant Cell 18, 792–803 (2006).
Gendron, J. M. & Staiger, D. New horizons in plant photoperiodism. Ann. Rev. Plant Biol. 74, 481–509 (2023).
Nusinow, D. A. et al. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475, 398–402 (2011).
Hazen, S. P. et al. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. PNAS. 102, 10387–10392 (2005).
Huang, H. et al. Identification of evening complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Mol. Cell. Proteom. 15, 201–217 (2016).
Chow, B. Y., Helfer, A., Nusinow, D. A. & Kay, S. A. ELF3 recruitment to the PRR9 promoter requires other Evening Complex members in the Arabidopsis circadian clock. Plant Signal Behav 7, 170–173 (2012).
Huang, H. & Nusinow, D. A. Into the evening: complex interactions in the Arabidopsis circadian clock. Trends Genet. 32, 674–686 (2016).
Pratyusha, D. S. & Sarada, D. V. L. MYB transcription factors-master regulators of phenylpropanoid biosynthesis and diverse developmental and stress responses. Plant Cell Rep. 41, 2245–2260 (2022).
Wang, Z. & Tobin, E. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207–1217 (1998).
Green, R. M. & Tobin, E. M. Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. PNAS. 96, 4176–4179 (1999).
Gray, J. A., Shalit-Kaneh, A., Chu, D. N., Hsu, P. Y. & Harmer, S. L. The REVEILLE clock genes inhibit growth of juvenile and adult plants by control of cell size. Plant Physiol. 173, 2308–2322 (2017).
Alabadí, D., Yanovsky, M. J., Más, P., Harmer, S. L. & Kay, S. A. Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr. Biol. 12, 757–761 (2002).
Rawat, R. et al. REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. PNAS. 106, 16883–16888 (2009).
Lu, S. X., Knowles, S. M., Andronis, C., Ong, M. S. & Tobin, E. M. Circadian clock-associated1 and late elongated hypocotyl function synergistically in the circadian clock of Arabidopsis. Plant Physiol. 150, 834–843 (2009).
Kidokoro, S. et al. Posttranslational regulation of multiple clock-related transcription factors triggers cold-inducible gene expression in Arabidopsis. PNAS. 118, e2021048118 (2021).
Fogelmark, K. & Troein, C. Rethinking transcriptional activation in the Arabidopsis circadian clock. PLoS Comput. Biol. 10, e1003705 (2014).
Sorkin, M. L. et al. COLD REGULATED GENE 27 and 28 antagonize the transcriptional activity of the RVE8/LNK1/LNK2 circadian complex. Plant Physiol. 192, 2436–2456 (2023).
Farinas, B. & Mas, P. Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J. 66, 318–329 (2011).
Hung, F.-Y. et al. The Arabidopsis LDL1/2-HDA6 histone modification complex is functionally associated with CCA1/LHY in regulation of circadian clock genes. Nucleic Acids Res. 46, 10669–10681 (2018).
Nakamichi, N. et al. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22, 594–605 (2010).
Wang, L., Kim, J. & Somers, D. E. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. PNAS. 110, 761–766 (2013).
Laosuntisuk, K., Elorriaga, E. & Doherty, C. J. The game of timing: circadian rhythms intersect with changing environments. Annu. Rev. Plant Biol. 74, 511–538 (2023).
Farré, E. M. & Liu, T. The PRR family of transcriptional regulators reflects the complexity and evolution of plant circadian clocks. Curr. Opin. Plant Biol. 16, 621–629 (2013).
Alabadi, D. et al. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293, 880–883 (2001).
Sanchez, S. E., Rugnone, M. L. & Kay, S. A. Light perception: a matter of time. Mol. Plant 13, 363–385 (2020).
Vu, L. D., Gevaert, K. & De Smet, I. Feeling the heat: searching for plant thermosensors. Trends Plant Sci. 24, 210–219 (2019).
Jung, J. H. et al. A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature 585, 256–260 (2020).
Bohn, L. et al. The temperature sensor TWA1 is required for thermotolerance in Arabidopsis. Nature 629, 1126–1132 (2024).
Chung, B. Y. W. et al. An RNA thermoswitch regulates daytime growth in Arabidopsis. Nat. Plants 6, 522–532 (2020).
Jung, J.-H. et al. Phytochromes function as thermosensors in Arabidopsis. Science 354, 886–889 (2016).
