Abstract
It has been recently suggested that the expression levels of mutant HSP110 could be a prognostic marker in colorectal cancer with a high level of microsatellite instability (MSI-H). The aim of our study was to validate the prognostic significance of HSP110 mutation using immunohistochemistry and DNA testing in MSI-H colorectal cancer. Wild-type HSP110 (HSP110wt)-specific immunohistochemistry was performed in 168 MSI-H colorectal cancer tissues, and their expression levels were evaluated using a four-tier scoring system (0/1+/2+/3+). Of these tissues, 167 cases were analyzed for HSP110 T17 deletion. Associations with clinicopathological, molecular and survival parameters were statistically analyzed. The low-level expression of HSP110wt (0/1+) was observed in 40 MSI-H colorectal cancers (24%) and was significantly related to large HSP110 T17 deletions (≥ 4 bp, P<0.001). In survival analysis, patients with low HSP110wt expression (0/1+) showed better disease-free survival compared with those with high expression (2+/3+; P=0.005). This significance in survival difference was maintained in patients with 5-fluorouracil-based chemotherapy-treated tumors (P=0.024) and in those with stage III/IV tumors (P=0.032). Multivariate analysis confirmed the role of HSP110wt expression as an independent prognostic factor (P=0.016, hazard ratio=4.32). In MSI-H colorectal cancer, a low expression of HSP110wt is associated with large HSP110 T17 deletions and better clinical outcome. Immunohistochemistry of HSP110wt can be a simple and valuable tool for the prognostic and therapeutic stratification of patients with MSI-H colorectal cancer.
Similar content being viewed by others
Main
It has been indicated that colorectal cancer with a high level of microsatellite instability (MSI-H) has distinct clinicopathological features, including a predilection for proximal tumor location, lower tumor stages, poor tumor differentiation, extracellular mucin production, tumor-infiltrating lymphocytes, Crohn’s-like lymphocytic reaction and BRAF V600E mutation.1, 2 More importantly, MSI-H colorectal cancer is known as a better prognostic phenotype than MSI-low and microsatellite-stable (MSS) colorectal cancers.3, 4, 5 Regarding the prognostic implications of MSI-H colorectal cancer, we previously reported an interesting result in which patients with CpG island methylator phenotype-high (CIMP-H) displayed worse survival than those with CIMP-low/negative (CIMP-L/0) in MSI-H colorectal cancer.6 Although the prognostic value of CIMP has been controversial in colorectal cancer,1, 6, 7, 8, 9 our previous study implied that MSI-H colorectal cancer is not a molecularly and prognostically homogeneous phenotype; therefore, it could be classified into prognostic subgroups depending on its underlying genetic or epigenetic status.
Although it has been proven that the survival of patients with MSI-H colorectal cancer is comparatively favorable, their adjuvant chemotherapeutic response remains controversial.3, 10 In contrast to MSS colorectal cancer, a trend toward chemoresistance, especially toward 5-fluorouracil (5-FU)-based chemotherapy, has been observed in MSI-H colorectal cancer.4, 11, 12, 13, 14, 15 In fact, there is a lack of evidence regarding molecular factors that might predict responses to chemotherapy or sophisticatedly stratify prognoses in MSI-H colorectal cancer.16 Although the BRAF V600E mutation has been thought to be related to a poor prognosis in colorectal cancer,1, 17 its prognostic effect was significant only in MSS colorectal cancer, not in MSI-H colorectal cancer.18, 19 In addition, the Beta2-microglobulin mutation, myosin 1A expression level, SMAD4 expression level and LINE-1 methylation level have been suggested to be associated with prognosis in patients with MSI-H colorectal cancer,20, 21, 22, 23, 24 but more investigations are needed to establish molecular prognostic markers specific to MSI-H colorectal cancer. Recently, Dorard et al25 published data indicating the mutant expression of heat shock protein 110 kDa (HSP110, also known as HSP105) as a prognostic and predictive marker in MSI-H colorectal cancer. This mutant HSP110, also expressed as HSP110ΔE9 because of its exon 9 skipping, may have an important role in sensitizing colorectal cancer cells to chemotherapy, and its overexpression may contribute to a better prognosis for MSI-H colorectal cancer. Interestingly, this study also showed that the HSP110ΔE9 expression levels were significantly associated with the size of HSP110 T17 mononucleotide repeat deletions, which are the causal mutations for HSP110ΔE9. HSP110 T17 deletion may frequently occur in MSI-H colorectal cancer because this mononucleotide repeat is a type of microsatellite and can be vulnerable to deletion under MSI-H conditions.
