Abstract
Suicide is a major public health concern, and the number of deaths by suicide has been increasing in recent years in the US. There are various biological risk factors for suicide, but causal molecular mechanisms remain unknown, suggesting that investigation of novel mechanisms and integrative approaches are necessary. Transfer (t)RNAs and their modifications, including cytosine methylation (m5C), have received little attention regarding their role in normal or diseased brain function, though they are dynamic mediators of protein synthesis. tRNA regulation is highly interconnected with proteomic and metabolomic outcomes, suggesting that investigating these multiple levels of molecular regulation together may elucidate more information on neural function and suicide risk. In the current study, we used an integrative ‘omics’ approach to probe tRNA dysregulation, including tRNA expression and tRNA m5C, proteomics, and amino acid metabolomics in prefrontal cortex from 98 subjects who died by suicide during an episode of major depressive disorder (MDD) and neurotypical controls. While no changes were detected in amino acid content, results showed increased tRNAGlyGCC expression in the suicide brain that is not driven by changes in m5C. Proteomics revealed increased expression of proteins with high glycine codon GGC content, demonstrating a strong association between isoacceptor-specific tRNA expression and proteomic outcomes in the suicide brain, which is in line with previous work linking tRNAGly with alterations in glycine-rich proteins in a translational rodent model of depression. Further, we confirmed using a rodent model that tRNAGlyGCC overexpression was sufficient to increase the expression of proteins with high glycine codon GGC content that were upregulated in the suicide brain. By characterizing the effects of MDD-suicide in human PFC tissue, we now begin to elucidate a novel molecular signature with downstream consequences for psychiatric outcomes.
Similar content being viewed by others
Introduction
Suicide significantly contributes to mortality among younger age groups, and the number of suicide deaths has been increasing in the past 20–30 years [1]. While most often associated with diagnoses of Major Depressive Disorder (MDD), the etiology of suicidal behavior is still unknown, although various molecular mechanisms have been investigated throughout the years in post-mortem brains of subjects who died by suicide [2, 3], including genetic and epigenetic underpinnings [2, 4, 5], neurotransmitter abnormalities [6], and alterations in neural circuitry [2]. However, no single mechanism has been sufficient to explain the complex cause of suicidal behavior, suggesting that novel types of molecular pathways should be investigated.
Transfer (t)RNAs are small 70–90 nucleotide clover-leaf shaped RNAs that serve as mediators of protein translation through complementary binding to mRNA molecules and subsequent deposition of an amino acid onto the growing peptide chain during protein synthesis. In addition to canonical functions in protein synthesis, tRNAs have been linked to several non-canonical functions mostly related to smaller RNA molecules derived from full length tRNAs, named tRNA fragments (tRFs), which have been linked to translational inhibition, gene silencing, and retroelement control, regulation of the immune response and cell death, and other functions [7]. Notably, tRNAs are unique in that they contain abundant epitranscriptomic modifications that exhibit dynamic patterns of regulation that contribute to tRNA stability and formation of tRFs.
One unexpected and surprising finding emerging from clinical genome sequencing is that many deleterious mutations affecting tRNAs and their structural and functional regulators are associated with predominantly neurological and even psychiatric phenotypes [8]. For example, some patients with heterozygous mutations in AARS2 alanyl-tRNA-synthetase initially develop symptoms including frontal lobe dysfunction, depression, and other psychiatric features while in their 20s and 30s [9]. Therefore, exploration of the tRNA regulome could open radically new avenues and treatment options for common mood and psychosis spectrum disorders. While tRNA modifications have not been explored in the human brain, even studies of full-length tRNA expression have been sparse and inconsistent, due to large input requirements and lack of specialized techniques for tRNA sequencing library preparations [10].
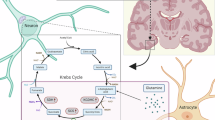
Our group recently identified a role for an abundant tRNA modification, cytosine methylation (m5C), in brain function and neuropsychiatric phenotypes using a mouse model. Specifically, knockout of the main mammalian tRNA methyltransferase, Nsun2, in the adult mouse brain reduced mature glycine tRNA (tRNAGly) expression and impaired translation of glycine-rich synaptic proteins while concurrently increasing glycine amino acid levels [11]. These findings align with previous work that identifies Nsun2-mediated tRNA methylation as a metabolic sensor of external cellular stress, providing signals to tightly regulate translation during oxidative stress [12,13,14]. Patients with a history of suicidal ideation show increased peripheral markers for oxidative stress [15] and proteomic abnormalities in the periphery [16, 17] and in the postmortem suicide brain [18,19,20] Our previous study also identified a bidirectional link between prefrontal cortex tRNA m5C levels, tRNA expression, and depressive-like behavior in mice [11], suggesting that there may be detectable changes to m5C and tRNA expression in brains of patients who died by suicide during an episode of MDD. Therefore, the current study aimed to elucidate the role of the tRNA regulome on proteomic outcomes associated with suicide by integrating multiple approaches of tRNA quantification, proteomics, and metabolomics to identify molecular changes in post-mortem dorsolateral prefrontal cortex (PFC) of subjects who died by suicide during an episode of MDD and healthy comparison subjects (see Fig. 1 for schematic).
Dorsolateral prefrontal cortex (DLPFC) was dissected from fresh frozen human postmortem brains. Samples included subjects who died by suicide during an episode of MDD (MDD suicide) and neurotypical comparison subjects. Tissue was used for four molecular assays, including tRNA sequencing, tRNA methylation profiling, proteomics, and amino acid metabolomics.
Methods
All methods were performed in accordance with the relevant guidelines and regulations.
