Abstract
Background/Objectives:
Adipose tissue is one of the main organs regulating energy homeostasis via energy storage as well as endocrine function. The adipocyte cell number is largely determined by adipogenesis. While the molecular mechanism of adipogenesis has been extensively studied, its role in dynamic DNA methylation plasticity remains unclear. Recently, it has been shown that Tet methylcytosine dioxygenase (TET) is catalytically capable of oxidizing DNA 5-methylcytosine (5-mC) to 5-hydroxymethylcytosine (5-hmC) toward a complete removal of the methylated cytosine. We investigate whether expression of the Tet genes and production of hydroxymethylcytosine are required for preadipocyte differentiation.
Subjects/Methods:
Murine 3T3-L1 preadipocytes were used to evaluate the role of Tet1 and Tet2 genes during adipogenesis. Changes in adipogenic ability and in epigenetic status were analyzed, with and without interfering Tet1 and Tet2 expression using small interfering RNA (siRNA). The adipogenesis was evaluated by Oil-Red-O staining and induced expression of adipogenic genes using quantitative polymerase chain reaction (qPCR). Levels of 5-hmC and 5-mC were measured by MassARRAY, immunoprecipitation and GC mass spectrometry at specific loci as well as globally.
Results:
Both Tet1 and Tet2 genes were upregulated in a time-dependent manner, accompanied by increased expression of hallmark adipogenic genes such as Pparγ and Fabp4 (P<0.05). The TET upregulation led to reduced DNA methylation and elevated hydroxymethylcytosine, both globally and specifically at the Pparγ locus (P<0.05 and P<0.01, respectively). Knockdown of Tet1 and Tet2 blocked adipogenesis (P<0.01) by repression of Pparγ expression (P<0.05). In particular, Tet2 knockdown repressed conversion of 5-mC to 5-hmC at the Pparγ locus (P<0.01). Moreover, vitamin C treatment enhanced adipogenesis (P<0.05), while fumarate treatment inhibited it (P<0.01) by modulating TET activities.
Conclusions:
TET proteins, particularly TET2, were required for adipogenesis by modulating DNA methylation at the Pparγ locus, subsequently by inducing Pparγ gene expression.
Similar content being viewed by others
Introduction
Adipose tissue has a key role in the regulation of energy homeostasis, not only through energy storage but also endocrine function such as adipokine secretion.1 Its dysfunction is closely associated with various metabolic disorders including obesity and type 2 diabetes. It is primarily composed of adipocytes, which differentiate from mesenchymal stem cells. The lineage commitment of mesenchymal stem cells to adipocytes is driven by the activation of master transcriptional regulators such as CCAAT/enhancer-binding proteins (C/EBPα, C/EBPβ and C/EBPδ) and peroxisome proliferator-activated receptor γ (PPARγ), resulting in subsequent expression of adipocyte-specific genes.2, 3 PPARγ is a member of the nuclear receptor superfamily of ligand-activated transcription factors,3, 4 cooperating with C/EBPα to act as a positive regulator in adipogenesis.5, 6 Heterozygous PPARγ-deficient mice showed the reduction in fat mass and adipocyte cell size,7 while overexpression of PPARγ in fibroblasts and preadipocytes was sufficient for adipogenesis, even in the absence of C/EBPα.8, 9 Consistently, pharmaceutical PPARγ ligands such as thiazolidinedione have been shown to have an impact on the regulation of gene expression in various metabolic pathways and function as an insulin sensitizer.10 These findings indicate that PPARγ-mediated control of adipogenesis may influence physiological body fat mass as well as metabolism and may be related with metabolic disorders such as obesity. Thus, understanding the molecular mechanisms by which PPARγ regulates adipogenesis may be critical to develop therapeutic strategies for the metabolic pathophysiology of obesity.