On the basis of these findings, we hypothesized that both HSP110 expression and the size of the HSP110 T17 deletion could be prognostic markers in MSI-H colorectal cancer. In this study, we evaluated wild-type HSP110 (HSP110wt) expression and HSP110 T17 deletion using immunohistochemistry and DNA testing, respectively, and provided the clinicopathological and prognostic implications of these factors in MSI-H colorectal cancer.
Materials and methods
Tissue Samples and MSI Determination
A total of 168 colorectal cancers with MSI-H were examined in this study. Formalin-fixed, paraffin-embedded tissues from all cases were obtained from the pathologic archives, which originated from colorectal cancer patients who underwent curative resection at Seoul National University Hospital, Seoul, Korea or at Seoul National University Bundang Hospital, Seongnam, Korea from 2004 to 2007. During this period, 1535 and 806 patients received surgical treatment for colorectal cancer at the surgery departments of Seoul National University Hospital and Seoul National University Bundang Hospital, respectively. The DNA extracted from tumor and normal colonic tissues of the resected specimens of these patients had previously been tested for the determination of MSI in the molecular pathology division of Seoul National University Hospital. Following these tests, 119 (7.8%) cases from Seoul National University Hospital and 68 (8.4%) cases from Seoul National University Bundang Hospital were determined as MSI-H. Of these total 187 cases, 168 cases were available for use on formalin-fixed, paraffin-embedded tissue blocks and were finally included in this study. The MSI status was determined using the NCI Bethesda recommended microsatellite markers (BAT-25, BAT-26, D5S346, D17S250 and D2S123).26 The clinical data of all patients, including age, gender, anatomic location of tumor, gross tumor type, American Joint Committee on Cancer (AJCC) TNM stage (seventh edition), adjuvant chemotherapy status and time of death or recurrence, were collected by reviewing their medical records. The Seoul National University Hospital Institutional Review Board approved this study (IRB no. H-1203-072-402).
Histopathological Assessment
Two pathologists (JHK and GHK) independently assessed the histopathological characteristics of 168 MSI-H colorectal cancers by microscopic examination of the hematoxylin and eosin-stained slides. The histological parameters included tumor differentiation, extracellular mucin pools, medullary carcinoma component and signet ring cell carcinoma component. Although MSI-H colorectal cancers are defined as low-grade tumors regardless of morphological degree of differentiation according to the latest edition (fourth edition) of the WHO classification of tumors of the digestive system,27 the histological differentiation of each MSI-H colorectal cancer was classified into one of three categories (well, moderately or poorly differentiated) based on the percentage of gland formation (>95%, 50–95% and 0–49%, respectively) in this study. The results that were discordant between the observers were reviewed and discussed, and finally, a consensus was reached.
Immunohistochemistry
A tissue microarray was constructed as previously described.6 Three different areas of carcinoma in formalin-fixed, paraffin-embedded tissue blocks of individual MSI-H colorectal cancer (n=168) were extracted as three tissue cores (2 mm in diameter) per each case and transferred to recipient tissue microarray blocks (Superbiochips Laboratories, Seoul, South Korea). Next, sections of tissue microarray blocks were immunostained with antibodies to MLH1 (DAKO, Glostrup, Denmark), MSH2 (Invitrogen, Camarillo, CA, USA), PMS2 (Ventana Medical Systems, Tucson, AZ, USA), MSH6 (Ventana Medical Systems) and HSP110 (Product name: NCL-HSP105, Leica Biosystems, Newcastle upon Tyne, UK). The immunogen of this anti-HSP110 antibody is the C-terminus of HSP110wt, and this site is specific for HSP110wt, but not HSP110ΔE9.
Interpretation of Immunohistochemistry
The expression of MLH1/MSH2/PMS2/MSH6 was assessed to be negative (loss) or positive (retained) by one pathologist (JHK). The positivity of these mismatch repair proteins was determined when nuclear staining pattern in tumor cells was observed, and the loss of expression was determined when all three tissue cores from a tumor specimen were completely negative. Semiquantitative measurement of HSP110wt expression was performed independently by two pathologists (JHK and GHK) who were blinded to clinicopathological and molecular information. A four-tier scoring system based on staining intensity was adopted, which included scores of completely negative (0), faintly positive (1+), intermediately positive (2+) and strongly positive (3+). Staining with a nuclear-to-cytoplasmic pattern in >5% of the tumor cells in the individual tissue cores was considered positive, and staining in <5% of tumor cells of each core was considered negative (0). Among the three tissue cores of each case, identical scores for two or more cores provided an overall score for the case. All of the cases evaluated by the individual pathologists were determined to belong to one of the four scores without any occurrence of cases showing all different scores in the three tissue cores. Discordant scores between the observers were reviewed and discussed, and finally, a consensus was reached.