Ethics approval and consent to participate
For human tissue, written informed consent for post-mortem brain donation was provided to the brain bank from the donors’ families. Samples were deidentified before being transferred to Icahn School of Mount Sinai and therefore exempt from the purview of the Institutional Review Board.
For animals, tRNAGly-high mice were generated at the Radboud University (Nijmegen, Netherlands) and all work was approved by the national Dutch ethics committee ‘Centrale Commissie Dierproeven’ (AVD1030020184826/2017-0067).
Human samples
Postmortem brain tissue was obtained from the Douglas-Bell Canada Brain Bank (Montreal, Canada). A total of 98 fresh frozen brain samples from the dorsolateral prefrontal cortex (Brodmann area 9) were received (n = 49 MDD suicide, 49 healthy comparison). Preliminary data from our lab for human brain cytosine methylation was initially used for statistical power calculations using G*power software. 28 samples per group was sufficient to provide an effect size (d) of 0.77 (from means and SDs of control and experimental groups), with alpha = 0.05 with a power of 0.8. Because we already obtained 49 samples per condition to further separate the suicide group by antidepressant treatment and/or childhood trauma for additional effects, we confirmed sufficient power for molecular analyses. Sample characteristics are presented in Table 1. We note that sample sizes differ slightly for each assay (presented in Results) as quality of RNA, protein, or sequencing library determined exclusion of certain samples from analysis. During all molecular assay preparation and initial data analysis, experimenter was blind to condition. Age, PMI, and pH between the cohort of suicide and control samples were compared using the Mann–Whitney U test. Gender, antidepressant use, and childhood abuse between the suicide and control groups were compared with the binomial test (Table 1).
RNA isolation
Fresh frozen post-mortem tissue was homogenized with a vibrating pestle and total RNA was extracted using Trizol reagent according to standard protocols. Total RNA was eluted in nuclease-free water, quantified with the Qubit fluorometer (Invitrogen, Carlsbad, CA), and stored at −80 °C for later processing.
tRNA bisulfite sequencing
tRNA bisulfite sequencing was performed as described in Blaze et al. [11, 21] and previously adapted from Bormann et al [22]. Total RNA was bisulfite converted with the EZ RNA Methylation kit (Zymo Research) according to manufacturer’s instructions but including an additional 2 PCR cycles to denature tRNAs completely, as previously reported [14]. We then performed cDNA synthesis on bisulfite-treated RNA using the reverse primer for the tRNA isodecoder of interest, GlyGCC-1 [11, 12, 21] followed by PCR amplification with the forward primer. tRNA amplicons were run on a 4% agarose E-gel (Invitrogen) and appropriately sized bands were extracted and purified (Qiaquick Gel Extraction Kit, Qiagen). Amplicon libraries were indexed (Nextera XT, Illumina) and pooled at equimolar concentrations followed by sequencing with 75 bp paired-end reads on the MiSeq (Illumina).
Data analysis
Raw fastq files from bisulfite sequencing data were analyzed using BisAmp [22], a publicly available web server for quantification of methylated reads after sequencing. For each cytosine site, methylation percentages were calculated and analyzed using two approaches. First, a t-test was applied to compare methylation proportions between control and suicide groups. To account for multiple comparisons, the false discovery rate (FDR) was controlled using the two-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli [23, 24]. To further assess the relationship between suicide and cytosine methylation levels, a generalized linear model (GLM) was employed using the following specification:
Here, the model included fixed effects for condition (control vs. suicide), gender, age, postmortem interval (PMI), and brain pH, along with interaction terms between condition and these covariates. The GLM was run using the default settings in R, which assume a Gaussian distribution and treat methylation percentages as continuous variables.
YAMAT/UMI sequencing
Y-shaped Adapter MAture tRNA (YAMAT) sequencing with unique molecular identifiers (UMIs) was performed as previously described [11, 21] and originally adapted from Shigematsu and colleagues [25]. 10 μg of total RNA was deacylated to remove amino acids from 3′ ends and pre-treated to remove m1A, m1C, and m3C modifications using a proprietary demethylation mix (Arraystar, Inc). Pre-treated RNA was purified and forked linkers were added followed by the addition of 10X annealing buffer (50 mM Tris HCl pH 8, 100 mM MgCl2, 5 mM EDTA) and incubated overnight with T4 RNA ligase 2. Linker-ligated RNA was incubated with RT Primer (TruSeq Small RNA Library Prep Kit, Illumina) and reverse transcribed with Superscript III RT (Invitrogen) followed by bead purification. Libraries were amplified using Phusion Hotstart II Polymerase (Thermo Scientific) with primers and indexes from the TruSeq Small RNA kit (Illumina) followed by bead purification and quantification on the Qubit Fluorometer (Invitrogen). Confirmation of library quality and size was performed on the Agilent Bioanalyzer (Agilent Technologies) and 8–10 libraries were pooled at equimolar concentrations to sequence with 75 bp paired-end reads on the MiSeq (Illumina).
Data analysis
Raw sequencing reads were initially processed with UMItools for UMI extraction, followed by Cutadapt to trim adapter sequences, and then deduplication based on the extracted UMIs [26]. Paired-end reads were subsequently merged using PEAR [27] and aligned against a human tRNA reference genome via Bowtie2 [28]. For downstream analyses, only tRNA isodecoders with a minimum summed read count of 30 from all samples were considered. Differential expression between the control and suicide groups was analyzed using limma [29], controlling for confounding variables such as gender, age, post-mortem interval, and pH. The False Discovery Rate (FDR) was estimated using bootstrap resampling, performed 10,000 times. tRNA isodecoders were identified as significantly differentially expressed with a raw or adjusted p-value of less than 0.1 and an absolute fold change of at least 1.5.
