Abstract
Regulatory T cells (Treg cells), which have abundant expression of the interleukin 2 receptor (IL-2R), are reliant on IL-2 produced by activated T cells. This feature indicates a key role for a simple network based on the consumption of IL-2 by Treg cells in their suppressor function. However, congenital deficiency in IL-2R results in reduced expression of the Treg cell lineage–specification factor Foxp3, which has confounded experimental efforts to understand the role of IL-2R expression and signaling in the suppressor function of Treg cells. Using genetic gain- and loss-of-function approaches, we found that capture of IL-2 was dispensable for the control of CD4+ T cells but was important for limiting the activation of CD8+ T cells, and that IL-2R-dependent activation of the transcription factor STAT5 had an essential role in the suppressor function of Treg cells separable from signaling via the T cell antigen receptor.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Sakaguchi, S., Sakaguchi, N., Asano, M., Itoh, M. & Toda, M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor α-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J. Immunol. 155, 1151–1164 (1995).
Hori, S., Nomura, T. & Sakaguchi, S. Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003).
Fontenot, J.D., Gavin, M.A. & Rudensky, A.Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4, 330–336 (2003).
Waldmann, T.A. The multi-subunit interleukin-2 receptor. Annu. Rev. Biochem. 58, 875–911 (1989).
Furtado, G.C., Curotto de Lafaille, M.A., Kutchukhidze, N. & Lafaille, J.J. Interleukin 2 signaling is required for CD4+ regulatory T cell function. J. Exp. Med. 196, 851–857 (2002).
Almeida, A.R., Legrand, N., Papiernik, M. & Freitas, A.A. Homeostasis of peripheral CD4+ T cells: IL-2R alpha and IL-2 shape a population of regulatory cells that controls CD4+ T cell numbers. J. Immunol. 169, 4850–4860 (2002).
Malek, T.R., Yu, A., Vincek, V., Scibelli, P. & Kong, L. CD4 regulatory T cells prevent lethal autoimmunity in IL-2Rβ-deficient mice. Implications for the nonredundant function of IL-2. Immunity 17, 167–178 (2002).
Fontenot, J.D., Rasmussen, J.P., Gavin, M.A. & Rudensky, A.Y. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat. Immunol. 6, 1142–1151 (2005).
Burchill, M.A., Yang, J., Vogtenhuber, C., Blazar, B.R. & Farrar, M.A. IL-2 receptor β-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J. Immunol. 178, 280–290 (2007).
Yao, Z. et al. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood 109, 4368–4375 (2007).
Malek, T.R. & Bayer, A.L. Tolerance, not immunity, crucially depends on IL-2. Nat. Rev. Immunol. 4, 665–674 (2004).
Vang, K.B. et al. IL-2, -7, and -15, but not thymic stromal lymphopoeitin, redundantly govern CD4+Foxp3+ regulatory T cell development. J. Immunol. 181, 3285–3290 (2008).
Chen, W. et al. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J. Exp. Med. 198, 1875–1886 (2003).
Malin, S. et al. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat. Immunol. 11, 171–179 (2010).
Barron, L. et al. Cutting edge: mechanisms of IL-2-dependent maintenance of functional regulatory T cells. J. Immunol. 185, 6426–6430 (2010).
Setoguchi, R., Hori, S., Takahashi, T. & Sakaguchi, S. Homeostatic maintenance of natural Foxp3+ CD25+ CD4+ regulatory T cells by interleukin (IL)-2 and induction of autoimmune disease by IL-2 neutralization. J. Exp. Med. 201, 723–735 (2005).
Rubtsov, Y.P. et al. Stability of the regulatory T cell lineage in vivo. Science 329, 1667–1671 (2010).
Komatsu, N. et al. Heterogeneity of natural Foxp3+ T cells: a committed regulatory T-cell lineage and an uncommitted minor population retaining plasticity. Proc. Natl. Acad. Sci. USA 106, 1903–1908 (2009).
Hori, S. Lineage stability and phenotypic plasticity of Foxp3+ regulatory T cells. Immunol. Rev. 259, 159–172 (2014).