DNA Extraction and HSP110 T17 Deletion Analysis
Genomic DNA extraction from formalin-fixed, paraffin-embedded tissues of colorectal cancers was performed as previously described.6 In 1 of the 168 colorectal cancer samples, an insufficient amount of genomic DNA was extracted; a total of 167 primary MSI-H colorectal cancer tissue samples were thus involved in the HSP110 deletion analysis. The size of the HSP110 T17 mononucleotide repeat deletion in genomic DNA extracted from primary MSI-H colorectal cancer tissues (n=167), as well as from normal colonic mucosal tissue (n=1), primary MSS colorectal cancer tissues (n=2), was detected by fluorescence capillary electrophoresis-based DNA fragment size analysis. To amplify the target DNA sequences, including the T17 mononucleotide repeat located in HSP110 intron 8, fluorescence-labeled primers were designed (forward, 5′-TGAAAACCCTGTCCATCCAT-3′; reverse, FAM-5′-CTTTAAATGCCGGGGAAAGT-3′). PCR was then performed using MightyAmp DNA polymerase Ver.2 (Takara, Shiga, Japan) under the following conditions: initial pre-denaturation at 98 °C for 2 min followed by 40 cycles of 98 °C for 10 s, 57 °C for 15 s and 68 °C for 30 s, and a final extension of 68 °C for 10 min. The PCR products were analyzed using an ABI DNA analyzer 3730xl (Applied Biosystems, Foster City, CA, USA) and Peak Scanner software v1.0 (Applied Biosystems). The presence of an altered peak indicated the deletion of the HSP110 T17 repeat, and the deletion sizes were measured by subtracting the DNA fragment sizes of an altered peak from those of a normal peak.
CIMP Determination and KRAS/BRAF Mutation Analyses
CIMP determination and KRAS/BRAF mutation analyses were conducted as previously described.6 The sodium bisulfite modification of genomic DNA and the following MethyLight technique were used to quantify the promoter CpG island methylation of eight markers (ie, NEUROG1, CRABP1, CACNA1G, CDKN2A (p16), MLH1, IGF2, SOCS1 and RUNX3). CIMP-H was defined as a status with promoter hypermethylation detected in five or more markers, and CIMP-L was defined as a status with hypermethylation observed in one to four markers. None of the eight markers were hypermethylated in CIMP-0. KRAS codons 12 and 13 and BRAF codon 600 mutations were analyzed using the PCR-restriction fragment length polymorphism and direct sequencing methods. Among 168 MSI-H colorectal cancers included in this study, six and one samples were excluded in KRAS and BRAF mutation analyses, respectively, because of their insufficient amount of extracted DNA.
Statistical Analysis
Comparisons of the clinicopathological and molecular parameters were conducted with the χ2 test or Fisher’s exact test. Interobserver reproducibility of HSP110wt scoring system was assessed by the percentage of agreement, and the Kappa statistic. The Kappa coefficient (κ) has been used to measure the inter-rater agreement for categorical data. The interpretation guideline for the Kappa statistic is as follows: the κ value between 0 and 0.20 is regarded as slight agreement, 0.21 and 0.40 as fair agreement, 0.41 and 0.60 as moderate agreement, 0.61 and 0.80 as substantial agreement, and 0.81 and 1.00 as very strong agreement.28 For survival analysis, disease-free survival was defined as the time from curative surgery to death or tumor recurrence or to the last clinical follow-up. The average time of disease-free survival was 1682 days (ranging from 31 to 3261 days) in this study. The Kaplan–Meier method with the log-rank test was used in the survival analysis. In addition, the multivariate Cox proportional hazards regression model was performed in order to examine the prognostic significance of HSP110wt expression after adjusting prognostic factors with P-value<0.05 in the univariate Cox proportional hazards model. All statistical analyses were performed using IBM SPSS Statistics version 20 (Chicago, IL, USA), and statistical significance was determined at P<0.05.