To analyze the effect of each confounding variable, log-transformed counts per million (CPM) values for each tRNA isodecoder were calculated to normalize the data. Permutational multivariate analysis of variance using the adonis function in R was employed to assess the effects of gender, age, postmortem interval (PMI), pH, antidepressant use, and childhood abuse on tRNA expression levels. The analysis was conducted with the following model:
The Euclidean distance metric was used, and 10,000 permutations were applied to ensure robust statistical inference.
Amino acid metabolomics
Unbiased metabolomic profiling of amino acids was performed as described previously [11]. Briefly, fresh frozen post-mortem tissue was homogenized with a vibrating pestle on ice in 80% methanol with heavy amino acid mix standards (Cambridge Isotope Laboratories MSK-A2-1.2 mix). After centrifugation, supernatant was placed into a new tube and dried using a SpeedVac before storage at -80 °C for later processing. Amino acid metabolomics and data analysis were performed at the Rockefeller Proteomics Resource Center (New York, NY) as previously described [11].
LC-MS/MS
Digestion
Fresh frozen postmortem tissue was homogenized with a vibrating pestle in buffer containing SDS, HEPES, sucrose, and protease inhibitors and quantified using a BCA assay (ThermoFisher). LC-MS/MS was performed at the Rockefeller Proteomics Resource Center (New York, NY) as previously described [11].
Data analysis
Raw data was searched using MaxQuant v. 1.6.6.0 using standard settings. Spectra were queried against the Homo sapien proteome (obtained from uniprot.org) with a false discovery rate (FDR) of 1% applied on PSM, peptide and protein level. Ratio-compression from co-fragmention was minimized by requiring a minimum peptide interference factor (PIF) of 0.75. Further data analysis was performed within the Perseus framework. Raw intensity data underwent differential analysis using limma [29]. Confounding variables including gender, age, post-mortem interval, and pH were accounted for when assessing differential expression between the control and suicide groups. The False Discovery Rate (FDR) was calculated using bootstrap resampling, conducted 10,000 times. Proteins were considered significantly differentially expressed if they had an adjusted p-value of less than 0.1 and an absolute fold change of at least 1.2.
Codon content analysis
A generalized linear model (GLM) was applied to assess the impact of codon usage on changes in protein expression:
Here, log(fold change) represents the log-transformed protein expression fold change between suicide and control groups, after controlling for gender, age, postmortem interval (PMI), and pH. Codon usage was defined as the frequency of each codon, calculated as the codon count divided by the total protein length. All codons, except the start and stop codons, were included as predictor variables in the model. For codons not present in a protein, a value of zero was assigned. The GLM was run using the default settings in R, assuming that the log-transformed fold change follows a normal distribution. All analyses were conducted in R.
tRNAGly-high mice
tRNAGly-high mice were generated by the Storkebaum lab as described previously [30]. Briefly, to generate this mouse line, a 1798 base-pair genomic DNA fragment containing the chr8.trna383-Gly-GCC and chr8.trna384-Gly-GCC genes was PCR amplified, cloned, purified, and microinjected into C57BL/6 zygotes. Mice with this allele express ~27 copies a genomic transgene containing two tRNA-GlyGCC genes and were compared to wild-type mice without the transgene. Genotype was confirmed with copy number assays using Taqman probes and quantitative real-time PCR as described previously [30]. For the current experiment, 10-month old mice (n = 3 male/3 female/genotype) were sacrificed with carbon dioxide and brains were immediately removed and flash frozen for later analysis. No experimental blinding was done.
Western blot
Mouse cortical tissue was homogenized using a vibrating pestle in buffer containing SDS, HEPES, sucrose, and protease/phosphatase inhibitors (Roche, cat# 04693159001 and cat# 04906845001) to obtain whole cell lysate and quantified using a BCA assay. Purified protein was denatured at 95 °C for 10 min and then electrophoresed on 4–12% NuPAGE Bis-Tris protein gels (Invitrogen) in Novex SDS running buffer (Invitrogen). Gels were transferred to nitrocellulose membranes using the Trans-blot Turbo Transfer system (Bio-Rad). Membranes were then incubated with blocking buffer (5% milk) followed by overnight incubation at primary antibody in blocking buffer. Primary antibodies were REEP1 (Proteintech, 1:500) and actin (Cell Signaling Technology, 1:5000). Membranes were washed three times and incubated with a fluorescent secondary antibody (Alexa Fluor 647-conjugated anti-rabbit IgG #711-605-152, Jackson ImmunoResearch Laboratories Inc.) for 1 h at room temperature, followed by another three washes. Bands were imaged on a ChemiDoc (BioRad) before quantification with NIH ImageJ software. An unpaired t-test was used to compare conditions and significance was denoted at p < 0.05.
Results
Increased levels of tRNAGly GCC are present in the MDD-suicide DLPFC but not associated with site-specific m5C
We first sought to determine if there were any changes in the tRNA regulome in the brains of subjects who died by suicide during an episode of MDD (MDD suicide) vs. healthy comparison subjects. To perform an unbiased screen of the tRNAome, we performed YAMATseq on total RNA from the DLPFC (Brodmann Area 9) of MDD suicide (n = 33) vs. neurotypical comparison subjects (n = 32). We reliably detected 138 isodecoders, which are tRNAs with the differing body sequences but the same anticodon, in all samples, and found one differentially expressed tRNA isodecoder: Gly-GCC-3 after FDR correlation (adjusted p < 0.1, abs (FC) ≥ 1.5) (Fig. 2; Supplementary Table 1). Due to the exploratory nature of this work, we also lowered the stringency of the analysis by identifying differentially expressed tRNAs using an unadjusted (raw) p value < 0.1 and found two additional differentially expressed tRNA isodecoders Gly-GCC-5, and Lys-TTT-3 (raw p < 0.1, abs (FC) ≥ 1.5) (Supplementary Fig. 1; Supplementary Table 1). While Gly-GCC-3 and Gly-GCC-5 were both significantly increased in suicide vs. comparison brains, Lys-TTT-3 was significantly decreased. This data was obtained after controlling for age, sex, post-mortem interval (PMI), and pH. We also employed permutational multivariate analysis of variance to check effects of each covariate including suicide, gender, age, PMI, pH, childhood abuse, and antidepressant at time of death on global tRNA expression (Supplementary Table 2), and only found a global effect of age on the tRNAome.