Recent evidence has shown that cell differentiation required proper epigenomic transition, besides serial coordination of adipogenic transcriptional factors.11 Epigenomic transition for cell differentiation and lineage decision takes place via changes of epigenetic patterns such as DNA methylation and histone modifications.11 DNA methylation is found in cytosines of the dinucleotide sequence CpG in mammals and has been demonstrated to be responsible for the stable maintenance of gene expression patterns during development or differentiation.12 Mammalian DNA methyltransferases (DNMTs) are responsible for the acquisition of DNA methylation during gametogenesis, embryogenesis and somatic tissue development,11, 13, 14 in which the pattern of DNA methylation changes and consequently differentiated cells develop a stable DNA methylation pattern that regulate tissue-specific gene transcription.15 Generally, DNA methylation regulates gene expression by recruiting proteins involved in gene repression or by inhibiting the binding of transcription factors to DNA.15, 16
Compared with establishment of DNA methylation, its removal process has not been well delineated. Recently, however, Tet methylcytosine dioxygenase (TET) protein family, composed of the three members TET1, TET2 and TET3, have the capacity to convert 5-methylcytosine (5-mC) to 5-hydroxymethylcytosine (5-hmC) of DNA that is an intermediate for oxidative demethylation.17, 18 They are Fe(II) and a-ketoglutarate (α-KG)-dependent dioxygenases that successively convert 5-mC to 5-hmC, 5-formylcytosine and 5-carboxycytosine of the DNA.17, 18 Genome-wide studies on embryonic stem (ES) cells have revealed that 5-hmC is enriched in gene bodies, promoter regions and CpG islands of actively transcribed genes in ES cells.19, 20
Since adipogenesis is one of the most dynamic processes of cellular differentiation, the analysis of genome-wide or gene-specific DNA demethylation pattern during this process may clarify its underlying mechanisms. The role of histone marks during adipocyte differentiation has been actively explored,21, 22, 23, 24, 25 but DNA methylation or demethylation remains poorly investigated. A few studies have indicated gene-specific DNA methylation changes during adipogenesis.26, 27 For example, the promoters of several adipogenic loci such as Pparγ, Leptin (Lep) and Glucose transporter 4 (Glut4) are hypomethylated during adipogenesis and becomes expressed.26, 27 How these genes become demethylated remains poorly investigated, but is likely mediated by 5-hmC. A genome-wide analysis of 5-hmC during adipogenesis suggested that the level of 5-hmC was more abundant in differentiated 3T3-L1 adipocyte compared to preadipocyte, and 5-hmC enrichment was closely associated with distal target regions of PPARγ.28 It suggests that TETs may have a crucial role in adipogenesis. In the current study, we closely investigate how the dynamics between DNA methylation and hydroxymethylation drives differentiation of 3T3-L1 cells to adipocytes.
Materials and methods
Cell culture and adipocyte differentiation
3T3-L1 preadipocyte cells (courtesy of Dr Yuri Kim lab at the Ewha Womans University) were maintained in Dulbecco’s modified Eagle’s medium (Welgene, Daegu, Korea) with 10% calf serum (Gibco, Gaithersburg, MD, USA), following the test for mycoplasma contamination. The cells were differentiated according to the protocol in Green et al.29 Briefly, the cells were continuously cultured for 2 days after reaching confluence and were stimulated with a differentiation cocktail containing 10 μg ml−1 insulin (Sigma-Aldrich, St Louis, MO, USA), 1 μM dexamethasone (Sigma-Aldrich) and 500 μM isobutyl-methyl xanthine (Sigma-Aldrich) in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum (ATLAS, Fort Collins, CO, USA), which were then incubated for 6–12 days with media replacement every 2 days.
Oil-Red-O staining
For Oil-Red-O staining, cells were fixed with 10% formalin and stained with Oil-Red-O solution (Sigma-Aldrich). For quantification of neutral triglycerides and lipids, Oil-Red-O staining was dissolved in 100% isopropanol (Junsei Chemical Co, Ltd, Tokyo, Japan) and the absorbance was measured at 510 nm.
Vitamin C and fumarate treatment
Confluent 3T3-L1 cells were treated with 100 μM diethyl fumarate (Sigma-Aldrich) or 50 μg ml−1 of ascorbic acid (Sigma-Aldrich). After 2 days, the cells were induced to differentiate into adipocyte with Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum containing the differentiation cocktail. For ascorbic acid treatment, we applied the differentiation cocktail without insulin in order to slow down differentiation so that a possible difference caused by enhanced activities of TET proteins could be detected. Treatment of diethyl fumarate and ascorbic acids was maintained while changing the medium for differentiation.
Gene silencing using small interfering RNA
For small interfering RNA (siRNA) knockdown of Tet1 or Tet2, transfections were performed with Dharmafect1 (Dharmacon, Lafayette, CO, USA), using siRNAs targeting Tet1 (Dharmacon; M062861-01), Tet2 (Dharmacon; M058965-01) or non-targeting control (Dharmacon; D001810-10). 3T3-L1 cells were transfected with control siRNA and Tet1- or Tet2-specific siRNAs before adipocyte differentiation. After 2 days, cells were induced to differentiate with a cocktail of insulin, dexamethasone and IBMX (DMI), and kept in culture for 6 days. After the siRNA transfection, the levels of Tet1 and Tet2 were quantified by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR).