Results
HSP110wt Expression Status in MSI-H Colorectal Cancer
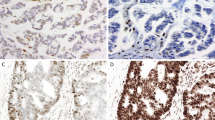
Immunohistochemistry for HSP110wt was performed in a total of 168 MSI-H colorectal cancer tissue samples, and the expression level of each case was determined according to one of four scores (Figure 1). There was 93.5% agreement for HSP110wt scoring results between two observers (JHK and GHK; κ=0.905, P<0.001). This κ value represents very strong agreement, and supports good reproducibility of our HSP110wt scoring system. The main hypothetical concept of this immunohistochemistry study is illustrated in Figure 2. Based on the earlier findings of the Duval group,25 it was predicted that HSP110wt expression would be inversely correlated with HSP110ΔE9 expression, HSP110 T17 deletion size and prognosis (Figure 2). The frequency of each of the four scores is shown in Figure 3a. Zero and 1+ scores represented tumors with low levels of HSP110wt expression, and this low expression phenotype (0 or 1+) was observed in 40 (24%) of the 168 MSI-H colorectal cancers (Figure 3a).
HSP110wt immunohistochemical staining of tissue microarray sections of 168 MSI-H colorectal cancers are semiquantitatively assessed. (a) A case showing the complete loss of HSP110wt expression is scored as 0. (b) A case showing weak staining of HSP110wt in tumor cells is scored as 1+. (c) A case showing intermediate staining of HSP110wt in tumor cells is scored as 2+. (d) A case showing strong staining of HSP110wt in tumor cells is scored as 3+.
In 167 MSI-H colorectal cancers, in addition to HSP110wt immunohistochemistry, HSP110 T17 deletion size analysis is performed. (a) Frequencies of HSP110wt immunohistochemistry scores in 168 MSI-H colorectal cancers are demonstrated. (b) Frequencies of HSP110 T17 deletion sizes in 167 MSI-H colorectal cancers are demonstrated. (c) A matched distribution between the HSP110wt immunohistochemistry scores and the HSP110 T17 deletion sizes in MSI-H colorectal cancers (n=167) is shown.
Relationship Between HSP110wt Expression and HSP110 T17 Deletion
To explore the clinicopathological and molecular features of MSI-H colorectal cancers according to their HSP110wt expression status, the tumors were divided into two expression groups: low (0/1+) or high (2+/3+). Correlations of HSP110wt expression with various factors were statistically analyzed, but there was no significant difference between the two groups except for CIMP status and HSP110 T17 deletion size (Table 1). A relatively lower frequency of CIMP-H status was observed in the low expression group than in the high expression group (P=0.015; Table 1).
The size of the HSP110 T17 deletion was determined by fluorescence capillary electrophoresis-based DNA fragment analysis. For preliminary testing, one normal colonic mucosal tissue adjacent to primary colorectal cancer tissue and two primary MSS colorectal cancer tissues were used as negative controls of the HSP110 T17 deletion, and 10 primary MSI-H colorectal cancer tissues, which were randomly selected from 167 MSI-H colorectal cancers collected in our study, were tested as positive controls. The results of this preliminary test corresponded with our expectations. None of the negative controls showed the HSP110 T17 deletion, and various sized deletions of HSP110 T17 repeat were detected in the positive controls (Supplementary Figure S1).
Next, the HSP110 T17 deletion size was analyzed in all MSI-H colorectal cancer tissues (n=167), and the deletion of the HSP110 T17 repeat was present in 147 MSI-H colorectal cancers (88%; Figure 3b). In contrast with a previous study by Dorard et al25, which reported that the HSP110 T17 deletion was observed in 100% of the tested MSI-H colorectal cancer samples with deletion sizes ranging from 3 to 8 bp, 20 cases (12%) showed no deletion, and the deletion sizes ranged from 1 to 7 bp in this study (Figure 3b). When the HSP110wt expression status and HSP110 T17 deletion size of each MSI-H colorectal cancer were matched, tumors with low expression of HSP110wt (0/1+) were concentrated in the larger deletion sizes group (≥4 bp) (Figure 3c). Tumors with high expression of HSP110wt (2+/3+) showed a dispersed distribution, but strongly positive expression (3+) was relatively more frequent in the tumors with smaller deletion sizes (<4 bp; Figure 3c). The low-level expression of HSP110wt (0/1+) was significantly associated with a large HSP110 T17 deletion size (≥4 bp; P<0.001; Table 1).