a YAMATseq was performed in DLPFC tissue, and we detected 138 unique isodecoders in MDD suicide (n = 33) vs. healthy comparison subjects (n = 32). Note the differentially expressed tRNA isodecoder, Gly-GCC-3, after FDR correction and controlling for gender, age, PMI, and pH (adj. p < 0.1, abs (FC) ≥ 1.5). b Bisulfite tRNA sequencing performed at the tRNAGlyGCC locus (left). Right, no significant differences between MDD suicide (n = 49) and comparison (n = 48) brains were found at any m5C site.
Our previous study in mouse cerebral cortex [11] showed a selective alteration in tRNAGly after the neuronal loss of Nsun2-mediated m5C, therefore we wondered whether the observed increase in tRNAGlyGCC isodecoders in the MDD-suicide PFC was perhaps driven by alterations in the level of m5C at the five known tRNAGly m5C sites (Fig. 2b, left; Supplementary Table 3). To explore, we performed bisulfite tRNA sequencing specifically at the tRNAGlyGCC locus and found no significant differences between MDD suicide (n = 49) and comparison (n = 48) brains after controlling for gender, age, PMI, and pH (Fig. 2b, right), suggesting another mechanism outside of cytosine methylation may be driving the increase in tRNAGlyGCC.
No change in glycine amino acid levels in the MDD suicide brain
We showed previously in mouse cerebral cortex that a change in glycine tRNA expression driven by Nsun2 deficit was associated with a dramatic, >2-fold increase in glycine amino acid levels [11]. Therefore, we hypothesized that the observed increase in tRNAGlyGCC in the PFC of MDD-suicide subjects may be accompanied by changes in glycine amino acid levels. To explore, we used an unbiased metabolomic approach and reliably detected 16 of the 20 canonical amino acids in all samples, including glycine. We found no significant changes in glycine amino acid levels between groups (n = 49/group) after controlling for confounding variables (p = 0.742) (Supplementary Fig. 2a, b; Supplementary Table 4). We likewise found no significant change in other amino acids after FDR correction. However, when lowering the stringency threshold (raw p values < 0.1), we still found no changes in glycine but found significant decreases in levels of phenylalanine (p = 0.039), tyrosine (p = 0.040), and alanine (p = 0.032), and a significant increase in aspartate (p = 0.0002) in MDD suicide brains vs. comparison (Supplementary Fig. 2a, b; Supplementary Table 4).
Proteomic changes in the MDD suicide brain are associated with glycine codon usage
To assess whether the significant increase in tRNAGlyGCC levels in the MDD-suicide PFC was associated with altered proteomic outcomes, we performed LC-MS/MS on MDD suicide and comparison DLPFC samples and detected 2188 proteins. Among the detected proteins, 36 proteins were significantly altered in the MDD suicide group (n = 43) vs. neurotypical comparison (n = 41), including 2 proteins decreased and 34 increased (adj. p < 0.1, abs (FC) ≥ 1.2) (Fig. 3a; Supplementary Table 5). We conclude that our diseased PFC specimens are not affected by gross abnormalities in the bulk proteome. Of the differentially expressed proteins, we identified the top 10 gene ontology categories using Ingenuity Pathway Analysis which included some involved in immune function, cell signaling, and amino acid degradation (Supplementary Fig. 3a). However, it has been established that tRNA abundance is a key determinant for translational elongation rates during protein biogenesis, including codon-specific variabilities in ribosome speed along mRNAs [31,32,33]. Therefore, we hypothesized that the increase in tRNAGlyGCC in the PFC of MDD-suicide brains, as reported here, could be associated with an increase in glycine codon usage and altered expression of Gly-rich proteins. To explore, we quantified proteome-wide codon usage (defined as the usage frequency of each codon or usage count/protein length) in our diseased and control brains. Thus, from a total of 61 anticodons, 12 showed significant disease-associated alterations in codon-usage (FDR adj. p < 0.05, Generalized linear model), with two out of four glycine codons showing a significant increase in codon usage (Fig. 3b). These included codon GGC, which codes for the tRNAGlyGCC anticodon, and codon GGG which codes for the tRNAGlyCCC anticodon (p = 0.043 and p = 0.018 respectively; Generalized linear model) (Fig. 3b; Supplementary Table 6). Given these findings, we hypothesized that transcripts with high GGC codon content may be particularly affected by the abnormal elevation of tRNAGlyGCC levels in the MDD-suicide PFC. Indeed, proteins in the 5th percentile as it pertains to GGC codon content showed a significant increase in expression in the MDD suicide group compared to the rest of the proteome (Mann-Whitney U test, one-tailed due to a priori hypothesis of GGC codon usage in this cohort, p = 0.03) (Fig. 3c). A representative example would be the REEP1 protein, a regulator of adrenergic receptor cell surface expression [34] and encoded by a gene ranking in the top 1% for GGC codon usage (GGC codon usage frequency >0.08 (Fig. 3a, d). We therefore performed a correlation of tRNAGly-GCC-3 expression vs. REEP1 expression for each subject (Supplementary Fig. 3b). We showed a weak positive correlation (r2 = 0.033, p = 0.186) which was strengthened when we excluded one statistical outlier detected using Grubb’s test (r2 = 0.056, p = 0.089). While we note that this relationship would likely also be strengthened by increasing the sample size, we now provide preliminary integration of tRNA and protein data confirming codon-specific changes in protein content relating to tRNA expression.
