Abstract
Multiple myeloma is a malignant hematopoietic neoplasm characterized by unclear molecular mechanisms and lack of highly effective targeted therapies for clinical application. Interferon regulatory factor 4 (IRF4) is a well-known core transcription factor that regulates the progression of myeloma, but the molecular mechanisms underlying its protein homeostasis regulation are unknown. Our research shows that lysosomal-associated membrane protein 5 (LAMP5) interacts with IRF4 and prevents its degradation through the autophagy-lysosome pathway, thereby facilitating the progression of myeloma. Additionally, LAMP5 enhances the interaction between IRF4 and the nuclear transport protein karyopherin α2 (KPNA2), facilitating the nuclear transport of IRF4 and preventing its cytoplasmic retention and subsequent autophagy-lysosome degradation. Nuclear IRF4 promotes the transcription of c-MYC, and the c-MYC protein positively feeds back to activate LAMP5 transcription. This vicious regulatory loop drives rapid progression of myeloma. High-throughput drug screening shows pyrazofurin that significantly disrupts the interaction between LAMP5 and IRF4, leading to the degradation of IRF4 and inhibition of myeloma progression. This study elucidates a novel mechanism underlying IRF4 protein homeostasis maintenance and provides a potential inhibitor for myeloma treatment.
Similar content being viewed by others
Introduction
Multiple myeloma (MM), a neoplastic disorder of plasma cells, constitutes approximately 10% of hematological cancers and is on the rise in the burgeoning elderly population [1]. Myeloma is frequently associated with skeletal complications, including bone lesions in over 80% of cases, which can lead to pathological fractures, severe pain, spinal cord compression, and hypercalcemia [2]. These skeletal-related events significantly diminish patients’ quality of life and lead to grave prognosis [3]. Despite the use of novel chemotherapeutic agents, such as bortezomib and lenalidomide, either in monotherapy or in combination regimens, the majority of patients experience relapse or develop refractory disease following high-dose chemotherapy, highlighting the imperative for the disease to remain incurable [4]. The quest for innovative therapies with distinct mechanisms of action is particularly urgent for patients with refractory and high-risk myeloma. Unraveling the intricate molecular pathways underlying myeloma is essential for the development of potentially efficacious therapeutics.
The origins of myeloma cells are enigmatic, with some studies hinting at a shared immunoglobulin gene rearrangement between the common precursor cells of T and B lymphocytes and myeloma cells. Despite their predominant B-cell and plasma cell-like characteristics, myeloma cells are believed to arise from an earlier malignancy of hematopoietic precursor cells than pre-B cells. However, the precise mechanisms underlying this early transformation remain obscure [5]. It has been hypothesized that factors such as radiation, chemicals, and viruses may induce genetic mutations or chromosomal translocations, thereby activating oncogenes. For example, mutations that activate H-ras or gene rearrangements that activate c-MYC are known to initiate tumorigenesis [1, 6]. A significant correlation has been observed between chromosomal translocations and the c-MYC oncogene, with over 90% of the mice exhibiting such translocations [7]. Similar aberrations of the c-MYC gene, including rearrangements and increased expression, have been documented in myeloma patients [7]. Additionally, oncogenic mutations in N-ras or K-ras have been identified in approximately 27% of myeloma cases at the initial diagnosis, increasing to 46% post-treatment [8, 9]. Furthermore, inactivation of the tumor suppressor genes retinoblastoma gene (Rb) and p53 [10], along with activation of the nuclear transcription factor-kappa B (NF-κB) pathway and aberrations in the Wnt pathway, are frequently observed in myeloma [11, 12].
LAMP5, also known as brain and dendritic cell-associated lysosome-associated membrane protein (BAD-LAMP), is a member of the LAMP family and plays critical roles in various cellular processes [13]. In mice, LAMP5 is predominantly expressed in neurons, and its absence in interneurons has been linked to neural network dysfunction in Alzheimer disease models [14]. In humans, LAMP5 is expressed in type I interferon (IFN)-producing plasmacytoid dendritic cells (pDCs), where it plays a pivotal role in activating NF-κB and promoting tumor necrosis factor (TNF) production upon DNA detection [15]. The LAMP5 expression patterns are particularly intriguing in the context of cancer. It is upregulated in metastatic gastric cancer tissues, and its knockdown inhibits gastric cancer cell proliferation, invasion, and migration, promotes apoptosis and cell cycle arrest, and reduces cancer stemness [16]. Furthermore, LAMP5 expression is significantly elevated in hematologic tumors, such as blastic plasmacytoid dendritic cell neoplasms [17]. It is highly and specifically expressed in mixed lineage leukemia-rearranged (MLL-r) leukemias, where it is associated with poor prognosis. The downregulation of LAMP5 in these leukemias leads to the inhibition of NF-κB signaling and increased activation of type 1 interferon signaling downstream of the Toll-like receptor/interleukin 1 receptor activation [17, 18]. Notably, LAMP5 is overexpressed in myeloma cells, and its high expression correlates positively with poor patient prognosis [19]. However, the mechanisms by which LAMP5 regulates myeloma progression and its potential as a therapeutic target in myeloma remain unclear.