Prognostic Significance of HSP110wt Expression in MSI-H Colorectal Cancer
For the survival analysis, the 167 MSI-H colorectal cancers tested for the HSP110 T17 deletion were first divided into two groups, that is, the large HSP110 T17 deletions group (HSP110-dL) or the small HSP110 T17 deletions group (HSP110-dS), with each group having a specific cutoff value for deletion size. Each of the seven cutoff values (1–7 bp) was used in the survival analysis, and no significant survival difference between HSP110-dL and HSP110-dS was observed, regardless of the cutoff values (Supplementary Figure S2).
The Kaplan–Meier survival analysis with the log-rank test demonstrated a significant survival difference among the four groups, depending on the HSP110wt immunohistochemistry score (P=0.036; Figure 4a), as well as between the low expression group (0/1+) and the high expression group (2+/3+) in 168 MSI-H colorectal cancers (P=0.005; Figure 4b). In agreement with our hypothesis, the low expression phenotype was significantly associated with favorable survival (Figures 4a and b). Moreover, HSP110wt expression was determined to be an independent prognostic factor for MSI-H colorectal cancer in the multivariate analysis (P=0.016, hazard ratio=4.32; Table 2).
Kaplan–Meier survival analysis with the log-rank test demonstrates the prognostic significance of HSP110wt expression in MSI-H colorectal cancers. (a) There are significant survival differences among the four groups, according to the HSP110wt immunohistochemistry scores of all MSI-H colorectal cancer patients (n=168; P=0.036). (b) Patients with low expression of HSP110wt (0/1+) show better disease-free survivals than do the high expression group (2+/3+) from all patients with MSI-H colorectal cancer (n=168; P=0.005). (c, d) The low expression group (0/1+) is also associated with better disease-free survival in 5-FU-based chemotherapy-treated (c, n=108) or AJCC TNM stage III/IV (d, n=60) subgroups of MSI-H colorectal cancer patients (P=0.024 and P=0.032, respectively).
Next, we analyzed the survival differences of HSP110wt expression phenotypes in the subsets of MSI-H colorectal cancer patients according to stage or chemotherapy status, as the study by Dorard et al25 reported significant survival effects of HSP110ΔE9 expression in stage III or chemotherapy-treated patients. In agreement with the previous results of Dorard et al, the low expression group showed a better prognosis in the subsets of 5-FU-based chemotherapy-treated patients (n=108) and in stage III/IV patients (n=60; P=0.024 and P=0.032, respectively; Figures 4c and d). However, in chemotherapy-untreated patients (n=59) and stage I/II patients (n=108), there was no prognostic significance for HSP110wt expression (Supplementary Figures S3a and b).
Finally, age-stratified survival analysis was also performed. Using the average age (59 years) of all MSI-H colorectal cancer patients, the younger age group (<59 years, n=85) and the older age group (≥ 59 years, n=83) were stratified. In both the younger and older age groups, the HSP110wt-low expression phenotype showed tendencies toward better disease-free survival with marginal significances (P=0.05 and P=0.058, respectively; Supplementary Figures S3c and d).
Discussion
HSPs are cellular stress proteins induced by physiologically and environmentally hazardous factors, such as hyperthermia, hypoxia, radiation, oxygen-free radicals and mechanical stresses, as well as pathological factors such as neoplasms, infections, cardiovascular diseases and neurodegenerative diseases.29, 30, 31, 32 The major role of HSPs in stressful conditions is known to be cytoprotective through two biological functions-molecular chaperone and anti-apoptotic effects.29, 33 In malignancy, HSPs can promote cancer cell survival and contribute to the growth, invasion and metastasis of tumors by preventing the degradation of oncogenic proteins and repressing cancer cell apoptosis.29, 33, 34 Previous studies have reported that the overexpression of HSPs was observed in tumor tissues or in the blood of patients with malignant tumors; furthermore, the prognostic significance of HSP expression has also been demonstrated in a variety of cancers, including prostate, liver cell, breast, bladder and colorectal cancer.29
HSPs have been classified into several families according to their molecular weight, such as small HSPs (including HSP27), HSP40, HSP60, HSP70, HSP90 and HSP110.35 Until recently, the expression status of HSPs and their prognostic and therapeutic implications for cancer have mainly focused on HSP27, HSP60, HSP70 and HSP90.29, 33 However, there have been insufficient investigations into the expression of HSP110, the mutations of HSPs and their clinical implications in cancer. Only a few previous studies, which were conducted with small colorectal cancer sample sizes, reported that HSP110 overexpression was associated with worse overall survival, advanced stage and nodal metastasis in colorectal cancer.36, 37 In these circumstances, the study by Dorard et al25 provided important clues about the effect of HSP110 mutations on colorectal cancer. It is reasonable that the increased expression of mutant HSP110 inhibits the normal functions of HSP, which are beneficial to cancer cell survival; therefore, this molecular alteration can result in positive effects on survival in patients with cancer. Based on the results of Dorard et al, we assumed that HSP110wt expression could also be a prognostic factor in MSI-H colorectal cancer because HSP110wt expression may be negatively correlated with that of mutant HSP110. Thus, it was expected that a decreased expression of HSP110wt would be related to large HSP110 T17 deletions and improved survival in patients with MSI-H colorectal cancer. Our results confirmed that this expectation was not incorrect, and a summary of our study is consistent with this hypothetical model (Figure 2).