a LC-MS/MS on MDD suicide (n = 43) and healthy comparison (n = 41) DLPFC samples. Among the 2188 detected proteins, 2 proteins were significantly decreased and 34 were significantly increased in the MDD suicide vs. comparison group (adj. p < 0.1, abs (FC) ≥ 1.2). b Proteome-wide codon usage in MDD suicide vs. comparison brains. 12 out of 61 total codons were significantly associated with MDD suicide (FDR adj. p < 0.05, Generalized linear model), including two glycine codons (codon GGC (p = 0.043) which codes for the tRNAGlyGCC anticodon, and codon GGG (p = 0.018) which codes for the tRNAGlyCCC anticodon) which showed a significant increase in codon usage in the MDD suicide group vs. comparison (Generalized linear model). c MDD suicide vs. comparison changes in expression of proteins with codon GGC usage frequency in the top 5% vs. remaining proteins (Mann-Whitney U test, one-tailed, p = 0.03). d Representative example REEP1 protein which ranks in the top 1% for GGC codon usage (GGC codon usage frequency >0.08).
We note that significantly increased proteins in the suicide brain were also enriched for codon AAA, which is associated with the decreased tRNA-Lys-TTT isodecoder (Fig. 3b; Supplementary Table 6). This is the opposite pattern we would expect, as loss of tRNA expression has been associated with depletion of proteins associated with specific tRNA isodecoders [11, 35, 36]. However, tRNALysTTT has 11 total isodecoders, so it likely that there may be compensation for the one dysregulated isodecoder. Conversely, tRNAGlyGCC has 5 total isodecoders, and increased expression of two out of five isodecoders seems to have a more potent effect on protein dysregulation in the suicide brain.
tRNAGly GCC overexpression increases the expression of protein with high glycine GGC codon content, REEP1
In the postmortem human suicide brain, we identified increased expression of proteins with high glycine codon GGC content which was associated with increased expression of tRNAGlyGCC, but we were thus far unable to identify if the tRNA change could be causally related to translational efficiency. Therefore, to infer a causal role of tRNA expression on expression of codon-biased proteins, we used a mouse model of tRNAGlyGCC overexpression in which mice harbor ~27 copies of a transgene expressing two tRNA-Gly-GCC genes, leading to an additional ~54 copies of tRNA-Gly-GCC (tRNAGly-high) compared to wild-type controls lacking the transgene [30] (Fig. 4a, b; n = 6 tRNAGly-high/6 WT; unpaired t-test, t(10) = 18.96, p < 0.0001, two-tailed; ****p < 0.0001). We then sought to determine whether mice with overexpression of tRNAGlyGCC have higher levels of REEP1, the protein that was increased in the human suicide brain and is in the top 1% of proteins for GGC codon content. Indeed, we identified a significant increase in REEP1 protein in tRNAGly-high cortex compared to wild-type controls (n = 6 tRNAGly-high/6 WT; unpaired t-test, t(10) = 2.492, p = 0.0319, two-tailed) (Fig. 4c), demonstrating that increased expression of REEP1 in the human suicide brain may be causally related to the selective increase in tRNAGlyGCC.
a Mouse model of tRNAGlyGCC overexpression in which mice harbor ~27 copies of a transgene expressing two tRNA-Gly-GCC genes, leading to an additional ~54 copies of tRNA-Gly-GCC (tRNAGly-high) compared to wild-type controls lacking the transgene. b Taqman copy number assay demonstrated a significant increase in transgene copy number in tRNAGly-high mice compared to WT control mice (n = 6 tRNAGly-high/6 WT; unpaired t-test, t(10) = 18.96, p < 0.0001, two-tailed; ****p < 0.0001). c Western blot for REEP1 protein expression in tRNAGly-high cortex compared to wild-type controls and using actin as a loading control (n = 6 tRNAGly-high/6 WT; unpaired t-test, t(10) = 2.492, p = 0.0319, two-tailed; *p < 0.05).
Discussion
Here, we used an integrative ‘omics’ approach applying tRNA-seq and tRNA methylation profiling together with proteomic and metabolomic assessments to explore the tRNA regulome in the PFC of MDD-suicide subjects and controls. Our study presents to the best of our knowledge the first example of how in the human brain, alterations in the tRNA pool could lead to codon-biased translation. Similar principles may apply to the mouse brain, as we demonstrate here that mice overexpressing tRNAGlyGCC show an enrichment of REEP1 protein levels in brain, directly modeling the changes seen in the suicide human brain. To our knowledge, aside from previous reports of codon-based translation in human peripheral tissue in cancer [37, 38], the only other investigation of this level of analysis in the brain is in the mouse hippocampus. It has been reported that deletion of tRNA-Arg-TCT-4-1 in mice, one out of four tRNAArgUCU isodecoders in the mouse genome, decreases tRNAArgUCU levels in the brain and impairs translational processes specifically at the AGA codon in brain ribosomes [35, 36], leading to altered neuronal function in the hippocampus [36].
tRNAs, although considered ‘basic’ elements in a cell’s translational machinery, bear promise for mechanistic insights and potential therapeutic options for neuropsychiatric disease. While we note that to our knowledge no previous studies directly link tRNA function with psychiatric outcomes as demonstrated here, but there is increasing evidence for tRNA pathways, particularly involving tRNA aminoacylation, being linked to psychopathology in the clinical population [8]. For example, serum level genetic determinants for 486 metabolites revealed a link between aminoacyl-tRNA biosynthesis and common risk variants associated with depression and ADHD [39], demonstrating that tRNAs are implicated in psychiatric disease using functional analyses of genome-wide association studies. Additionally, a recent study exploring metabolomic profiles in rodent models for stress-induced depression identified tRNA charging among the top 5 ranking pathways significantly affected in the depressed brain [40]. Our study now adds to this growing body of literature showing that tRNA function is involved in psychopathology and adds the crucial component of direct measurement of the tRNAome in the human brain.