Through a combination of in vitro, in vivo, and patient’ sample studies, we found that LAMP5 plays a unique role in myeloma progression. Our innovative research uncovered a critical role for LAMP5 in the pathogenesis of myeloma. We found that LAMP5 is markedly upregulated in myeloma cells and acts as a potent driver of cell proliferation. Mechanistically, LAMP5 engages with IRF4, impeding its lysosomal degradation and preserving it for nuclear translocation facilitated by KPNA2. This orchestrated process results in the transcriptional activation of the oncogenic c-MYC gene by nuclear IRF4, which in turn amplifies LAMP5 expression, establishing a self-perpetuating loop that accelerates myeloma progression. Moreover, our high-throughput drug screening identified pyrazofurin as a potent LAMP5 inhibitor that effectively disrupted the LAMP5-IRF4 interaction. This disruption triggers IRF4 degradation, thereby inhibiting the oncogenic potential of the myeloma cells. Our findings not only elucidate a novel regulatory mechanism in myeloma but also position LAMP5 as a promising therapeutic target. The implications of our research are profound, offering new avenues for intervention and potentially transformative treatment of patients afflicted with this devastating disease.
Materials and methods
Cell lines and primary myeloma cells
Myeloma cell lines ARP-1 and LP-1 were sourced from Dr. Zhiqiang Liu at Shandong First Medical University and Shandong Academy of Medical Sciences. Additional cell lines, including IM-9, Jurkat, HEK293T, and Jeko-1 were acquired from the American Type Culture Collection (ATCC). Primary myeloma cells were extracted from the bone marrow of the patients using anti-CD138 magnetic beads (Miltenyi Biotec). Myeloma and lymphoma cell lines were maintained in RPMI1640 medium with 10% fetal bovine serum (FBS). HEK293T cells were cultured in Dulbecco’s modified Eagle medium (DMEM) with 10% FBS. Cell lines cultured for over six months underwent STR profiling authentication via Biowing Biotech (Shanghai, China). Experiments utilized cells confirmed to be mycoplasma- free through testing with the Universal Mycoplasma Detection Kit (ATCC, Manassas, VA, USA). Patient samples were collected at the First Affiliated Hospital of Xiamen University. Signed informed consent was obtained from all participating individuals prior to participation in the study. The study was approved by the Xiamen University Ethics Committee and adhered to the World Medical Association Declaration of Helsinki guidelines.
Plasmid construction, antibodies, and reagents
Human LAMP5, IRF4, HA-tagged LAMP5, HA-tagged IRF4, and HA-tagged KPNA2 were subcloned into the pLVx-AcGFP-N1 vector (NovoPro, #V012707). Short hairpin RNAs (shRNAs) targeting LAMP5 and c-MYC, as well as a non-targeting control shRNA, were sourced from Sigma-Aldrich and cloned into the pLKO.1 lentiviral vector. The primers used for the amino acid deletion fragment of IRF4 are listed in Supplementary Table 3. Except where specified, all chemicals were purchased from Sigma-Aldrich, and all antibodies for western blot analysis were purchased from Cell Signaling Technology.
Stable cell line construction
For transient transfection of HEK 293T cells, we employed polyethyleneimine (PEI) as the transfection reagent in OPTI-MEM medium (Life Technologies), maintaining a DNA-to-PEI ratio of 1:4. To generate viral particles, HEK 293T cells were co-transfected with the following plasmids: 4 μg of pMD2.G (#12259, Addgene), 6 μg of psPAX2 (#12260, Addgene), and 8 μg of lentiviral expression vectors carrying either human LAMP5, pLKO.1-based shRNAs targeting LAMP5, or c-MYC constructs. Viral supernatants were harvested 48 h post-transfection and concentrated 100-fold using Polyethylene glycol 8000 (Sigma-Aldrich). Myeloma cell infections were performed via spinfection at 800 × g for 30 min at 37 °C with concentrated viral particles in polybrene-containing medium. Following 12 h of incubation, the medium was refreshed, and cells were maintained for an additional 48-h period before subsequent experiments. Stable cell lines were established through puromycin selection (2 μg/mL; #540222, Sigma-Aldrich) in the culture medium.
Western blot analysis
The cells were harvested and lysed with 1× lysis buffer (Cell Signaling Technology). Cell lysates were subjected to SDS-PAGE, transferred to nitrocellulose membranes, and immunoblotted with antibodies against GAPDH (#5174), β-Tubulin (#2146), c-MYC (#5605), p62 (#39749), Hsc70 (#4872), IRF4 (#62834), LC3B (#3868), KPNA2 (#14372) and HA (#3724) (Cell Signaling Technology). The LAMP5 antibody (#sc-398190) was purchased from Santa Cruz Biotechnology.
Quantitative real-time PCR of mRNAs
Total RNA was isolated from the samples using the RNeasy extraction kit from QIAGEN. One microgram of the purified RNA was subjected to reverse transcription using the SuperScript II RT-PCR kit provided by Invitrogen. For the quantitative real-time PCR analysis, SYBR Green Master Mix from Life Technologies was employed, and the reactions were performed on a QuantStudio 3 Real-Time PCR System, also from Life Technologies. The PCR protocol included an initial step of denaturation at 95 °C for 10 min, followed by 40 cycles consisting of 95 °C for 15 s and 60 °C for 60 s. GAPDH was employed as the endogenous control for normalization. The specific primer sequences utilized in the reactions are detailed in Supplementary Table 4.
Cell viability and ELISA analysis
For assessing cell viability, cells were seeded at a density of 10,000 cells per well in triplicate. The Cell Counting Kit-8 from Dojindo was utilized for the assays. Cell proliferation was evaluated using an EdU assay kit from RiboBio, following the manufacturer’s protocol. ELISA kits were purchased from Thermo Fisher Scientific and Immunodiagnostic Systems.