Fujii, Y. et al. Phototropin perceives temperature based on the lifetime of its photoactivated state. Proc. Natl. Acad. Sci. USA 114, 9206–9211 (2017).
Legris, M. et al. Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354, 897–900 (2016).
Ezer, D. et al. The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat. Plants 3, 17087 (2017).
Box, M. S. et al. ELF3 controls thermoresponsive growth in Arabidopsis. Curr. Biol. 25, 194–199 (2015).
Jones, M. A., Morohashi, K., Grotewold, E. & Harmer, S. L. Arabidopsis JMJD5/JMJ30 acts independently of LUX ARRHYTHMO within the plant circadian clock to enable temperature compensation. Front. Plant Sci. 10, 57 (2019).
Rockwell, N. C., Su, Y.-S. & Lagarias, J. C. Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57, 837–858 (2006).
Pham, V. N., Kathare, P. K. & Huq, E. Phytochromes and phytochrome interacting factors. Plant Physiol. 176, 1025–1038 (2018).
Jiang, B. et al. Cold-induced CBF-PIF3 interaction enhances freezing tolerance by stabilizing the phyb thermosensor in Arabidopsis. Mol. Plant 13, 894–906 (2020).
Proveniers, M. C. G. & van Zanten, M. High temperature acclimation through PIF4 signaling. Trends Plant Sci. 18, 59–64 (2013).
Gangappa, S. N., Berriri, S. & Kumar, S. V. PIF4 coordinates thermosensory growth and immunity in Arabidopsis. Curr. Biol.27, 243–249 (2017).
Li, N., Bo, C., Zhang, Y. & Wang, L. Phytochrome interacting factors PIF4 and PIF5 promote heat stress induced leaf senescence in Arabidopsis. J. Exp. Bot. 72, 4577–4589 (2021).
Zhu, J.-Y., Oh, E., Wang, T. & Wang, Z.-Y. TOC1-PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nat. Commun. 7, 13692 (2016).
Fiorucci, A.-S. et al. PHYTOCHROME INTERACTING FACTOR 7 is important for early responses to elevated temperature in Arabidopsis seedlings. N. Phytol. 226, 50–58 (2020).
Portolés, S. & Más, P. The functional interplay between protein kinase CK2 and CCA1 transcriptional activity is essential for clock temperature compensation in Arabidopsis. PLoS Genet. 6, e1001201 (2010).
Li, W. et al. A competition-attenuation mechanism modulates thermoresponsive growth at warm temperatures in plants. N. Phytol. 237, 177–191 (2023).
Tian, Y. Y. et al. REVEILLE 7 inhibits the expression of the circadian clock gene EARLY FLOWERING 4 to fine-tune hypocotyl growth in response to warm temperatures. J. Integr. Plant Biol. 64, 1310–1324 (2022).
Salomé, P. A., Weigel, D. & McClung, C. R. The role of the Arabidopsis morning loop components CCA1, LHY, PRR7, and PRR9 in temperature compensation. Plant Cell 22, 3650–3661 (2010).
Dikaya, V. et al. Insights into the role of alternative splicing in plant temperature response. J. Exp. Bot. 72, 7384–7403 (2021).
Martín, G., Márquez, Y., Mantica, F., Duque, P. & Irimia, M. Alternative splicing landscapes in Arabidopsis thaliana across tissues and stress conditions highlight major functional differences with animals. Genome Biol. 22, 35 (2021).
Haltenhof, T., Preußner, M. & Heyd, F. Thermoregulated transcriptomics: the molecular basis and biological significance of temperature-dependent alternative splicing. Biochem. J. 481, 999–1013 (2024).
James, A. B. et al. REVEILLE2 thermosensitive splicing: a molecular basis for the integration of nocturnal temperature information by the Arabidopsis circadian clock. N. Phytol. 241, 283–297 (2024).
Calixto, C. P. G., Simpson, C. G., Waugh, R. & Brown, J. W. S. Alternative splicing of barley clock genes in response to low temperature. PLoS ONE. 11, e0168028 (2016).
Dantas, L. L. B. et al. Alternative splicing of circadian clock genes correlates with temperature in field-grown sugarcane. Front. Plant Sci. 10, 1614 (2019).
Vitoriano, C. B. & Calixto, C. P. G. Reading between the lines: RNA-seq data mining reveals the alternative message of the rice leaf transcriptome in response to heat stress. Plants 10, 1647 (2021).