The striking specificity of the tRNAGlyGCC alteration in our PFC MDD-suicide samples is interesting from the perspective of recent mouse work in which depletion of the tRNA methyltransferase NSUN2 produced a specific deficit in expression of multiple tRNAGly isodecoders, including tRNAGlyGCC, while the majority of the tRNAome remained intact [11]. Mice with depleted tRNAGly displayed decreased depressive-like behavior in tests of behavioral despair [11], which when combined with our current study suggests that depressive-like behavior in both human subjects and mouse models may depend on a delicate balance of levels of tRNAGly. We note that mice with a selective deficit in tRNAGly show an anti-depressant-like phenotype, while the MDD suicide brain is linked to an increase in tRNAGly, providing a potentially interesting inverse correlation of tRNAGly with depressive behavior in humans and mice.
Of note, tRNA stability and function (including translation) in the brain and other tissues are heavily regulated by a large number of site- and residue-specific covalent modifications [41, 42]. Therefore, while we did not detect any changes in methyl m5C levels at tRNAGlyGCC in the MDD suicide brain, it remains to be determined whether there is a dysregulation for any other chemical modifications at tRNAGlyGCC including m2G, pseudouridine, and dihydrouridine [43]. It is noteworthy that brain function and behavior could also be impacted by altered chemical modifications of additional, non-glycine tRNAs. For example, wybutosine and 2′-O-methylation at the phenylalanine tRNA (tRNAPhe) reportedly deplete the tRNAPhe tRNA pool and disrupts translation efficiencies for phenylalanine codons, producing memory deficits and increased anxiety in mice [44].
A surprising finding from our integrative ‘omics’ study was the absence of glycine amino acid alteration in the MDD suicide brain. Alterations in tRNA expression have been associated with large changes in amino acid content [11, 36], suggesting that changes in tRNAGly in the suicide brain may produce a change in glycine amino acid, but no changes to glycine were detected. However, there is evidence for considerable individual variation in amino acid levels in various tissues (including the brain) among human subjects [45], and concentrations of amino acids have been primarily measured in depressed patients through peripheral measures, including cerebrospinal fluid or interstitial fluid (CSF or ISF) [46, 47] and blood [48,49,50,51]. We note a recent study, assessing plasma in patients with suicidal ideation has revealed alterations in amino acids including glycine [52].
Although we did not detect changes in glycine, our metabolomics assay identified three other amino acids that were decreased in the MDD suicide brain, including phenylalanine and tyrosine. Interestingly, these changes are also linked to an increase in PCBD1 in the suicide brain, a protein identified in our unbiased LC-MS/MS screen. PCBD1 is an enzyme involved in phenylalanine hydroxylation and its conversion to tyrosine [53], and we demonstrate here a parallel between differential expression of the protein and its amino acid products in the MDD suicide brain. Additionally, among the proteins differentially expressed in the MDD suicide vs. comparison brain, we identified an increase in CACNG8, a neurotransmitter receptor regulator protein, which is in line with previous work from another group showing an increase in CACNG8 gene expression in the dmPFC of subjects who died by suicide [54].
Limitations
Although we show here a promising link between tRNA expression and codon-specific proteomic outcomes in patients with MDD who died by suicide, we note that there are some limitations to the current work. First, we have a large cohort of MDD suicide and comparison subjects, but there is the possibility that the occurrence of a depressive episode in the absence of suicide is of mechanistic importance. Along the same lines of disentangling MDD from the act of suicide, there may likewise be importance of subjects who died by suicide who were not diagnosed with MDD but perhaps another psychiatric disorder such as psychosis. There is a major difficulty in obtaining a sufficient n of either of these cohorts (MDD without suicide or suicide without MDD), especially when considering post-mortem interval (PMI) and age of subjects compared to the MDD suicide and comparison groups. Nevertheless, obtaining these extra cohorts will be crucial in the future for disentangling the molecular changes identified in the MDD suicide brain and attributing these changes to suicidal behavior or MDD diagnosis. Another limitation of the current study lies in use of heterogeneous cell types in the human brain tissue used for molecular studies. Due to input restrictions for tRNA sequencing, we were unable to isolate neurons and glia before performing ‘omics’ assays and therefore cannot make claims regarding cell-type specificity or cell density of different cell types in the suicide vs. comparison brain.
Conclusions
By using an integrative approach, we have now demonstrated a link between neuropsychiatric disease, namely suicide with MDD, and the tRNA regulome of the human brain, and provide here a potential mechanism using an animal model of tRNA overexpression. Future work will link the tRNA regulome, including codon content and proteomics to translational efficiency in diseased brain tissue, and aim to decipher the role of antidepressant medication as opposed to the primary disease process (depression) and, separately, the risk for suicide. Nevertheless, the current findings provide a new avenue for investigating the pathophysiology of depression and suicide and potential novel therapeutic targets via tRNA regulation.
Code availability
Code for ‘omics’ data analysis available upon request.
References
Suicide Mortality in the United States, 2001–2021. In: National Center for Health S (ed). https://doi.org/10.15620/cdc:125705: Hyattsville, MD: 2023.