Immunoprecipitation and pull-down assays
Cells were lysed and placed on ice for 15 min to optimize the lysis process. The lysates containing total protein were then processed for immunoprecipitation using antibodies coupled to agarose beads, which were performed at 4 °C overnight. Next, the beads were washed extensively six times and resuspended in 30 μl of 1× SDS buffer. They were briefly boiled for 5 min, and the samples were loaded onto an SDS-PAGE gel for electrophoresis, including a 5% input control. The proteins were transferred to a PVDF membrane for western blot analysis. IgG was used as a negative control, whereas total cell lysate served as a positive control for the input. For the pull-down assay, HEK293T cells were transfected with a plasmid expressing the HA-tagged protein. The lysates precipitated from these cells using HA beads were mixed with lysates from cells transfected with full-length or truncated IRF4. Both immunoprecipitated complexes and whole cell lysates were analyzed by western blotting. Non-transfected cells or cells transfected with control plasmids, along with whole cell lysates, were used as negative controls.
ChIP assay
Following fixation with 4% formaldehyde solution, the cells were sonicated to produce chromatin fragments that were appropriate for analysis. The samples were then subjected to immunoprecipitation at 4 °C for 3 h using an antibody specific to c-MYC, with IgG acting as a negative control. Subsequently, the DNA from both the immunoprecipitated complexes and the total chromatin was reverse cross-linked, purified, and analyzed by PCR using primers designed to target the promoter regions of the genes of interest. The primer sequences are listed in Supplementary Table 5.
Fluorescent staining
Myeloma cells were fixed with 4% formaldehyde and permeabilized with 0.3% Triton X-100 in 1× PBS buffer. After blocking with 2% goat serum, the cells were stained with a Quadruple-Fluorescence immunohistochemical mouse/rabbit kit (ImmunoWay) following the manufacturer’s instructions. Immunofluorescence images were acquired using an IX71 confocal microscope (Olympus). Antibodies against IRF4 (66451-1-Ig), Ki67 (27309-1-AP), and KPNA2 (10819-1-AP) were purchased from Proteintech. The CD138 antibody (#AF2780) was purchased from R&D Systems. The LAMP5 antibody (#sc-398190) was purchased from Santa Cruz Biotechnology. Formalin-fixed, paraffin-embedded sections of bone marrow biopsy samples obtained from myeloma patients were deparaffinized and stained with anti-LAMP5, IRF4 or anti-CD138 antibodies. Subcutaneous tissue samples extracted from the mice were fixed, sectioned, and processed similarly. The quantitative results were summarized in Supplementary Fig. 8.
Whole-transcriptome sequencing
RNAs were isolated from IM-9 or LP-1 (Vec, LAMP5) and purified using VAHTSTM DNA Clean Beads (Vazyme). RNA-sequencing (RNA-seq) libraries were prepared using the VAHTS Universal V6 RNA-seq Library Prep Kit from Vazyme. The pooled libraries were quantified using the Qubit TM dsDNA HS Assay Kit from Thermo Fisher Scientific, and their size distribution was assessed using the Fragment Analyzer from Advanced Analytical. The libraries were then sequenced on an MGI Tech DNBSEQ-T7 Sequencer.
Mass spectrometry
HA-LAMP5 overexpressed IM-9 cells were lysed and incubated on ice for 15 min to allow efficient protein extraction. The cell lysate was then immunoprecipitated using an HA-specific antibody bound to agarose beads at 4 °C overnight. After six washes, the precipitated proteins were prepared for analysis via nano-liquid chromatography-tandem mass spectrometry (nano-LC/MS/MS), which was performed using equipment from Thermo Fisher Scientific and interfaced with an Agilent Technologies 1100 HPLC system. MS/MS data were analyzed using the SEQUEST algorithm within the BioWorks Browser software suite, version 3.3.1, to search against the NCBI protein database. Control samples were obtained from IM-9 cells transfected with an empty vector. The mass spectrometry proteomics data were deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD055882.
Luciferase assay in vitro
The construct covering the full-length (LAMP5-Full), and mutated forms (LAMP5-Mut1 and LAMP5-Mut2) were subcloned into the pGL2 vector, and their transcriptional activities in HEK293T cells were examined using a Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer instructions. Primers used for subcloning are listed in Supplementary Table 6.
High-throughput virtual screening (HTVS)
Structure-based high-throughput virtual screening (HTVS) was performed using Schrödinger Maestro 11.4. The human LAMP5 structure (AlphaFold ID: AF-Q9UJQ1-F1) was retrieved from the AlphaFold database. The MCE Bioactive Compounds Library Plus (71678 compounds) underwent energy optimization via Schrödinger’s LigPrep module, followed by molecular docking through a tiered workflow: initial HTVS screening (top 10% selected), Standard Precision (SP) docking of HTVS hits (top 10% retained), and Extra Precision (XP) refinement. Four compounds with the highest absolute docking scores (indicative of stronger predicted binding affinity) were prioritized for experimental validation. Compound PKG inhibitor peptide (#HY-P1292), pyrazofurin (#HY-122502), parishin B (#HY-N2124), and D-Gluconic acid calcium hydrate (#HY-Y0569B) were purchased from MedChemExpress (Monmouth Junction, NJ, USA). Three-dimensional binding poses were visualized using PyMol.