Pandiyan, P., Zheng, L., Ishihara, S., Reed, J. & Lenardo, M.J. CD4+CD25+Foxp3+ regulatory T cells induce cytokine deprivation-mediated apoptosis of effector CD4+ T cells. Nat. Immunol. 8, 1353–1362 (2007).
Thornton, A.M., Donovan, E.E., Piccirillo, C.A. & Shevach, E.M. Cutting edge: IL-2 is critically required for the in vitro activation of CD4+CD25+ T cell suppressor function. J. Immunol. 172, 6519–6523 (2004).
Barthlott, T. et al. CD25+CD4+ T cells compete with naive CD4+ T cells for IL-2 and exploit it for the induction of IL-10 production. Int. Immunol. 17, 279–288 (2005).
Busse, D. et al. Competing feedback loops shape IL-2 signaling between helper and regulatory T lymphocytes in cellular microenvironments. Proc. Natl. Acad. Sci. USA 107, 3058–3063 (2010).
Yamaguchi, T. et al. Construction of self-recognizing regulatory T cells from conventional T cells by controlling CTLA-4 and IL-2 expression. Proc. Natl. Acad. Sci. USA 110, E2116–E2125 (2013).
Smigiel, K.S. et al. CCR7 provides localized access to IL-2 and defines homeostatically distinct regulatory T cell subsets. J. Exp. Med. 211, 121–136 (2014).
Zambrowicz, B.P. et al. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of β-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl. Acad. Sci. USA 94, 3789–3794 (1997).
Onishi, M. et al. Identification and characterization of a constitutively active STAT5 mutant that promotes cell proliferation. Mol. Cell. Biol. 18, 3871–3879 (1998).
Boyman, O., Kovar, M., Rubinstein, M.P., Surh, C.D. & Sprent, J. Selective stimulation of T cell subsets with antibody-cytokine immune complexes. Science 311, 1924–1927 (2006).
Chen, Y. et al. Foxp3+ regulatory T cells promote T helper 17 cell development in vivo through regulation of interleukin-2. Immunity 34, 409–421 (2011).
Cong, Y., Feng, T., Fujihashi, K., Schoeb, T.R. & Elson, C.O. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc. Natl. Acad. Sci. USA 106, 19256–19261 (2009).
Kim, J.M., Rasmussen, J.P. & Rudensky, A.Y. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 8, 191–197 (2007).
Su, L.K. et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science 256, 668–670 (1992).
Arvey, A. et al. Inflammation-induced repression of chromatin bound by the transcription factor Foxp3 in regulatory T cells. Nat. Immunol. 15, 580–587 (2014).
Levine, A.G., Arvey, A., Jin, W. & Rudensky, A.Y. Continuous requirement for the TCR in regulatory T cell function. Nat. Immunol. 15, 1070–1078 (2014).
Vahl, J.C. et al. Continuous T cell receptor signals maintain a functional regulatory T cell pool. Immunity 41, 722–736 (2014).
Tadokoro, C.E. et al. Regulatory T cells inhibit stable contacts between CD4+ T cells and dendritic cells in vivo. J. Exp. Med. 203, 505–511 (2006).
Lio, C.W. & Hsieh, C.S. A two-step process for thymic regulatory T cell development. Immunity 28, 100–111 (2008).
Feng, Y. et al. Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell 158, 749–763 (2014).
Tran, D.Q. et al. Analysis of adhesion molecules, target cells, and role of IL-2 in human FOXP3+ regulatory T cell suppressor function. J. Immunol. 182, 2929–2938 (2009).
Rudra, D. et al. Runx-CBFbeta complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat. Immunol. 10, 1170–1177 (2009).
Sojka, D.K., Bruniquel, D., Schwartz, R.H. & Singh, N.J. IL-2 secretion by CD4+ T cells in vivo is rapid, transient, and influenced by TCR-specific competition. J. Immunol. 172, 6136–6143 (2004).
Shanmugasundaram, R. & Selvaraj, R.K. Regulatory T cell properties of chicken CD4+CD25+ cells. J. Immunol. 186, 1997–2002 (2011).
Andersen, K.G., Nissen, J.K. & Betz, A.G. Comparative genomics reveals key gain-of-function events in Foxp3 during regulatory T cell evolution. Front. Immunol. 3, 113 (2012).