Quantitative RT-PCR analysis
RNA was isolated using the RNeasy mini kit (QIAGEN, Hiden, Germany). After incubating with DNase I (Thermo Scientific, Waltham, MA, USA), complementary DNA was synthesized using RevertAid reverse transcriptase (Thermo Scientific). qPCR was performed using SYBR green mix (QIAGEN), and data were normalized to the Nono gene expression level, which has been reported to be a proper internal control during 3T3-L1 differentiation according to Arsenijevic et al. (2012).30 The primer pairs used are available in Table 1.
MassARRAY quantitative analysis
Genomic DNA from 3T3-L1 cells was isolated using the DNeasy Blood and Tissue Kit (QIAGEN). Bisulfite conversion of the genomic DNA was performed with the EZ DNA CT Conversion Reagent (Zymo Research Corporation, Irvine, CA, USA). PCRs were carried out with bisulfite-treated template DNA using Hotstart PCR polymerase (QIAGEN). Bisulfite amplification primers for specific regions of Pparγ, Cebpα and Fabp4 were designed using the EpiDesigner application (Sequenom Inc., San Diego, CA, USA). The primer pairs contain a T7-promoter tag to allow further in vitro transcription. Information of primer pairs used is available in Table 1. By sequencing analysis, partially methylated CpG sites were identified and quantified using the MassARRAY Compact System following the MassCLEAVE protocol (Sequenom Inc.). The resulting methylation calls were analyzed with the EpiTYPER software (Sequenom Inc.) to generate quantitative CpG methylation results, presented in the figures as the fraction of methylation. Importantly, DNA methylation analysis using bisulfite conversion,for example, applied in the MassARRAY methodology, does not distinguish between 5-mC and 5-hmC, but rather represents a summary of both marks. However, it is well documented that 5-mC is several orders of magnitude more common than 5-hmC.
Immunoprecipitation of methylated or hydroxymethylated DNA (MeDIP and hMeDIP)
Genomic DNA from cultured 3T3-L1 cells was isolated using the DNeasy Blood and Tissue Kit (QIAGEN) and sonicated to produce fragments ranging from 150 to 180 bp. Two micrograms of fragmented DNA were immunoprecipitated by methylated DNA-specific immunoprecipitation (MeDIP kit; Diagenode, Belgium, Europe) or hydroxymethylated DNA-specific immunoprecipitation (hMeDIP Kit; Diagenode). qPCR was used for quantifying the enrichment of methylated or hydroxymethylated DNA at the Pparγ locus. The sequence of the primer pair is available in Table 1.
Gas chromatography mass spectrometry detecting cytosine, methylcytosine and hydroxymethylcytosine
Isolated DNA (1 μg per sample) was hydrolyzed in 90% (V/V) formic acid at 100 °C for 1 h. Formic acid was removed under vacuum at 45 °C and samples were sealed air tight and stored at −80 °C until derivatization. Derivatization was performed as previously described.31 Briefly, dried samples were derivatized by adding 20 μl of BSTFA+TCMS (99:1; Sigma Aldrich; 3-3148, Lot: LC05232V) and 20 μl acetonitrile (VWR International, Darmstadt, Germany) at 30 °C for 30 min in a glass micro insert. Five microliters aliquots were directly subjected to gas chromatography mass spectrometry (GC–MS) analysis. The analysis was carried out on an Agilent 6890 GC/ 5973 MS System. Samples were injected into a HP-5 MS-fused silica column (30 m × 0.25 mm; 0.25 μm film thickness of the 5% phenyl 95% dimethylpolysiloxane stationary phase; Agilent J&W Scientific, Waldbronn, Germany) in pulsed splitless mode. The MS was operated in 70 eV electron impact selected ion monitoring mode for analysis. Dwell times for all ions were set to 200 ms. Characteristic ions of 5-hmC were determined by collecting a scan spectrum within the range of 50–400 m/z.
Statistical analysis
The data, except the results from GC–MS, were presented as means±s.d. values of the duplicates, triplicates and sextuplets for expression analysis, methylation analysis and Oil-Red-O staining, respectively, and were analyzed by two-sided Student’s t-test. P<0.05 was considered statistically significant. The statistical analysis for the results from GC–MS was omitted since the analysis was based on the pooled samples.