In vivo mouse experiments
6 weeks old female NSG mice purchased from the Shanghai Model Organisms Center, Shanghai, China, were maintained in Xiamen University Animal Care-accredited facilities. Mouse studies were approved by the Institutional Animal Care and Use Committee of Xiamen University, and all protocols followed conformed with the Guidelines of Ethical Conduct in the Care and Use of Nonhuman Animals in Research. To demonstrate the effect of LAMP5 on myeloma cell growth, NSG mice were randomly assigned to groups in a blinded manner and subcutaneously injected with myeloma cell lines ARP-1 or IM-9 [nontargeted shRNA (shCtrl), and LAMP5 shRNA (shLAMP5)] (1× 106 cell/mouse) (n = 3 mice/group). Tumor diameters were measured every other day. After 3 weeks, the mice were sacrificed and tumor tissues were photographed and weighed. Tumor tissues were fixed in 10% neutral-buffered formalin, and sections of them were stained with Ki67 (Cell Signaling Technology) following standard protocols. To identify the effect of pyrazofurin in vivo, myeloma cells (ARP-1 or IM-9) (1× 106 cell/mouse) were subcutaneously injected into NSG mice, and intraperitoneal administration of pyrazofurin (5 mg/kg bodyweight) three times per week for a duration of two weeks, beginning one week after cell injection (n = 3 mice/group). Tumor diameters were measured every other day. After 3 weeks, the mice were sacrificed and tumor tissues were photographed. Subcutaneous tissue samples extracted from the mice were fixed, sectioned, and stained with Ki67 or IRF4 antibodies. No animals were excluded from the analysis.
6-week-old female C57BL/6J mice were randomly assigned to groups in a blinded manner and intravenously injected with vk12598 cells, followed by intraperitoneal administration of pyrazofurin (5 mg/kg bodyweight) three times per week for a duration of two weeks, beginning two weeks after cell injection. Mice injected with PBS buffer were used as controls (n = 9 mice/group). After four weeks, we measured the establishment of myeloma by detecting increased levels of circulating IgG2b. The spleens were collected and subjected to immunofluorescence analysis. No animals were excluded from the analysis.
Statistical analysis
The determination of sample size, replication structure, and the chosen intermediate endpoint was informed by prior experience [2, 3, 20], in which the power of the study was 80%, and the level for type I error was 5% (P < 0.05). Statistical significance was determined using the Graphpad software (Version 9.0). For the comparison between two groups, two-tailed unpaired Student t test were employed. When comparing more than two groups, one-way ANOVA with Tukey post hoc test for multiple comparisons was applied. The variance was similar between groups that were being statistically compared. A P value of less than 0.05 was set as the threshold for statistical significance.
Results
LAMP5 is critical for myeloma disease progression
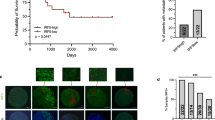
By utilizing the Broad Institute Cancer Cell Line Encyclopedia (CCLE) database, our analysis indicated that LAMP5 exhibits the highest expression in myeloma cells among numerous types of tumors and its expression level is also significantly higher than that in other tissues or organs of the human body (Fig. 1A, B). We also analyzed the alteration of gene expression in plasma cells of myeloma patients compared to plasma cells of healthy donors using published GEO datasets (GSE2658, GSE5900 and GSE6477). LAMP5 expression was much higher in myeloma cells than that in normal plasma cells (Fig. 1C). Quantitative real-time PCR results revealed elevated LAMP5 expression in the majority of bone marrow samples obtained from primary myeloma cells and human myeloma cell lines. In contrast, significantly reduced expression was observed in normal plasma cells (Fig. 1D). Normal plasma cells evolve into monoclonal gammopathy of undetermined significance (MGUS) through genetic alterations. Secondary mutations and interactions with the microenvironment drive progression to smoldering multiple myeloma (sMM). Subsequent genomic instability and immune dysregulation lead to active multiple myeloma [4]. We further analyzed the expression changes of LAMP5 during myeloma progression. The results demonstrated that LAMP5 expression gradually increases with disease progression (Supplementary Fig. 1A, B), highlighting its critical role in the progression of myeloma. Furthermore, analysis of the GSE6477 dataset revealed that LAMP5 expression is significantly higher in relapsed myeloma compared to newly diagnosed cases (Supplementary Fig. 1C), suggesting its potential involvement in drug resistance. Additionally, we collected primary myeloma cells from 20 patients with 1q21 amplification and 20 patients without 1q21 amplification. Quantitative real-time PCR analysis confirmed that LAMP5 expression is elevated in the 1q21-amplified group compared to the non-amplified group (Supplementary Fig. 1D). These results further underscore the clinical relevance of LAMP5 in myeloma pathogenesis and its association with genetic alterations such as 1q21 amplification. Moreover, published single-cell sequencing data indicated that myeloma cells (CD138+ cells) express higher levels of LAMP5 (Fig. 1E). Immunofluorescence analysis of bone marrow samples from myeloma patients indicated higher expression of LAMP5 in myeloma cells (Fig. 1F). In our subsequent analysis examining the correlation between LAMP5 and myeloma progression using data from the Multiple Myeloma Research Foundation (MMRF) CoMMpass study IA15, we observed that patients with elevated LAMP5 expression in myeloma cells had a reduced overall survival rate compared to those with lower expression levels (Fig. 1G). We additionally collected 100 cases of primary myeloma cells categorized into high or low LAMP5 expression groups. The clinical characteristics analysis showed that compared to myeloma patients with low LAMP5 expression, those with high LAMP5 expression have faster disease progression, higher levels of lactate dehydrogenase in their serum, and a lower proportion of achieving complete remission after initial treatment (Supplementary Table 1).
A, B LAMP5 mRNA expression in myeloma and other indicated cancer cell lines or other tissues, according to the Cancer Cell Line Encyclopedia (CCLE) dataset. C LAMP5 mRNA levels in plasma cells from myeloma patients (n = 559) compared to normal plasma cells from healthy donors (n = 22) (GEO: GSE2658 and GSE5900) (left). LAMP5 mRNA levels in plasma cells from myeloma patients (n = 73) compared to normal plasma cells from healthy donors (n = 14) (GEO: GSE6477) (right). P value was determined by unpaired two-tailed t test. D Quantitative real-time PCR analysis shows LAMP5 expression in normal plasma cells (Healthy, n = 10), malignant plasma cells (Pt MM, n = 20), and human myeloma cell lines (MM cell lines, n = 6). GAPDH served as loading control. P values were determined using one-way ANOVA. E Representative two patients of UMAP plot of CD138 (left) and LAMP5 (right) expression in scRNA-seq data (GSE161801). F Immunofluorescent staining of healthy of myeloma patients bone marrow samples with DAPI and antibodies against LAMP5 and CD138 (n = 5). Scale bar, 10 μm. G Overall survival (OS) of patient’s myeloma cells with high LAMP5 (High, n = 200) and low LAMP5 (Low, n = 200) expression in MMRF CoMMpass study IA15.