Rubtsov, Y.P. et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 28, 546–558 (2008).
Anders, S. et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 8, 1765–1786 (2013).
Bolger, A.M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Love, M.I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Tarca, A.L. et al. A novel signaling pathway impact analysis. Bioinformatics 25, 75–82 (2009).
Maere, S., Heymans, K. & Kuiper, M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 (2005).
Merico, D., Isserlin, R., Stueker, O., Emili, A. & Bader, G.D. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 5, e13984 (2010).
Acknowledgements
We thank T. Kitamura (The University of Tokyo) for the mSTAT5b-CA vector (pMX-STAT5B(1*6)); K. Rajewsky (Max Delbrück Center) for the Rosa26 construct; and L. Henninghausen (US National Institutes of Health) for Stat5a-Stat5bfl mice. Supported by the Japan Society for the Promotion of Science, Strategic Young Researcher Overseas Visits Program for Accelerating Brain Circulation (T.C.), Lucille Castori Center for Microbes, Inflammation & Cancer (T.C.), the US National Institutes of Health Medical Scientist Training Program (T32GM07739 to the Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD-PhD Program, for A.G.L. and X.F.), the US National Institutes of Health (P30 CA008748 to Memorial Sloan Kettering Cancer Center Core Facilities; and R37 AI034206 to A.Y.R.), DFG Emmy Noether programme (G.G.), the Ludwig Center at Memorial Sloan Kettering Cancer Center (A.Y.R.), Hilton-Ludwig Cancer Prevention Initiative funded by the Conrad N. Hilton Foundation and Ludwig Cancer Research (A.Y.R.), and the Howard Hughes Medical Institute (A.Y.R.).
Author information
Authors and Affiliations
Contributions
T.C., J.D.F. and A.Y.R. conceived of the project, designed the experiments and wrote and edited the manuscript.; T.C., A.K.K., A.G.L., X.F., Y.Z., G.G. and Y.F. conducted experiments; U.K. generated the Il2rbfl allele; and J.D.F. generated the Il2rafl allele.
Corresponding authors
Ethics declarations
Competing interests
A.K.K. and J.D.F. were employees of Biogen during the completion of this work; A.K.K. is an employee of AbbVie; and J.D.F. is an employee of Juno Therapeutics.
Integrated supplementary information
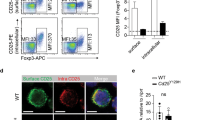
Supplementary Figure 1 IL-2R signaling is indispensable for the maintenance and function of Treg cells.
(a) The analysis shown in Fig. 1j performed by gating on CD62LhiCD44lo (upper panels) and CD62LloCD44hi (lower panels) YFP+Foxp3+ Treg cell subsets. (b) The expressions of CD62L and CD44 in CD3+CD4+Foxp3+ (upper panels) and frequencies of Foxp3+ cells among CD3+CD4+ cells (lower panels) in the spleen and small intestine lamina propria lymphocytes (SILPL) of the indicated mice (5 wk old). The right graph shows the summary data. (c) The expression of indicated markers by splenic CD3+CD4+Foxp3+ cells in the indicated mice (5 wk old). (d) Lifespan of Foxp3CreIl2rafl/fl (solid; n=25) and Foxp3CreIl2rafl/wt (dotted; n=20) mice. (e) Analysis of LN cellularity, Foxp3 expression levels (MFI) and frequencies of Foxp3+ Treg cells among CD3+CD4+ cells (upper panels) and pro-inflammatory cytokine production by CD4+Foxp3− and CD8+Foxp3− cells (lower panels) in the indicated mice (4 wk old). (f) Histopathology of the indicated organs of 4-wk-old Foxp3CreIl2rafl/fl mice. Scale bar, 100 μm. (g) The expression of Foxp3 and CD25 in LN CD4+ T cells in the indicated mice (6 wk old). The lower histogram represents the expression levels of CD25 in Foxp3+ cells shown in the upper panels. (h) The expression of CD62L and CD44 in LN CD3+CD4+Foxp3− (upper panels) and CD3+CD8+Foxp3− (lower panels) cells. (i, j) Cytokine production by splenic CD4+Foxp3− (i) and CD8+Foxp3− (j) cells isolated from indicated mice and in vitro stimulated with anti-CD3/CD28 for 5 hr. (k) Histopathology of the indicated organs of 4-wk-old Foxp3CreStat5a/bfl/fl mice. Scale bar, 100 μm. Sex and age matched mice were analyzed. Cells were analyzed by flow cytometry. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (two-tailed unpaired Student’s t test). Data are from one experiment representative of three (a) or two (b, c, e) independent experiments with similar results with three or more mice per group in each experiment (a, b, c, e; each dot represents a single mouse; mean ± s.e.m.) or representative data of n=6 (b), n=3 (f), n=8 (g, h, i, j), n=5 (k) mice per group analyzed are shown.