Mann JJ, Rizk MM. A brain-centric model of suicidal behavior. Am J Psychiatry. 2020;177:902–16.
Turecki G, Brent DA. Suicide and suicidal behaviour. Lancet. 2016;387:1227–39.
Nagy C, Suderman M, Yang J, Szyf M, Mechawar N, Ernst C, et al. Astrocytic abnormalities and global DNA methylation patterns in depression and suicide. Mol Psychiatry. 2015;20:320–8.
Haghighi F, Xin Y, Chanrion B, O’Donnell AH, Ge Y, Dwork AJ, et al. Increased DNA methylation in the suicide brain. Dialogues Clin Neurosci. 2014;16:430–8.
Sequeira A, Mamdani F, Ernst C, Vawter MP, Bunney WE, Lebel V, et al. Global brain gene expression analysis links glutamatergic and GABAergic alterations to suicide and major depression. PLoS ONE. 2009;4:e6585.
Su Z, Wilson B, Kumar P, Dutta A. Noncanonical roles of tRNAs: tRNA fragments and beyond. Annu Rev Genet. 2020;54:47–69.
Blaze J, Akbarian S. The tRNA regulome in neurodevelopmental and neuropsychiatric disease. Mol Psychiatry. 2022;27:1–10.
Dallabona C, Diodato D, Kevelam SH, Haack TB, Wong LJ, Salomons GS, et al. Novel (ovario) leukodystrophy related to AARS2 mutations. Neurology. 2014;82:2063–71.
Torres AG, Reina O, Stephan-Otto Attolini C, Ribas de Pouplana L. Differential expression of human tRNA genes drives the abundance of tRNA-derived fragments. Proc Natl Acad Sci USA. 2019;116:8451–6.
Blaze J, Navickas A, Phillips HL, Heissel S, Plaza-Jennings A, Miglani S, et al. Neuronal Nsun2 deficiency produces tRNA epitranscriptomic alterations and proteomic shifts impacting synaptic signaling and behavior. Nat Commun. 2021;12:4913.
Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, et al. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol. 2012;19:900–5.
Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, et al. Stem cell function and stress response are controlled by protein synthesis. Nature. 2016;534:335–40.
Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33:2020–39.
Odebrecht Vargas H, Vargas Nunes SO, Pizzo de Castro M, Cristina Bortolasci C, Sabbatini Barbosa D, Kaminami Morimoto H, et al. Oxidative stress and lowered total antioxidant status are associated with a history of suicide attempts. J Affect Disord. 2013;150:923–30.
Brunner J, Bronisch T, Uhr M, Ising M, Binder E, Holsboer F, et al. Proteomic analysis of the CSF in unmedicated patients with major depressive disorder reveals alterations in suicide attempters. Eur Arch Psychiatry Clin Neurosci. 2005;255:438–40.
Semančíková E, Tkáčiková S, Talian I, Bencková M, Pálová E, Sabo J. In search of possible peripheral biomarkers for suicide: similarities between platelet and cerebrospinal fluid proteome (Preliminary Results). Eur Psychiatry. 2017;41:S638–S638.
Zhou Y, Lutz P-E, Ibrahim EC, Courtet P, Tzavara E, Turecki G, et al. Suicide and suicide behaviors: a review of transcriptomics and multiomics studies in psychiatric disorders. J Neurosci Res. 2020;98:601–15.
Schlicht K, Büttner A, Siedler F, Scheffer B, Zill P, Eisenmenger W, et al. Comparative proteomic analysis with postmortem prefrontal cortex tissues of suicide victims versus controls. J Psychiatr Res. 2007;41:493–501.
Kékesi KA, Juhász G, Simor A, Gulyássy P, Szegő ÉM, Hunyadi-Gulyás É, et al. Altered functional protein networks in the prefrontal cortex and amygdala of victims of suicide. PLoS ONE. 2012;7:e50532.
Blaze J, Browne CJ, Futamura R, Javidfar B, Zachariou V, Nestler EJ, et al. tRNA epitranscriptomic alterations associated with opioid-induced reward-seeking and long-term opioid withdrawal in male mice. Neuropsychopharmacology. 2024;49:1276–84.
Bormann F, Tuorto F, Cirzi C, Lyko F, Legrand C. BisAMP: a web-based pipeline for targeted RNA cytosine-5 methylation analysis. Methods. 2019;156:121–7.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300.
Benjamini Y, Krieger AM, Yekutieli D. Adaptive linear step-up procedures that control the false discovery rate. Biometrika. 2006;93:491–507.
Shigematsu M, Honda S, Loher P, Telonis AG, Rigoutsos I, Kirino Y. YAMAT-seq: an efficient method for high-throughput sequencing of mature transfer RNAs. Nucleic Acids Res. 2017;45:e70–e70.
Smith T, Heger A, Sudbery I. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 2017;27:491–9.
Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate illumina paired-end reAd mergeR. Bioinformatics. 2014;30:614–20.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47–e47.
Zuko A, Mallik M, Thompson R, Spaulding EL, Wienand AR, Been M, et al. tRNA overexpression rescues peripheral neuropathy caused by mutations in tRNA synthetase. Science. 2021;373:1161–6.
Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol. 2009;16:274–80.
Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, Kooistra SM, et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014;158:1281–92.
Gorochowski TE, Ignatova Z, Bovenberg RAL, Roubos JA. Trade-offs between tRNA abundance and mRNA secondary structure support smoothing of translation elongation rate. Nucleic Acids Res. 2015;43:3022–32.
Björk S, Hurt CM, Ho VK, Angelotti T. REEPs are membrane shaping adapter Proteins that modulate specific G protein-coupled receptor trafficking by affecting ER cargo capacity. PLoS ONE. 2013;8:e76366.