To elucidate the role of LAMP5 in oncogenic processes, we used small hairpin RNAs (shRNAs) to silence LAMP5 in ARP-1 and IM-9 myeloma cell lines and introduced exogenous LAMP5 complementary DNA (cDNA) into LP-1 and IM-9 cells to achieve overexpression (Supplementary Fig. 2A, B). Subsequently, we conducted a comprehensive transcriptome RNA sequencing (RNA-seq) analysis to discern the transcriptional changes in LAMP5-overexpressing myeloma cells relative to control cells. Computational analysis, leveraging the Molecular Signatures Database (MSD) hallmark gene sets from the Broad Institute, revealed a pronounced enrichment of proliferation-associated genes, including those involved in the MAPK, PI3K-Akt, and NF-κB signaling pathways (Fig. 2A, B). In proliferation assays, myeloma cells with elevated LAMP5 expression exhibited enhanced growth compared to those with LAMP5 knockdown (Fig. 2C, D, Supplementary Fig. 3A, B). EdU staining assays corroborated these findings, demonstrating reduced clonogenic potential in shLAMP5 myeloma cells compared to control cells (Fig. 2E). To further validate our observations, we utilized a humanized mouse model engrafted intrahepatically with CD138+ primary myeloma cells isolated from patients (Fig. 2F). These cells were derived from two distinct cases in Fig. 1D: one displaying high LAMP5 expression and the other with low LAMP5 expression in myeloma cells. Serum collected from the mice underwent ELISA to quantify human λ light chains, a biomarker reflecting tumor burden. Notably, mice injected with LAMP5-high myeloma cells exhibited significantly elevated λ light chain levels compared to those receiving LAMP5-low cells (Fig. 2G). In addition, In vivo experiments involving the subcutaneous injection of ARP-1 or IM-9 cells (shCtrl and shLAMP5) into NSG mice confirmed that LAMP5 suppression significantly impeded tumor progression in the host mice compared to the control group (Fig. 2H–K). Collectively, these data underscore the pivotal role of LAMP5 in facilitating myeloma cell proliferation.
A, B RNA-seq analysis of pathway enrichment in LAMP5-overexpressing myeloma cells (LAMP5) compared to control cells (Vec). C, D Proliferation of myeloma cells with reduced (shLAMP5) or increased (LAMP5) LAMP5 expression over time, as determined by CellTiter-Glo Luminescent Cell Viability Assay. Nontargeted shRNA (shCtrl) or control vector (Vec) served as control (n = 3 biological replicates). E Representative images and the percentage of EdU-positive cells of ARP-1 or IM-9 cells (shCtrl or shLAMP5), as determined by EdU staining assay. Scale bar, 40 μm. (n = 3 biological replicates). F Schematic diagram showing primary patient-derived mouse model generation and analyses performed. Mice were injected with primary myeloma cells that had high or low expression of LAMP5. G ELISA analysis shown the concentrations of human λ chains in mouse serum (n = 5 mice/group). H Photographic images of tumors in NSG mice after implantation of ARP-1 or IM-9 (shCtrl or shLAMP5) cells (n = 3 mice/group). Time course of tumor volume (I) and tumor weight (J). K Immunofluorescent staining of Ki67 expression in tumor tissues. Scale bar, 40 μm. Data are averages ±SD. G, J: P values were determined by unpaired two-tailed t test; C, D, I: P values were determined using two-way ANOVA.
LAMP5 interacts with IRF4 to maintain its protein stability
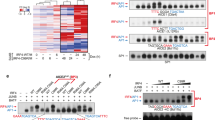
In our subsequent exploration of the regulatory mechanism of LAMP5 in myeloma cell proliferation, we attempted to delineate the interacting proteins of LAMP5 within myeloma cells. Commencing with immunoprecipitation followed by mass spectrometry analysis, we identified IRF4 as a specific binding partner of LAMP5 (Fig. 3A, Supplementary Table 2). IRF4, the cornerstone of the interferon regulatory family of transcription factors, plays an indispensable role in the etiology of myeloma [21]. Predominantly expressed in B and plasma cells, IRF4 has a significant influence on the differentiation of B cells into plasma cells and class switching of immunoglobulins. Overexpression of IRF4, a frequent observation in cells from myeloma patients, is often attributed to activating mutations or chromosomal translocations, which are pivotal for cell survival [21]. To elucidate the interplay between endogenous LAMP5 and IRF4 within myeloma cells, we conducted immunoprecipitation assays using specific antibodies against LAMP5 or IRF4 in myeloma cell lysates. Subsequent analysis of these lysates confirmed the presence of both proteins in the precipitates, thereby substantiating the endogenous association of the LAMP5/IRF4 complex in myeloma cells (Fig. 3B). Co-immunoprecipitation experiments were performed, in which HEK293T cells were co-transfected with plasmids encoding LAMP5 and IRF4. After immunoprecipitation, the precipitates were probed for the presence of either protein, which revealed a specific interaction between LAMP5 and IRF4 (Fig. 3C). Further pulldown studies were performed by mixing lysates from HEK293T cells expressing HA-tagged LAMP5 with those expressing IRF4, and precipitating the complexes using an anti-HA antibody. Our observations unequivocally demonstrated a direct interaction between LAMP5 and IRF4 (Fig. 3D). Immunofluorescence staining further corroborated the interaction between LAMP5 and IRF4 in myeloma cell lines and primary patient samples (Fig. 3E, F). Based on previous research, IRF4 harbors two conserved domains [22]: the N-terminal DNA-binding domain (DBD) and C-terminal IRF association domain (IAD) (Fig. 3G). To pinpoint the domain of IRF4 mediating interaction with LAMP5, we selectively ablated these domains (ΔDBD or ΔIAD) (Fig. 3H). Pulldown assays revealed that LAMP5 engages with IRF4 via its DNA-binding domain (Fig. 3I).