Results
DNA methylation gradually decreased at the Pparg locus during adipogenesis
First, we established the 3T3-L1 adipogenesis model according to the custom standard protocol32 and confirmed differentiation using the mRNA expression level of adipogenic molecular markers (Figures 1a–c). mRNA expression of Pparγ, Cebpα and Fabp4 genes was significantly induced compared to the preadipocyte at the latest as differentiation d3 (P<0.01). To monitor DNA methylation changes during adipogenesis, we first measured the global level of DNA methylation by quantifying DNA methylation at diagnostic repetitive elements (LINE, SINE B1, IAP and microsatellite), which represent a reliable surrogate measure of heterochromatic sites subject to extensive DNA methylation. Those loci did not show any significant change in DNA methylation during the process of adipogenic differentiation in 3T3-L1 cells (data not shown). Next, the gene-specific DNA methylation was analyzed using MassARRAY assays at major adipogenic genes including Cebpα, Pparγ and Fabp4. The locations of DNA methylation analysis were selected for putative regulatory sequences such as the putative promoter and enhancer based on genome-wide identification with histone markers. The DNA methylation at the putative enhancer of the Pparγ locus marginally decreased at differentiation d6 (P=0.08) and significantly decreased at differentiation d9 and d12 (Figure 1d, P<0.01). The average percentage of DNA methylation was 93% in preadipocyte, while that in adipocyte differentiation day 12 was decreased to 44%. The methylation of the Fabp4 gene also significantly decreased at differentiation d9 and d12 (Figure 1f, P<0.01); however, the magnitude of the changes was relatively small. The methylation of Cebpα, however, showed consistent levels during adipogenesis (Figure 1e).
Changes in relative expression of Pparγ, Cebpα and Fabp4 genes and DNA methylation at their loci during adipogenic differentiation of 3T3 -L1 cells. DNA and RNA samples were collected after adipogenic differentiation of 3T3 -L1 cells at different time points. Relative expression levels of adipogenic markers, Pparγ, Cebpα and Fabp4 genes against Nono gene measured by qPCR in triplicates during differentiation (a–c). DNA methylation at the Pparγ, Cebpα and Fabp4 loci quantified by MassARRAY in duplicates during differentiation (d–f), presented as a fraction of methylation. The data were analyzed by Student's t-test and **P<0.01 indicates significance at days versus preadipocytes (pre); d0, d3, d6, d9, d12 indicate days after adipogenesis induction.
Tet genes contribute to the demethylation of the Pparg locus during adipogenesis
To investigate which adipogenesis-related factor contributes to the demethylation of Pparγ region, we obtained a publically available gene expression data set upon 3T3-L1 differentiation from Serandour et al.28 The gene set enrichment and functional annotation analysis using DAVID bioinformatics resources (version 6.7) indicated Tet1 as one of the highly upregulated candidate genes in adipogenesis (data not shown). Among TET family proteins, TET3 is lowly expressed in adipose tissue, while TET1 and TET2 are highly expressed according to The Human Protein Atlas (www.proteinatlas.org). Gene expression of Tet1 and Tet2 genes was confirmed in our adipogenesis model with 3T3-L1. Both genes were significantly induced during adipocyte differentiation from differentiation d6 (P<0.05), similar to the pattern of other adipogenic molecular markers such as Pparγ or Cebpα (Figures 2a and b). As TET proteins convert 5-mC in DNA into 5-hmC, we quantified the global level of 5-hmC during adipogenesis using GC–MS. The global level of 5-hmC tended higher in differentiated cells compared to preadipocytes (Figure 2c), although the level was not directly correlated with the expression level of Tet genes. Since we observed a significant drop in crude DNA methylation at the Pparγ locus (Figure 1d, P<0.05), we then measured the 5-mC and the 5-hmC levels specific at the Pparγ locus with MeDIP and hMeDIP using 5-mC and 5-hmC antibodies, respectively. While 5-mC decreased (Figure 2d), 5-hmC significantly increased at the Pparγ locus after differentiation d12 (Figure 2e, P<0.01).
Changes in relative expression of Tet1 and Tet2 genes and 5-mC and 5-hmC in the global and Pparγ locus-specific levels during adipogenic differentiation of 3T3-L1 cells. Relative expression of Tet1 and Tet2 genes against the Nono gene measured by qPCR in triplicates during differentiation (a, b). Global 5-hmC level during differentiation measured by GC–MS (c). A locus-specific 5-mC and 5-hmC at the Pparγ locus quantified by MeDIP (d) and hMeDIP (e), respectively, in duplicates during adipogenesis. The data were analyzed by Student's t-test, and *P<0.05 and **P<0.01 indicate significance at days versus preadipocytes (pre), preadipocyte; d0, d3, d6, d9, d12 indicate days after adipogenesis induction.