Broft, P., Rosenkranz, R., Schleiff, E., Hengesbach, M. & Schwalbe, H. Structural analysis of temperature-dependent alternative splicing of HsfA2 pre-mRNA from tomato plants. RNA Biol. 19, 266–278 (2022).
Filichkin, S. A. et al. Environmental stresses modulate abundance and timing of alternatively spliced circadiantranscripts in Arabidopsis. Mol. Plant 8, 207–227 (2015).
Nibau, C., Gallem, M., Dadarou, D., Doonan, J. H. & Cavallari, N. Thermo-sensitive alternative splicing of FLOWERING LOCUS M is modulated by cyclin-dependent kinase G2. Front. Plant Sci. 10, 1680 (2019).
Meyer, M., Plass, M., Pérez-Valle, J., Eyras, E. & Vilardell, J. Deciphering 3’ss selection in the yeast genome reveals an RNA thermosensor that mediates alternative splicing. Mol. Cell 43, 1033–1039 (2011).
Healy, T. M. & Schulte, P. M. Patterns of alternative splicing in response to cold acclimation in fish. J. Exp. Biol. 222, jeb193516 (2019).
Martin Anduaga, A. et al. Thermosensitive alternative splicing senses and mediates temperature adaptation in Drosophila. Elife 8, e44642 (2019).
Haltenhof, T. et al. A conserved kinase-based body-temperature sensor globally controls alternative splicing and gene expression. Mol. Cell 78, 57–69.e4 (2020).
Preußner, M. et al. Body temperature cycles control rhythmic alternative splicing in mammals. Mol. Cell 67, 433–446.e4 (2017).
Wilkinson, M. E., Charenton, C. & Nagai, K. RNA splicing by the spliceosome. Annu. Rev. Biochem. 89, 359–388 (2020).
Saldi, T., Riemondy, K., Erickson, B. & Bentley, D. L. Alternative RNA structures formed during transcription depend on elongation rate and modify RNA processing. Mol. Cell 81, 1789–1801.e5 (2021).
Catalan-Moreno, A. et al. RNA thermoswitches modulate Staphylococcus aureus adaptation to ambient temperatures. Nucleic Acids Res. 49, 3409–3426 (2021).
Loh, E., Righetti, F., Eichner, H., Twittenhoff, C. & Narberhaus, F. RNA thermometers in bacterial pathogens. Microbiol. Spectr. 6, 10 (2018).
Thomas, S. E., Balcerowicz, M. & Chung, B. Y. RNA structure-mediated thermoregulation: What can we learn from plants. Front. Plant Sci. 13, 938570 (2022).
Johansson, J. et al. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110, 551–561 (2002).
Schlaen, R. G. et al. The spliceosome assembly factor GEMIN2 attenuates the effects of temperature on alternative splicing and circadian rhythms. PNAS. 112, 9382–9387 (2015).
Marshall, C. M., Tartaglio, V., Duarte, M. & Harmon, F. G. The Arabidopsis sickle mutant exhibits altered circadian clock responses to cool temperatures and temperature-dependent alternative splicing. Plant Cell 28, 2560–2575 (2016).
Sanchez, S. E. et al. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 468, 112–116 (2010).
Butler, W. L. Energy distribution in the photochemical apparatus of photosynthesis. Annu. Rev. Plant Physiol. 29, 345–378 (1978).
Zhang, S. et al. Recognition of CCA1 alternative protein isoforms during temperature acclimation. Plant Cell Rep. 40, 421–432 (2021).
Seo, P. J. et al. A self-regulatory circuit of circadian clock-associated1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 24, 2427–2442 (2012).
James, A. B. et al. Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell 24, 961–981 (2012).
Zhang, X. et al. Constitutive expression of CIR1 (RVE2) affects several circadian-regulated processes and seed germination in Arabidopsis. Plant J. 51, 512–525 (2007).
Acknowledgements
This work was supported by UKRI/BBSRC (BB/S005404/1, BB/Z514469/1). Z.G.M. and C.S. are grateful for PhD studentships supported by the University of Glasgow, CN Seeds Ltd and the Perry Foundation.
Author information
Authors and Affiliations
Contributions
C.S., Z.G.M., M.F.P. and M.A.J. wrote the paper and approved the final draft. C.S. and M.A.J. created figures.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sharples, C., McFarlane, Z.G., Pinheiro, M.F. et al. Integrating temperature into the Arabidopsis circadian system. npj Biol Timing Sleep 2, 35 (2025). https://doi.org/10.1038/s44323-025-00055-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s44323-025-00055-z