A Analysis of immunoprecipitates of HA-tagged LAMP5 in IM-9 cells with mass spectrometry. The proteins identified are indicated. B Co-immunoprecipitation of LAMP5 with IRF4 in ARP-1 or IM-9 cells. C Co-immunoprecipitation of LAMP5 with IRF4 in HEK293T cells co-transfected with LAMP5 and IRF4 plasmids. D Pull-down of HA-LAMP5 with IRF4 in HEK293T cells. E Immunofluorescent staining of ARP-1 or IM-9 cells with DAPI and antibodies against LAMP5 and IRF4. Scale bar, 10 μm. F Immunofluorescent staining of myeloma patients bone marrow samples with DAPI and antibodies against CD138, LAMP5 and IRF4 (n = 5). Scale bar, 10 μm. G Schematic of the truncations including ΔDBD and ΔIAD fragments. H Western blotting showing different truncations of IRF4 (ΔDBD and ΔIAD) in HEK293T cells. I Pull-down of LAMP5 with different truncations of IRF4 (ΔDBD and ΔIAD) in HEK293T cells. J–M The expression of IRF4 or c-MYC in myeloma cells with reduced (shLAMP5) or increased (LAMP5) LAMP5 expression. Nontargeted shRNA (shCtrl) or control vector (Vec) served as control. N, O The expression of LDHA or KLF2 in myeloma cells with reduced (shLAMP5) LAMP5 expression. Nontargeted shRNA (shCtrl) served as control. P, Q Immunofluorescent staining of ARP-1 or IM-9 cells (shCtrl and shLAMP5) with DAPI and antibody against LAMP5 and IRF4. Scale bar, 10 μm. R Immunofluorescent staining of tumor tissues in Fig. 2F with DAPI and antibodies against LAMP5 and IRF4. Scale bar, 20 μm. S Correlation coefficient of the mRNA levels of LAMP5 and mRNA levels of IRF4 or c-MYC in myeloma patients (n = 559). The correlations were evaluated using Pearson coefficient. r, correlation coefficient. B–D, H, I, and M are representative of two independent experiments. E, J–L, and N–R are representative of three independent experiments. Data are averages ±SD. P value was determined by Pearson coefficient. J, K, L, N, O P values were determined using one-way ANOVA.
To elucidate the effect of LAMP5 on IRF4 expression and protein homeostasis in myeloma cells, we conducted a series of experiments. Initially, we observed that neither the knockdown nor overexpression of LAMP5 influenced the transcriptional levels of IRF4 mRNA in myeloma cells (Fig. 3J, K). However, it significantly affected the mRNA levels of c-MYC, a direct downstream target of IRF4 (Fig. 3L). Western blot analysis revealed that knockdown of LAMP5 in myeloma cells reduced the protein levels of both IRF4 and c-MYC, whereas overexpression of LAMP5 increased their protein levels (Fig. 3M). Quantitative real-time PCR results demonstrated that the downregulation of LAMP5 significantly repressed the expression of IRF4 target genes, lactate dehydrogenase A (LDHA) and krüppel-like factor 2 (KLF2) (Fig. 3N, O). These findings suggest that LAMP5 may modulate the protein homeostasis of IRF4. Furthermore, immunofluorescence staining confirmed that the knockdown of LAMP5 decreased IRF4 protein levels in both myeloma cell lines and mouse xenograft tumors (Fig. 3P–R). Given the critical role of IRF4 in myeloma cell survival, we further explored whether the interaction between LAMP5 and IRF4 could modulate the sensitivity of myeloma cells to the chemotherapeutic agent bortezomib. Our findings revealed that the overexpression of LAMP5 in myeloma cells significantly mitigated bortezomib-induced apoptosis, suggesting a potential role for LAMP5 in chemoresistance (Supplementary Fig. 4). Furthermore, correlation analysis of primary myeloma cells indicated no correlation between LAMP5 expression and IRF4 mRNA levels; however, a correlation with c-MYC mRNA levels was observed (Fig. 3S). Collectively, these results demonstrate that LAMP5 interacts with IRF4 and maintains the protein homeostasis of IRF4, thereby promoting the expression of its downstream target genes.