TETs are required for adipogenesis
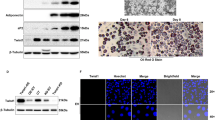
We continued by testing whether TET1 and TET2 are required for adipogenesis by depleting them with specific siRNAs in 3T3-L1 preadipocytes prior to differentiation. The knockdown of Tet1 or Tet2 expression led to significantly reduced levels of lipid droplet measured by Oil-Red-O staining, compared to cells with non-target control siRNA treatment (Figure 3a). To confirm knockdown efficiency, their expression levels were estimated by quantitative RT-PCR. As expected, the mRNA levels of Tet1 and Tet2 were markedly reduced down to less than 20% upon siRNA treatment (Figures 3b and c, P<0.01). To check whether the knockdown of Tet1 or Tet2 was sufficient to downregulate the expression of adipogenic genes, the mRNA levels of Pparγ and Fabp4 were also measured by quantitative RT-PCR analysis at the differentiated adipocyte stage (day 6). Indeed, the expression of Pparγ was significantly reduced both after the knockdown of Tet1 and Tet2; P<0.01 and P<0.05 respectively (Figure 3d). Furthermore, the decreased expression of Pparγ subsequently resulted in the significant decline of Fabp4 expression; P<0.01 (Figure 3e). The expression levels of Cebpα (which acts upstream of PPARγ) were not significantly changed throughout 6 days after the adipogenic induction in Tet1- or Tet2-silenced 3T3-L1 cells (data not shown).
Impact of Tet1 and Tet2 knockdown on adipogenesis via 5-hmC-mediated Pparg regulation. The amount of triglycerides measured by Oil-Red-O staining in sextuplets upon Tet1 or Tet2 knockdown followed by the induction of adipogenesis (a). Tet1 and Tet2 knockdown efficiency based on Tet1 and Tet2 mRNA levels measured by qPCR in triplicates (b, c). Pparγ and Fabp4 expression levels, measured by qPCR in triplicates, in adipogenic induced 3T3-L1 cells after Tet1 and Tet2 knockdown (d, e). Global 5-hmC level during differentiation measured by GC–MS (f) and a locus-specific 5-mC and 5-hmC at the Pparγ locus quantified by MeDIP (g) and hMeDIP (h), respectively, in duplicates upon Tet1 or Tet2 knockdown followed by the induction of adipogenesis. The data were analyzed by Student's t-test, and *P<0.05 and **P<0.01 indicate significance between the knockdown versus the control; ‘ns’ presents ‘not significant’; d0, d3, d6, d9, d12 indicate days after adipogenesis induction.
We next analyzed whether the effect of Tet1 and Tet2 knockdown during adipogenesis was via changes in the 5-hmC level rather than other indirect effects, both globally and at the Pparγ locus. Mass spectrometry analysis indicated that both Tet1 and Tet2 knockdown efficiently reduced the global 5-hmC level on day 6 (Figure 3f). More importantly, 5-hmC levels at the Pparγ locus (as measured by hMeDIP) markedly decreased upon the knockdown of Tet2 (P<0.05), and the reduction was inversely correlated with an increase in the 5-mC level at the locus (P<0.01), measured by MeDIP (Figures 3g and h). The knockdown of Tet1 did not significantly change the 5-hmC or 5-mC level at the locus.
Modulation of TET activity via metabolites influences adipogenesis
We next modulated TET activity with metabolites and monitored adipogenesis and Pparγ gene expression. Vitamin C enhances the activity of Fe (II) and α-KG-dependent dioxygenases, including the TET proteins, presumably by maintaining the ferrous state of iron.33 Fumarate, in contrast, leads to inhibition of the TET proteins by competing with α-KG, an essential TET co-factor.34 Here we investigated whether the treatment with vitamin C or fumarate altered adipogenesis by changing DNA methylation status during adipogenesis. 3T3-L1 cells were treated with 50 μg ml−1 ascorbic acid or 100 μM diethyl fumarate during adipogenesis for 6 days of induced adipogenesis. Lipid accumulation, a hallmark of adipocyte formation, in cells treated with vitamin C was significantly increased, while that with fumarate was significantly decreased when stained with Oil-Red-O staining (Figures 4a and e; P<0.05 and P<0.01, respectively). The mRNA level of Pparγ, measured by quantitative RT-PCR, significantly increased after vitamin C treatment (Figure 4b, P<0.05) and decreased after fumarate treatment (Figure 4f, P<0.05). The decreased expression of Pparγ subsequently resulted in the significant decline of Fabp4 expression in fumarate-treated cells (Figure 4f, P<0.05). We, furthermore, analyzed the global levels of 5-mC and 5-hmC to determine whether the treatment of those two metabolites was sufficient to alter TET protein activity. Upon vitamin C treatment, the level of 5-mC decreased and 5-hmC increased as measured by GC–MS (Figure 4c). In contrast, 3T3-Ll cells dramatically lost 5-hmC and gained 5-mC after fumarate treatment (Figure 4h). These results were confirmed by additional independent analysis using dotblots (data not shown). More importantly, we observed a significant decrease from of 5-mC at the Pparγ locus after vitamin C treatment (Figure 4d, P<0.01), and, in contrast, significantly increased after fumarate treatment (Figure 4h, P<0.01).