Supplementary Figure 2 Restoration of the suppressor activity of IL-2Rα-deficient Treg cells by expression of an active form of STAT5.
(a) The schematic of Rosa26Stat5bCA allele. CAG, cytomegalovirus early enhancer element with the promoter, the first exon and the first intron of chicken beta-actin gene and the splice acceptor of the rabbit beta-globin gene; IRES, internal ribosome entry site; EGFP, enhanced green fluorescent protein (b, c) Photographs of 4-wk-old mice. (d) The expression of Foxp3 and CD25 in CD3+CD4+ cells in the LNs and spleens of the indicated mice (4 wk old). (e) Intracellular phospho-STAT5 levels in Treg cells from the indicated mice in vitro stimulated with rmIL-2 (1,000 U/ml) for 20 min. (f) The expression of CD62L and CD44 in CD3+CD4+Foxp3− and CD3+CD8+Foxp3− cells in the LNs of the indicated mice. (g) Cytokine production by splenic CD4+Foxp3− cells stimulated for 5 hr with anti-CD3/CD28. (h) The numbers of CD3+CD4+Foxp3−IFN-γ+ (left graph) and CD3+CD8+Foxp3−IFN-γ+ (right graph) cells in the LNs of the indicated mice (4 wk old). (i) The frequencies CD44hi cells among CD4+Foxp3− cells (upper left graph) and the frequencies of CD62LloCD44hi and CD62LhiCD44hi cells among CD8+Foxp3− cells (upper right graph) in the LNs of Foxp3CreIl2rafl/wt, Foxp3CreIl2rafl/wtRosa26Stat5bCA, Foxp3CreIl2rafl/fl, and Foxp3CreIl2rafl/flRosa26Stat5bCA mice at the indicated ages. Lower graphs show the numbers of each cell subset in the LNs of indicated mice. (j) Lifespan of Foxp3CreIl2rafl/fl (solid; n=25) and Foxp3CreIl2rafl/flRosa26Stat5bCA (dotted; n=21) mice. ***, P < 0.001 (two-tailed unpaired Student’s t test). Data are from one experiment representative of two (e, i) or three (h) independent experiments with similar results with three or more mice per group in each experiment (h, i; mean ± s.e.m.) or representative data of more than six mice per group analyzed are shown (d, f, g).
Supplementary Figure 3 Effects of in vivo neutralization of IL-2 on the activation of CD4+ or CD8+ cells.
(a) Representative flow cytometric analyses for the experiment shown in Fig. 2e. (b) LN cells of Foxp3Cre (upper 6 panels) and Foxp3CreIl2rbfl/fl (lower 8 panels) mice were unstimulated (US) or stimulated with rmIL-2 (1,000 or 10 U/ml) for 20 min, and intracellular levels of phospho-STAT5 in Treg (CD4+YFP+CD25hi), T naive (YFP−CD44loCD25lo; CD4+ and CD8+), and Teff (YFP−CD44hi; CD25lo and CD25hi; CD4+ and CD8+) cells were analyzed by flow cytometry. Data are representative of two independent experiments with three biological replicates in each experiment.
Supplementary Figure 4 Characterization of mice with Treg cells expressing an active form of STAT5.