Although the mRNA expression level of HSP110ΔE9 was tested as a prognostic marker in the study of Dorard et al25, we applied an immunohistochemistry-based approach using formalin-fixed, paraffin-embedded tissues to assess HSP110 expression. We adopted an HSP110wt-specific antibody to evaluate altered HSP110 expression in MSI-H colorectal cancer because there was no commercially available HSP110ΔE9-specific antibody. As the C-terminus of HSP110wt is truncated in HSP110ΔE9 because of exon 9 skipping,25 HSP110wt expression can be specifically detected using C-terminus-targeting antibodies. Although it seems difficult to develop HSP110ΔE9-specific antibodies because of overlapping between the whole structure of HSP110ΔE9 and the partial structure of HSP110wt,25 it is expected that HSP110ΔE9-specific immunohistochemistry can produce clearer and more correct results as a prognostic marker in MSI-H colorectal cancer. However, according to our results, HSP110wt-specific immunohistochemistry was successfully validated as a method to identify prognostic subgroups in MSI-H colorectal cancer. In the immunohistochemistry analysis, the low-level expression of HSP110wt (completely negative or faintly positive) was easily and distinctly recognized by experienced pathologists (Figure 1). We are convinced that this method is simple and reproducible, and it will therefore prove very useful in application to daily clinical and pathological practices.
MSI-H colorectal cancers with low expression of HSP110wt (0/1+) were specifically localized in large HSP110 T17 deletions (≥4 bp; Figure 3c and Table 1). However, tumors with high expression of HSP110wt (2+/3+) were dispersed throughout all deletion sizes (0–7 bp) (Figure 3c). Moreover, survival analysis showed that HSP110wt expression was a significant prognostic factor in MSI-H colorectal cancers but that HSP110 T17 deletion size was not (Figure 4; Table 2 and Supplementary Figure S2). From these findings, we infer that HSP110 T17 deletions of large sizes are necessary but not sufficient for the low-level expression of HSP110wt and that the expression status of HSP110wt is a more decisive factor on the prognostic effects than HSP110 T17 deletion size itself for MSI-H colorectal cancers.
Interestingly, in this study, the low expression of HSP110wt was correlated with a low frequency of CIMP-H status (Table 1). Therefore, we suspected that the favorable survival of patients with low expression of HSP110wt might be associated with the prognostic effect of CIMP status. In fact, we previously reported that the CIMP-H status was related to an adverse prognosis in patients with MSI-H colorectal cancer.6 However, prognostic features and chemotherapy responses according to CIMP status in colorectal cancer were controversial.1, 7, 9, 38, 39, 40, 41, 42 More importantly, although HSP110wt expression was an independent prognostic factor in the present analysis, CIMP status was not (Table 2). In addition, it is difficult to find a molecular mechanistic connection between the HSP110wt expression level and the CIMP status because HSP110 mutation is a genetic alteration, whereas CIMP is an epigenetic change. Collectively, it seems that the relationship between HSP110wt expression and CIMP status is a relatively stochastic event and that the prognostic significance of HSP110wt expression is independent of CIMP status in MSI-H colorectal cancer.
In conclusion, we identified that the decreased expression of HSP110wt, as well as the increased expression of HSP110ΔE9 from the report of the Duval group25 could be a marker that indicates better prognostic subgroups in MSI-H colorectal cancers. Together, this study and the previous results of the Duval group indicate that the HSP110 mutation occurred in most MSI-H colorectal cancers, but its prognostic effects varied according to the expression level of wild-type or mutant HSP110. It is anticipated that the HSP110 mutation will have an important role not only in the prediction of a clinical course and therapeutic response but also in the future applications of targeted therapy in colorectal cancers.