Modulation of TETs via ascorbic acid and fumarate during adipogenesis. The amount of triglycerides measured by Oil-Red-O staining in sextuplets after adipogenesis with and without ascorbic acid, an activator of TETs, and fumarate, a repressor of TETs (a, e). Pparγ and Fabp4 expression levels, measured by qPCR in triplicates, in adipogenic induced 3T3-L1 cells with ascorbic acid (b) and with fumarate (f). The global 5-hmC level during differentiation measured by GC–MS (c, g) and a locus-specific 5-mC at the Pparγ locus quantified by MassARRAY in duplicates (d, h). The data were analyzed by Student's t-test, and *P<0.05 and **P<0.01 indicate significance between the treated samples versus the NT ones; d0, d3, d6, d9, d12 indicate days after adipogenesis induction. FT, fumarate-treated; NT, nontreated; VT, vitamin C-treated.
Discussion
Adipogenesis is one of the most well-studied systems of cellular differentiation, and the understanding of how it is orchestrated is of major relevance to the development of human metabolic disease, including obesity and insulin resistance. During the pathological process, uncontrolled adipogenesis may be a major cause of altered uptake and storage of energy through an increase in both hyperplasia and hypertrophy of adipocytes. Previous studies have reported that several transcriptional factors including CEBPα and PPARγ have the primary role as the key drivers of adipogenesis.3, 4, 5, 6 In this study, we investigated whether the dynamics of DNA methylation drives differentiation of 3T3-L1 cells to adipocytes and how the DNA methylation status influence adipogenesis. We have demonstrated that expression of TETs are induced upon adipogenesis and are required for adipogenesis via locus-specific demethylation of the Pparγ locus, subsequently turning on its expression.
It is widely recognized that DNA methylation placement is essential to the chromatin during cellular differentiation.35 The acquisition and removal of DNA methylation at regulatory elements of the genome determine lineage fate by regulating the gene expression. A study using 5-aza-2'-deoxycytidine, an inhibitor of the DNA methyltransferase DNMT1, suggested that DNA methylation might be involved in adipogenesis in a differentiation stage-dependent manner.36 It is, however, known that the inhibitor is useful to reveal the active placement of DNA methylation, but not that of its removal. In recent years, oxidation of the methylated cytosines by the TET dioxygenases has been recognized as an intermediate step toward complete removal of methylated cytosines.37 Importantly, 5-hmC correlates with expression levels of pluripotency-related genes in undifferentiated ES cells.19, 20 A few studies on the relationship between 5-hmC and adipocyte differentiation have been reported.28, 38, 39 Genome-wide 5-hmC profiling identified high levels of 5-hmC in differentiated adipocytes, in particular at PPARγ-binding sites, for example, the Fabp4 locus.28 It has been speculated that PPARγ directed DNA demethylation around PPAR response elements via either poly(ADP-ribosyl)ation of PPARγ38 or by recruitment of CTCF.39 While these studies indicate that hydroxymethylation at PPARγ-targeted genes influenced the function of PPARγ protein during adipogenesis,37, 38 in the present study we instead investigated whether transcriptional activity of the Pparγ locus is directly regulated by DNA demethylation. First, we demonstrate the role of TETs in turning on the expression of Pparγ via hydroxymethylcytosine-mediated regulation of the Pparγ locus during induced adipogenesis. Increased expression of Tet1 and Tet2 in differentiated adipocytes resulted in high level of 5-hmC. In particular, loss of 5-mC and gain of 5-hmC were concerted at the Pparγ locus, but not at the Cebpα or Fabp4 loci.
The functional analysis using siRNA knockdown revealed that TET1 and TET2 might not target the same loci during adipogenesis. Knockdown of Tet1 and Tet2 led to a decreased 5-hmC at the global level (Figure 3f); Tet2 repression led to a stronger depletion of 5-hmC at the Pparγ locus than the Tet1 knockdown (Figure 3h). Both TET proteins carry an oxygenase domain, but only TET1 and not TET2 possess a CXXC domain that binds unmethylated CpG sequences,40 possibly differentiating targets in the genome. In mouse ES cells, TET1 and TET2 have differential roles in lineage determination during development, where Tet1 loss-of-function in ES cells results in developmental skewing toward endoderm/mesoderm lineage, whereas Tet2 loss-of-function maintains a tendency toward a neuroectoderm lineage.41 Moreover, the enrichment for 5-hmC by two TETs appears distinct genome wide.42 Likewise, Fujiki et al.38 observed that TET2 did not directly interact with PPARγ, unlike TET1. Taken together, TET1 and TET2 might have distinct and/or complementary roles in active demethylation at specific regions of the genome or during specific stages during adipogenesis.