LAMP5 inhibits the degradation of IRF4 protein via the autophagy-lysosome pathway
Upon treatment with cycloheximide, a potent inhibitor of eukaryotic protein synthesis [23], we observed a marked acceleration in the degradation of IRF4 protein in LAMP5 knockdown myeloma cells compared to that in control cells (Fig. 4A, B). This finding prompted us to hypothesize that LAMP5 may stabilize IRF4 protein, shielding it from proteolytic degradation. Given the central roles of proteasomes and lysosomes in maintaining intracellular protein homeostasis, we treated the cultures with MG-132, a specific proteasome inhibitor [24], or leupeptin [25], a lysosomal protease inhibitor. Our results indicated that in the absence of LAMP5, leupeptin, but not MG-132, led to the accumulation of IRF4 protein in myeloma cells (Fig. 4C, D), suggesting that LAMP5 preserves IRF4 protein stability by diverting it from lysosomal degradation pathways. Sequestosome 1/ubiquitin-binding protein p62 (p62) and heat shock cognate protein 70 (Hsc70) are two key molecules responsible for autophagy-mediated protein degradation [26]. Co-immunoprecipitation assays showed that p62 bound to the IRF4 protein (Fig. 4E, F). To explore the potential role of LAMP5 in the interaction between IRF4 and p62, the myeloma cells were transduced with a lentiviral vector expressing shLAMP5. Subsequent immunoprecipitation using an anti-IRF4 antibody, followed by immunoblot analysis with an anti-p62 antibody, revealed a notable increase in the presence of p62 in precipitates from shLAMP5-transduced cells compared to those from shCtrl-transduced cells (Fig. 4G, H). This enhancement of the IRF4-p62 complex upon LAMP5 knockdown suggests a negative regulatory function of LAMP5 in this interaction.
A, B Time course of IRF4 protein in ARP-1 or IM-9 cells (shCtrl or shLAMP5) treated with 100 μM cycloheximide (CHX). C, D IRF4 protein levels in ARP-1 or IM-9 (shCtrl or shLAMP5) treated with 2.5 μM MG-132 or 2.5 μM leupeptin for 7 h. E, F Co-immunoprecipitation of IRF4 with p62 or Hsc70 in ARP-1 or IM-9 cells. G, H Co-immunoprecipitation of IRF4 with p62 in ARP-1 or IM-9 cells (shCtrl or shLAMP5). I LC3B-I/II and p62 protein levels in shCtrl or shLAMP5 myeloma cells. J, K Immunofluorescent staining of IM-9 cells (shCtrl or shLAMP5) with DAPI and antibodies against IRF4, LC3B, or p62. Scale bar, 5 μm. L Co-immunoprecipitation of lysates from IM-9 cells (shCtrl or shLAMP5) treated with or without 20 μM chloroquine (CQ) for 24 h. A–I, and L are representative of two independent experiments. J and L are representative of three independent experiments.
In addition, to investigate the regulatory function of LAMP5 in autophagy within myeloma cells, we observed a significant upregulation in LC3B-II levels (Fig. 4I) and an increase in autophagic puncta (Fig. 4J) upon LAMP5 knockdown. This suggests that LAMP5 may play a role in the inhibition of autophagy in these cells. Furthermore, we noted an increase in the co-localization of IRF4 with p62 in myeloma cells expressing shLAMP5, which was not observed in control cells (Fig. 4K). Given that p62 binds ubiquitinated protein aggregates and interacts with LC3 for autophagic clearance, we hypothesized that LAMP5 modulates the interaction between IRF4 and p62, thereby influencing IRF4 protein turnover. To test this hypothesis, ARP-1 cells were transduced with lentivirus carrying shCtrl or shLAMP5 and then treated with the autophagy inhibitor chloroquine to block autophagosome-lysosome fusion. Cell lysates were subjected to immunoprecipitation, and western blot analysis revealed a substantial increase in IRF4 polyubiquitination, and p62 and LC3B-II protein levels in shLAMP5 cells compared with shCtrl cells (Fig. 4L). These findings suggest that LAMP5 suppression enhances the binding of IRF4 to p62, which may subsequently promote p62-mediated IRF4 protein degradation, implicating a novel regulatory mechanism by which LAMP5 influences autophagic flux and protein homeostasis in myeloma cells.
LAMP5 promotes the nuclear import of IRF4 regulated by KPNA2
Our experiments provided evidence that LAMP5 plays a role in inhibiting the degradation of IRF4 through the autophagy-lysosome pathway in the cytoplasm of myeloma cells, thus maintaining its homeostasis. Given that IRF4 is a transcription factor that primarily exerts its transcriptional regulatory functions after translocation into the nucleus, our future research aims to explore whether LAMP5 aids in the nuclear translocation of IRF4 and its subsequent transcriptional activity. By using mass spectrometry analysis, karyopherin α2 (KPNA2) was identified as an additional component of the LAMP5/IRF4 complex (Fig. 5A). KPNA2 is known to mediate the nucleocytoplasmic transport of proteins [27], suggesting that LAMP5 may also regulate IRF4 transcriptional activity by influencing its nuclear import. Next, we explored the role of LAMP5 in nuclear translocation of IRF4. To elucidate the molecular mechanisms underlying this process, we conducted a series of co-immunoprecipitation assays. HEK293T cells were co-transfected with plasmids encoding HA-tagged KPNA2 and either IRF4 or LAMP5. After transfection, the cells were lysed and the lysates were subjected to immunoprecipitation using an anti-HA antibody. The precipitated complexes were probed for the presence of IRF4 or LAMP5, revealing a specific and direct interaction between KPNA2 and both IRF4 and LAMP5 (Fig. 5B, C). To further substantiate this interaction, we performed additional pulldown experiments using lysates from HEK293T cells expressing HA-tagged KPNA2 and IRF4. The complexes were precipitated using an anti-HA antibody, and the subsequent western blot analysis confirmed the presence of IRF4 in the precipitates, indicating a physical association with KPNA2 (Fig. 5D). Additionally, pulldown studies further demonstrated direct interactions between LAMP5 and IRF4 proteins (Fig. 5E), and revealed that KPNA2 interacts with IRF4 protein through its DNA-binding domain (Fig. 5F).