Ishimura R, Nagy G, Dotu I, Zhou H, Yang XL, Schimmel P, et al. RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science. 2014;345:455–9.
Kapur M, Ganguly A, Nagy G, Adamson SI, Chuang JH, Frankel WN, et al. Expression of the neuronal tRNA n-Tr20 regulates synaptic transmission and seizure susceptibility. Neuron. 2020;108:193–208.e199.
Pinzaru AM, Tavazoie SF. Transfer RNAs as dynamic and critical regulators of cancer progression. Nat Rev Cancer. 2023;23:746–61.
Goodarzi H, Nguyen HCB, Zhang S, Dill BD, Molina H, Tavazoie SF. Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell. 2016;165:1416–27.
Yang J, Yan B, Zhao B, Fan Y, He X, Yang L, et al. Assessing the causal effects of human serum metabolites on 5 major psychiatric disorders. Schizophrenia Bull. 2020;46:804–13.
Tian L, Pu J, Liu Y, Gui S, Zhong X, Song X, et al. Metabolomic analysis of animal models of depression. Metab Brain Dis. 2020;35:979–90.
Blaze J, Akbarian S. The tRNA regulome in neurodevelopmental and neuropsychiatric disease. Mol Psychiatry. 2022;27:3204–13.
Burgess RW, Storkebaum E. tRNA dysregulation in neurodevelopmental and neurodegenerative diseases. Annu Rev Cell Dev Biol. 2023;39:223–52.
Lei H-T, Wang Z-H, Li B, Sun Y, Mei S-Q, Yang J-H, et al. tModBase: deciphering the landscape of tRNA modifications and their dynamic changes from epitranscriptome data. Nucleic Acids Res. 2023;51:D315–D327.
Nagayoshi Y, Chujo T, Hirata S, Nakatsuka H, Chen CW, Takakura M, et al. Loss of Ftsj1 perturbs codon-specific translation efficiency in the brain and is associated with X-linked intellectual disability. Sci Adv. 2021;7:eabf3072.
Banay-Schwartz M, DeGuzman T, Faludi G, Lajtha A, Palkovits M. Alteration of protease levels in different brain areas of suicide victims. Neurochem Res. 1998;23:953–9.
Ishiwata S, Hattori K, Sasayama D, Teraishi T, Miyakawa T, Yokota Y, et al. Cerebrospinal fluid D-serine concentrations in major depressive disorder negatively correlate with depression severity. J Affect Disord. 2018;226:155–62.
Kofler M, Schiefecker AJ, Gaasch M, Sperner-Unterweger B, Fuchs D, Beer R, et al. A reduced concentration of brain interstitial amino acids is associated with depression in subarachnoid hemorrhage patients. Sci Rep. 2019;9:2811.
Woo H-I, Chun M-R, Yang J-S, Lim S-W, Kim M-J, Kim S-W, et al. Plasma amino acid profiling in major depressive disorder treated with selective serotonin reuptake inhibitors. CNS Neurosci Therapeutics. 2015;21:417–24.
Altamura C, Maes M, Dai J, Meltzer HY. Plasma concentrations of excitatory amino acids, serine, glycine, taurine and histidine in major depression. Eur Neuropsychopharmacol. 1995;5:71–75.
Xu H-B, Fang L, Hu Z-C, Chen Y-C, Chen J-J, Li F-F, et al. Potential clinical utility of plasma amino acid profiling in the detection of major depressive disorder. Psychiatry Res. 2012;200:1054–7.
Mauri MC, Boscati L, Volonteri LS, Scalvini ME, Steinhilber CPC, Laini V, et al. Predictive value of amino acids in the treatment of major depression with fluvoxamine. Neuropsychobiology. 2001;44:134–8.
Group VAM-AMW. Amino acids as biomarker candidates for suicidality in male OEF/OIF veterans: relevance to NMDA receptor modulation and nitric oxide signaling. Military Med. 2014;179:486–91.
Thöny B, Neuheiser F, Kierat L, Blaskovics M, Arn PH, Ferreira P, et al. Hyperphenylalaninemia with high levels of 7-biopterin is associated with mutations in the PCBD gene encoding the bifunctional protein pterin-4a-carbinolamine dehydratase and transcriptional coactivator (DCoH). Am J Hum Genet. 1998;62:1302–11.
Dóra F, Renner É, Keller D, Palkovits M, Dobolyi Á. Transcriptome profiling of the dorsomedial prefrontal cortex in suicide victims. Int J Mol Sci. 2022;23:7067.
Acknowledgements
We thank the subjects and their families for their contributions to this study. This work was supported by the American Foundation for Suicide Prevention (AFSP) Young Investigator Grant (YIG-1-013-21) to JB, NIH postdoctoral fellowship (F32MH115565) to JB, and NIH grants (U01DA048279 and R01DA047880) to SA.
Author information
Authors and Affiliations
Contributions
JB and SA designed and conceptualized the study. GT provided post-mortem brain samples from Douglas Bell Canada Brain Bank. CP extracted RNA from tissue samples. JB processed tissue for molecular assays, conducted tRNA sequencing and bisulfite sequencing, and analyzed data. SC and HG analyzed data and performed codon content analyses. SH, HA, and HM performed proteomics and metabolomics assays and provided preliminary data analysis. MPL and ES provided the tRNAGly-high mouse tissue. JB and SA wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Blaze, J., Chen, S., Heissel, S. et al. Altered tRNA expression profile associated with codon-specific proteomic changes in the suicide brain. Mol Psychiatry 30, 2871–2879 (2025). https://doi.org/10.1038/s41380-025-02891-8
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41380-025-02891-8