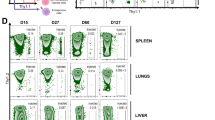
(a) The frequencies of STAT5b-CA-expressing CD4+Foxp3+ cells among total CD4+Foxp3+ cells in the spleen of Foxp3Cre-ERT2Rosa26Stat5bCA mice after a single tamoxifen gavage on day 0. (b) The frequencies of STAT5b-CA-expressing CD4+Foxp3+ cells among total CD4+Foxp3+ cells in the indicated organs of Foxp3Cre-ERT2Rosa26Stat5bCA mice three months after a single tamoxifen gavage. (c) TCR Vβ usage of the Treg cells in various tissues 2 months after tamoxifen gavage. MLNs, mesenteric lymph nodes; PPs, Peyer’s patches. (d) Changes in body weights after tamoxifen gavage. 4-month-old Foxp3Cre-ERT2 (black, n=7) and Foxp3Cre-ERT2Rosa26Stat5bCA (blue, n=7) mice were gavaged with tamoxifen and body weights were monitored the following 4 months. (e) Serum chemistry profiles 4.5 months after tamoxifen gavage. (f–h) General characterization of Treg cells of Foxp3Cre-ERT2 (black) and Foxp3Cre-ERT2Rosa26Stat5bCA (blue) mice three months after a single tamoxifen gavage. (f) The expression levels of indicated molecules on CD4+Foxp3+ cells in the indicated organs. (g) The frequencies of Foxp3+ cells among CD3+CD4+ cells (upper graph) and the expression levels of Foxp3 in the CD3+CD4+Foxp3+ cells (lower graph) in the indicated organs. (h) The frequencies of Foxp3+ cells among CD3+CD8+ cells in the indicated organs. (i) In vitro suppressor activity of Treg cells. Treg cells were isolated from Foxp3Cre-ERT2 (control) and Foxp3Cre-ERT2Rosa26Stat5bCA (STAT5b-CA) mice and co-cultured with T naive cells (responder cells). The proliferative activities of Treg and responder cells were determined by flow cytometry based on the dilution of CellTrace Violet (CTV) fluorescence intensity. Typical dye dilution patterns of T naive cells at a 4:1 responder vs. Treg cell ratio are shown in the left two panels. Summary graphs showing the proliferation of co-cultured responder T cells (Foxp3−) and Treg cells (Foxp3+) are shown in the right two panels. CTV MFI of cells inversely correlates with cell division. Cells were analyzed by flow cytometry (a–c, f–i). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (two-tailed unpaired Student’s t test). Data are from one experiment representative of two (c, d, i) or three or more (a, b, f–h) independent experiments with similar results with three or more biological replicates per group in each experiment. Each dot represents a single mouse (b, e–h). mean ± s.e.m. (a, b, d–i).
Supplementary Figure 5 Systemic reduction in the Teff cell population in the presence of STAT5b-CA+ Treg cells.
(a–d) Analysis of Foxp3Cre-ERT2 (black dots) and Foxp3Cre-ERT2Rosa26Stat5bCA (blue dots) mice three months after a single tamoxifen treatment. (a, b) The frequencies of Ki-67+ (upper graphs), CD62LhiCD44lo (middle; % T naive), and CD62LloCD44hi (lower; % Teff) cells among CD4+Foxp3− (a) and CD8+Foxp3− (b) cells of the indicated organs. (c) Cytokine production by CD4+Foxp3− cells in the spleen and mesenteric LN cells stimulated with anti-CD3/CD28 for 5 hr. (d) Serum Ig levels determined by ELISA. (e) The numbers and sizes of polyps in the distal small intestines of 5-month-old Foxp3Cre-ERT2ApcMin/+ and Foxp3Cre-ERT2Rosa26Stat5bCAApcMin/+ mice treated with tamoxifen at 4 wk of age. Cells were analyzed by flow cytometry (a–c). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (two-tailed unpaired Student’s t test). Data are from one experiment representative of three or more independent experiments with similar results with three or more mice per group in each experiment (a–c) or pooled samples with n=10 or more were analyzed (d, e). Each dot represents a single mouse (a–e). mean ± s.e.m. (a–e).