References
Ogino S, Nosho K, Kirkner GJ et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut 2009;58:90–96.
Jenkins MA, Hayashi S, O'Shea AM et al. Pathology features in Bethesda guidelines predict colorectal cancer microsatellite instability: a population-based study. Gastroenterology 2007;133:48–56.
de la Chapelle A, Hampel H . Clinical relevance of microsatellite instability in colorectal cancer. J Clin Oncol 2010;28:3380–3387.
Popat S, Hubner R, Houlston RS . Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol 2005;23:609–618.
Guastadisegni C, Colafranceschi M, Ottini L et al. Microsatellite instability as a marker of prognosis and response to therapy: a meta-analysis of colorectal cancer survival data. Eur J Cancer 2010;46:2788–2798.
Bae JM, Kim MJ, Kim JH et al. Differential clinicopathological features in microsatellite instability-positive colorectal cancers depending on CIMP status. Virchows Arch 2011;459:55–63.
Kim JH, Shin SH, Kwon HJ et al. Prognostic implications of CpG island hypermethylator phenotype in colorectal cancers. Virchows Arch 2009;455:485–494.
Barault L, Charon-Barra C, Jooste V et al. Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Res 2008;68:8541–8546.
Dahlin AM, Palmqvist R, Henriksson ML et al. The role of the CpG island methylator phenotype in colorectal cancer prognosis depends on microsatellite instability screening status. Clin Cancer Res 2010;16:1845–1855.
Boland CR, Goel A . Microsatellite instability in colorectal cancer. Gastroenterology 2010;138:2073–2087 e2073.
Ribic CM, Sargent DJ, Moore MJ et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med 2003;349:247–257.
Carethers JM, Smith EJ, Behling CA et al. Use of 5-fluorouracil and survival in patients with microsatellite-unstable colorectal cancer. Gastroenterology 2004;126:394–401.
Benatti P, Gafa R, Barana D et al. Microsatellite instability and colorectal cancer prognosis. Clin Cancer Res 2005;11:8332–8340.
Des Guetz G, Schischmanoff O, Nicolas P et al. Does microsatellite instability predict the efficacy of adjuvant chemotherapy in colorectal cancer? A systematic review with meta-analysis. Eur J Cancer 2009;45:1890–1896.
Sargent DJ, Marsoni S, Monges G et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol 2010;28:3219–3226.
Pritchard CC, Grady WM . Colorectal cancer molecular biology moves into clinical practice. Gut 2011;60:116–129.
Farina-Sarasqueta A, van Lijnschoten G, Moerland E et al. The BRAF V600E mutation is an independent prognostic factor for survival in stage II and stage III colon cancer patients. Ann Oncol 2010;21:2396–2402.
Samowitz WS, Sweeney C, Herrick J et al. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res 2005;65:6063–6069.
Roth AD, Tejpar S, Delorenzi M et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol 2010;28:466–474.
Kloor M, Michel S, Buckowitz B et al. Beta2-microglobulin mutations in microsatellite unstable colorectal tumors. Int J Cancer 2007;121:454–458.
Tikidzhieva A, Benner A, Michel S et al. Microsatellite instability and Beta2-microglobulin mutations as prognostic markers in colon cancer: results of the FOGT-4 trial. Br J Cancer 106:1239–1245 2012.
Mazzolini R, Dopeso H, Mateo-Lozano S et al. Brush border myosin Ia has tumor suppressor activity in the intestine. Proc Natl Acad Sci USA 2012;109:1530–1535.
Isaksson-Mettavainio M, Palmqvist R, Dahlin AM et al. High SMAD4 levels appear in microsatellite instability and hypermethylated colon cancers, and indicate a better prognosis. Int J Cancer 2012;131:779–788.
Rhee YY, Kim MJ, Bae JM et al. Clinical outcomes of patients with microsatellite-unstable colorectal carcinomas depend on L1 methylation level. Ann Surg Oncol 2012;19:3441–3448.
Dorard C, de Thonel A, Collura A et al. Expression of a mutant HSP110 sensitizes colorectal cancer cells to chemotherapy and improves disease prognosis. Nat Med 2011;17:1283–1289.
Boland CR, Thibodeau SN, Hamilton SR et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998;58:5248–5257.