Fewer studies were conducted regarding nutrients affecting DNA demethylation, whereas those regarding DNA methylation have been heavily investigated in the past decades. Vitamin C and citric acid cycle metabolites have been shown to have enhancing and inhibitory effect on TET proteins,34, 43 possibly modulating the availability of TET cofactors, Fe(II) and α-KG. Vitamin C treatment in ES cells resulted in global increase in 5-hmC in DNA by promoting the activity of TET enzymes.33 Fumarate also has been reported to induce the accumulation of α-KG by inhibiting α-KG dehydrogenase in citric acid cycle, which subsequently activates α-KG-dependent dioxygenase including TET enzymes.44 We applied vitamin C and fumarate as an activator and a repressor of TETs, and the results further confirmed that TET-mediated hydroxymethylcytosine was indeed a key factor to modulate adipogenesis.
In conclusion, our study has identified functional roles of TET1 and TET2 in transcriptional regulation of Pparγ-driving adipocyte differentiation. Further study is required to elucidate how TET proteins are directed to Pparγ, and the distinct roles TET1 and TET2 have in driving adipogenesis.
References
Galic S, Oakhill JS, Steinberg GR . Adipose tissue as an endocrine organ. Mol Cell Endocrinol 2010; 316: 129–139.
Christy RJ, Yang VW, Ntambi JM, Geiman DE, Landschulz WH, Friedman AD et al. Differentiation-induced gene expression in 3T3-L1 preadipocytes: CCAAT/enhancer binding protein interacts with and activates the promoters of two adipocyte-specific genes. Genes Dev 1989; 3: 1323–1335.
Rosen ED, Sarraf P, Troy AE, Bradwin G, Moore K, Milstone DS et al. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell 1999; 4: 611–617.
Rosen ED, Spiegelman BM . PPARgamma: a nuclear regulator of metabolism, differentiation, and cell growth. J Biol Chem 2001; 276: 37731–37734.
Rosen ED, Hsu CH, Wang X, Sakai S, Freeman MW, Gonzalez FJ et al. C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway. Genes Dev 2002; 16: 22–26.
Tang QQ, Zhang JW, Daniel Lane M . Sequential gene promoter interactions of C/EBPbeta, C/EBPalpha, and PPARgamma during adipogenesis. Biochem Biophys Res Commun 2004; 319: 235–239.
Kubota N, Terauchi Y, Miki H, Tamemoto H, Yamauchi T, Komeda K et al. PPAR gamma mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance. Mol Cell 1999; 4: 597–609.
Tontonoz P, Hu E, Spiegelman BM . Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell 1994; 79: 1147–1156.
Wu Z, Rosen ED, Brun R, Hauser S, Adelmant G, Troy AE et al. Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol Cell 1999; 3: 151–158.
Knouff C, Auwerx J . Peroxisome proliferator-activated receptor-gamma calls for activation in moderation: lessons from genetics and pharmacology. Endocr Rev 2004; 25: 899–918.
Jaenisch R, Bird A . Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 2003; 33: 245–254.
Holliday R, Pugh JE . DNA modification mechanisms and gene activity during development. Science 1975; 187: 226–232.
Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L et al. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell 2008; 2: 160–169.
Robertson KD . DNA methylation, methyltransferases, and cancer. Oncogene 2001; 20: 3139–3155.
Moore LD, Le T, Fan G . DNA methylation and its basic function. Neuropsychopharmacology 2013; 38: 23–38.
Crider KS, Yang TP, Berry RJ, Bailey LB . Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate's role. Adv Nutr 2012; 3: 21–38.
Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009; 324: 930–935.
Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011; 333: 1300–1303.
Wu H, D'Alessio AC, Ito S, Wang Z, Cui K, Zhao K et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev 2011; 25: 679–684.
Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 2011; 473: 398–402.
Steger DJ, Grant GR, Schupp M, Tomaru T, Lefterova MI, Schug J et al. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev 2010; 24: 1035–1044.
Wakabayashi K, Okamura M, Tsutsumi S, Nishikawa NS, Tanaka T, Sakakibara I et al. The peroxisome proliferator-activated receptor gamma/retinoid X receptor alpha heterodimer targets the histone modification enzyme PR-Set7/Setd8 gene and regulates adipogenesis through a positive feedback loop. Mol Cell Biol 2009; 29: 3544–3555.
Lee J, Saha PK, Yang QH, Lee S, Park JY, Suh Y et al. Targeted inactivation of MLL3 histone H3-Lys-4 methyltransferase activity in the mouse reveals vital roles for MLL3 in adipogenesis. Proc Natl Acad Sci USA 2008; 105: 19229–19234.