Supplementary Figure 6 RNA-seq analysis (supplementary information for Fig. 5)
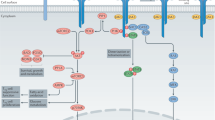
(a) Plot of gene expression (as log2 normalized read count) in control T naive versus STAT5b-CA T naive cells (i.e., naive CD4+ T cells from Foxp3Cre-ERT2Rosa26Stat5bCA mice). The diagonal lines indicate fold change of at least 1.5× or 0.67× fold. Significantly up- and down-regulated genes (defined as genes with at least 1.5× or 0.67× fold change, adjusted P-value ≤ 0.05, and expression above a minimal threshold based on the distribution of all genes) are colored red or blue, respectively, and their numbers are shown. (b) Volcano plot showing log10 FDR-adjusted P-values versus log2 fold change between STAT5b-CA and control Treg cells. Genes that fall outside of the x- or y-axis range of this plot are shown on the axes as empty triangles. The vertical and horizontal gray lines indicate 1.5× or 0.67× fold change (±log2 1.5 = ±0.58) and P = 0.05 (−log10 0.05 = 1.3), respectively. (c) Network analysis of GO term enrichment among significantly downregulated genes in STAT5b-CA expressing vs. control Treg cells. Downregulated genes were analyzed for over-represented GO terms using BiNGO in Cytoscape, and the resulting network was calculated and visualized using EnrichmentMap. Groups of similar GO terms were manually circled. Edge thickness and color are proportional to the similarity coefficient between connected gene sets. Node color is proportional to the FDR-adjusted P-value of the enrichment. Node size is proportional to gene set size.
Supplementary Figure 7 Strategies for targeting Il2rbfl and Rosa26Stat5bCA alleles.
(a) Generation of a conditional Il2rb allele and Il2rb targeting strategy. The targeting vector was constructed such that upon Cre-mediated deletion, the promoter region and exon 2 which comprises the first ATG of Il2rb were deleted with simultaneous activation of eGFP expression. Shown from top to bottom i) the Il2rb locus with the promoter region, exons and translational start site in exon 2 (E2); ii) the targeting vector comprising an eGFP, a triple SV40 polyA site (tpA), a PGK neopA cassette, a PGK promoter (Pr.) downstream of exon 2, a TK gene, and loxP and frt sites; arrows denote the orientation; iii) the targeted Il2rb locus. Restriction sites, probes used for detection and the expected fragments detected by Southern blot analysis are indicated. Correctly targeted embryonic stem (ES) cell lines were identified by Southern blot analysis of XbaI-digested DNA that displayed the 4.0 kb band of the integrated transgene along with the 14.0 kb wild-type band. Co-integration of the 3’ loxP site was verified by PCR analysis using primers that hybridize in a unique region spanning the PGK promoter and the 3’ frt site (forward primer) and in a region upstream of intron 3 of Il2rb (reverse primer). (b) The schematic of, and targeting strategy for Rosa26Stat5bCA allele. The targeting vector was constructed such that CAG promoter driven STAT5b-CA is expressed upon Cre-mediated deletion of a STOP cassette. Correctly targeted ES cell lines were identified by Southern blot analysis of EcoRI-digested DNA that displayed the 5.9 kb (probe A; 5’ side) and 11.6 kb (probe F; 3’ side) bands of the integrated transgene along with the 15.6 kb wild-type band (probe A and F; both sides).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Tables 1 and 2 (PDF 3584 kb)
Rights and permissions
About this article
Cite this article
Chinen, T., Kannan, A., Levine, A. et al. An essential role for the IL-2 receptor in Treg cell function. Nat Immunol 17, 1322–1333 (2016). https://doi.org/10.1038/ni.3540
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ni.3540
This article is cited by
-
Thymic gene expression analysis reveals a potential link between HIF-1A and Th17/Treg imbalance in thymoma associated myasthenia gravis
Journal of Neuroinflammation (2024)
-
Targeting immune cell recruitment in atherosclerosis
Nature Reviews Cardiology (2024)
-
AIH Therapy: Beyond First-Line
Current Hepatology Reports (2024)
-
Evaluation of regulatory T-cells in cancer immunotherapy: therapeutic relevance of immune checkpoint inhibition
Medical Oncology (2024)
-
New insights into the stemness of adoptively transferred T cells by γc family cytokines
Cell Communication and Signaling (2023)