Bosman FT World Health Organization., International Agency for Research on Cancer WHO Classification of Tumours of the Digestive System 4th edn International Agency for Research on Cancer: Lyon, 2010.
Landis JR, Koch GG . The measurement of observer agreement for categorical data. Biometrics 1977;33:159–174.
Seigneuric R, Mjahed H, Gobbo J et al. Heat shock proteins as danger signals for cancer detection. Front Oncol 2011;1:37.
Goldstein MG, Li Z . Heat-shock proteins in infection-mediated inflammation-induced tumorigenesis. J Hematol Oncol 2009;2:5.
Madrigal-Matute J, Martin-Ventura JL, Blanco-Colio LM et al. Heat-shock proteins in cardiovascular disease. Adv Clin Chem 2011;54:1–43.
Adachi H, Katsuno M, Waza M et al. Heat shock proteins in neurodegenerative diseases: pathogenic roles and therapeutic implications. Int J Hyperthermia 2009;25:647–654.
Jego G, Hazoume A, Seigneuric R et al. Targeting heat shock proteins in cancer. Cancer Lett 332:275–285 2013.
Calderwood SK, Ciocca DR . Heat shock proteins: stress proteins with Janus-like properties in cancer. Int J Hyperthermia 2008;24:31–39.
Kampinga HH, Hageman J, Vos MJ et al. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones 2009;14:105–111.
Slaby O, Sobkova K, Svoboda M et al. Significant overexpression of Hsp110 gene during colorectal cancer progression. Oncol Rep 2009;21:1235–1241.
Hwang TS, Han HS, Choi HK et al. Differential, stage-dependent expression of Hsp70, Hsp110 and Bcl-2 in colorectal cancer. J Gastroenterol Hepatol 2003;18:690–700.
Van Rijnsoever M, Elsaleh H, Joseph D et al. CpG island methylator phenotype is an independent predictor of survival benefit from 5-fluorouracil in stage III colorectal cancer. Clin Cancer Res 2003;9:2898–2903.
Min BH, Bae JM, Lee EJ et al. The CpG island methylator phenotype may confer a survival benefit in patients with stage II or III colorectal carcinomas receiving fluoropyrimidine-based adjuvant chemotherapy. BMC Cancer 2011;11:344.
Jover R, Nguyen TP, Perez-Carbonell L et al. 5-Fluorouracil adjuvant chemotherapy does not increase survival in patients with CpG island methylator phenotype colorectal cancer. Gastroenterology 2011;140:1174–1181.
Shen L, Catalano PJ, Benson AB 3rd et al. Association between DNA methylation and shortened survival in patients with advanced colorectal cancer treated with 5-fluorouracil based chemotherapy. Clin Cancer Res 2007;13:6093–6098.
Han SW, Lee HJ, Bae JM et al. Methylation and microsatellite status and recurrence following adjuvant FOLFOX in colorectal cancer. Int J Cancer 2013;132:2209–2216.
Acknowledgements
We thank Professors Woo Ho Kim, Tae-You Kim and Seung-Yong Jeong, from the Departments of Pathology, Internal Medicine and Surgery, respectively, of Seoul National University College of Medicine, for their insightful comments and advice. This study was supported by a grant from the National R&D Program for Cancer Control, Ministry of Health and Welfare, Republic of Korea (0720540), a grant from Korea Healthcare Technology R&D Project, Ministry for Health, Welfare & Family Affairs, Republic of Korea (A091081), Priority Research Centers Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (MEST; 2009-0093820), and the Mid-career Researcher Program through an NRF grant funded by MEST (2011-0015646).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Rights and permissions
About this article
Cite this article
Kim, J., Kim, KJ., Rhee, YY. et al. Expression status of wild-type HSP110 correlates with HSP110 T17 deletion size and patient prognosis in microsatellite-unstable colorectal cancer. Mod Pathol 27, 443–453 (2014). https://doi.org/10.1038/modpathol.2013.160
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/modpathol.2013.160
Keywords
This article is cited by
-
Consequences of the Hsp110DE9 mutation in tumorigenesis and the 5-fluorouracil-based chemotherapy response in Msh2-deficient mice
Cellular and Molecular Life Sciences (2022)
-
SMOC2, an intestinal stem cell marker, is an independent prognostic marker associated with better survival in colorectal cancers
Scientific Reports (2020)
-
Systematic review of the predictive effect of MSI status in colorectal cancer patients undergoing 5FU-based chemotherapy
BMC Cancer (2015)