Wang L, Xu S, Lee JE, Baldridge A, Grullon S, Peng W et al. Histone H3K9 methyltransferase G9a represses PPARgamma expression and adipogenesis. EMBO J 2013; 32: 45–59.
Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 2012; 483: 474–478.
Yokomori N, Tawata M, Onaya T . DNA demethylation modulates mouse leptin promoter activity during the differentiation of 3T3-L1 cells. Diabetologia 2002; 45: 140–148.
Noer A, Sorensen AL, Boquest AC, Collas P . Stable CpG hypomethylation of adipogenic promoters in freshly isolated, cultured, and differentiated mesenchymal stem cells from adipose tissue. Mol Biol Cell 2006; 17: 3543–3556.
Serandour AA, Avner S, Oger F, Bizot M, Percevault F, Lucchetti-Miganeh C et al. Dynamic hydroxymethylation of deoxyribonucleic acid marks differentiation-associated enhancers. Nucleic Acids Res 2012; 40: 8255–8265.
Green H, Meuth M . An established pre-adipose cell line and its differentiation in culture. Cell 1974; 3: 127–133.
Arsenijevic T, Gregoire F, Delforge V, Delporte C, Perret J . Murine 3T3-L1 adipocyte cell differentiation model: validated reference genes for qPCR gene expression analysis. PloS one 2012; 7: e37517.
Tang X, Zhen Q, Fan Z, Feng C, Ding M . [Determination of homocysteine in plasma by precolumn derivatization-high performance liquid chromatography with fluorescence detection]. Se Pu 2012; 30: 613–617.
Green H, Kehinde O . An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell 1975; 5: 19–27.
Blaschke K, Ebata KT, Karimi MM, Zepeda-Martinez JA, Goyal P, Mahapatra S et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 2013; 500: 222–226.
Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev 2012; 26: 1326–1338.
Bernstein BE, Meissner A, Lander ES . The mammalian epigenome. Cell 2007; 128: 669–681.
Sakamoto H, Kogo Y, Ohgane J, Hattori N, Yagi S, Tanaka S et al. Sequential changes in genome-wide DNA methylation status during adipocyte differentiation. Biochem Biophys Res Commun 2008; 366: 360–366.
Wu H, Zhang Y . Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev 2011; 25: 2436–2452.
Fujiki K, Shinoda A, Kano F, Sato R, Shirahige K, Murata M . PPARgamma-induced PARylation promotes local DNA demethylation by production of 5-hydroxymethylcytosine. Nat Commun 2013; 4: 2262.
Dubois-Chevalier J, Oger F, Dehondt H, Firmin FF, Gheeraert C, Staels B et al. A dynamic CTCF chromatin binding landscape promotes DNA hydroxymethylation and transcriptional induction of adipocyte differentiation. Nucleic Acids Res 2014; 42: 10943–10959.
Ko M, An J, Bandukwala HS, Chavez L, Aijo T, Pastor WA et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature 2013; 497: 122–126.
Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 2011; 8: 200–213.
Huang Y, Chavez L, Chang X, Wang X, Pastor WA, Kang J et al. Distinct roles of the methylcytosine oxidases Tet1 and Tet2 in mouse embryonic stem cells. Proc Natl Acad Sci USA 2014; 111: 1361–1366.
Loenarz C, Schofield CJ . Expanding chemical biology of 2-oxoglutarate oxygenases. Nature chemical biology 2008; 4: 152–156.
Salminen A, Kauppinen A, Hiltunen M, Kaarniranta K . Krebs cycle intermediates regulate DNA and histone methylation: epigenetic impact on the aging process. Ageing research reviews 2014; 16: 45–65.
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) (2014R1A1A2058942 to YJP and 2014R1A1A2058964 to AML). YY and JHP were supported by the Brain Korea 21 Plus Project in 2013 (22A20130012143).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Yoo, Y., Park, J., Weigel, C. et al. TET-mediated hydroxymethylcytosine at the Pparγ locus is required for initiation of adipogenic differentiation. Int J Obes 41, 652–659 (2017). https://doi.org/10.1038/ijo.2017.8
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ijo.2017.8
This article is cited by
-
Cell identity and 5-hydroxymethylcytosine
Epigenetics & Chromatin (2025)
-
A negative feedback loop between TET2 and leptin in adipocyte regulates body weight
Nature Communications (2024)
-
Targeted erasure of DNA methylation by TET3 drives adipogenic reprogramming and differentiation
Nature Metabolism (2022)
-
miR-410-3P inhibits adipocyte differentiation by targeting IRS-1 in cancer-associated cachexia patients
Lipids in Health and Disease (2021)
-
The trans-ancestral genomic architecture of glycemic traits
Nature Genetics (2021)