A Analysis of immunoprecipitates of HA-tagged LAMP5 in IM-9 cells with mass spectrometry. The proteins identified are indicated. B, C Co-immunoprecipitation of KPNA2 with IRF4 or LAMP5 in HEK293T cells co-transfected with HA-KPNA2 and IRF4 or LAMP5 plasmids. D Pull-down of HA-KPNA2 with IRF4 in HEK293T cells. E Pull-down of HA-KPNA2 with LAMP5 in HEK293T cells. F Pull-down of KPNA2 with different truncations of IRF4 (ΔDBD and ΔIAD) in HEK293T cells. G Co-immunoprecipitation of KPNA2 with IRF4 in ARP-1 or IM-9 cells. H Co-immunoprecipitation of KPNA2 with IRF4 in ARP-1 or IM-9 cells (shCtrl or shLAMP5). I The levels of IRF4 protein in the cytoplasm and nucleus of shCtrl or shLAMP5 myeloma cells. J, K Immunofluorescent staining of ARP-1 or IM-9 cells (shCtrl or shLAMP5) with DAPI and antibodies against IRF4 or KPNA2. Scale bar, 6 μm. L Immunofluorescent staining of tumor tissues in Fig. 2H with DAPI and antibodies against KPNA2 and IRF4. Scale bar, 20 μm. B-I are representative of two independent experiments. J–L are representative of three independent experiments.
To assess the endogenous interaction between KPNA2 and IRF4 in myeloma cells, we performed immunoprecipitation assays using specific antibodies against KPNA2 or IRF4 in myeloma cell lysates. Analysis of these precipitates confirmed the presence of both proteins, thereby validating the endogenous complex formation of KPNA2 and IRF4 in these cells (Fig. 5G). In addition, to explore the potential role of LAMP5 in the interaction between IRF4 and KPNA2, myeloma cells were transduced with a lentiviral vector expressing shLAMP5. Subsequent immunoprecipitation using an anti-IRF4 antibody, followed by immunoblot analysis with an anti-KPNA2 or IRF4 antibody, revealed a notably reduced presence of KPNA2 in the precipitates from shLAMP5-transduced cells compared to those from shCtrl-transduced cells (Fig. 5H). Moreover, we performed nuclear-cytoplasmic fractionation followed by immunoblot analysis. The results demonstrate that knockdown of LAMP5 in myeloma cells significantly reduces the nuclear translocation of IRF4 protein (Fig. 5I). These data further support our hypothesis that LAMP5 facilitates IRF4 nuclear transport. Immunofluorescence assays further substantiated that the depletion of LAMP5 leads to a significant reduction in IRF4 expression, the interaction between KPNA2 and IRF4, and the nuclear translocation of IRF4 in both myeloma cell lines and mouse xenograft tumors (Fig. 5J–L). This observation opens new avenues for investigating the role of LAMP5 in the nuclear translocation and transcriptional function of IRF4, which could have significant implications for understanding the molecular mechanisms underlying myeloma pathogenesis.
c-MYC positively feedbacks to activate the transcription of LAMP5
c-MYC is recognized as a principal transcriptional target of IRF4, and plays a pivotal role in the pathogenesis and progression of myeloma [28]. In our forthcoming research endeavors, we aimed to elucidate whether c-MYC exerts a feedback regulatory effect on LAMP5 transcription. We found that c-MYC knockdown significantly repressed LAMP5 expression (Fig. 6A–C). ChIP assays further confirmed the binding of c-MYC to the promoter of LAMP5 (Fig. 6D). Luciferase activity assays demonstrated that c-MYC could bind to the LAMP5 promoter at the -1083 bp binding site to enhance its transcription (Fig. 6E, F). Our experiments demonstrated that in myeloma cells, LAMP5 interacts with IRF4 to inhibit its degradation via the autophagy-lysosome pathway, thereby maintaining its stability. Furthermore, LAMP5 facilitates the nuclear translocation of IRF4 by interacting with KPNA2, thereby enabling its transcriptional regulatory activity. Intriguingly, we also observed that the downstream target genes of IRF4, c-MYC, could feedback and activate the transcription of the LAMP5 gene. This suggests the formation of a malignant regulatory loop that continually promotes the progression of myeloma (Fig. 6G).
A–C The expression of c-MYC or LAMP5 in ARP-1 or IM-9 cells (shCtrl or shLAMP5). Nontargeted shRNA (shCtrl) served as control. D ChIP assay showing c-MYC enrichment in the LAMP5 promoter in myeloma cell lines (ARP-1 or IM-9). E, F Schematic of the LAMP5 promoter luciferase reporter. Solid boxes: promoter region of LAMP5; red crosses: mutations of nucleotides. The luciferase activity of LAMP5-Full constructs was set at 1. G Depiction of LAMP5-mediated signaling pathways in myeloma cells. A, B, D, and F are representative of three independent experiments. C is representative of two independent experiments. Data are averages ±SD. A, B, D P values were determined by unpaired two-tailed t test.
LAMP5 inhibitor reduces myeloma progression
Currently, there are no commercially LAMP5 inhibitors available. Therefore, we embarked on a structure-based high-throughput virtual screening (HTVS) campaign, scrutinizing a library of 71678 compounds to identify prospective LAMP5 inhibitors (Fig. 7A) [29]. Utilizing a tiered approach involving successive standard precision (SP) and extra precision (XP) docking analyses, we shortlisted four compounds with the most favorable docking scores for experimental validation (Fig. 7B). In silico docking studies revealed that pyrazofurin engages in a complex with LAMP5, mediated by four hydrogen bonds (H-bonds) with Pro69, Tyr145, and Ser158 (Fig. 7C). Cell proliferation experiments demonstrated that pyrazofurin significantly inhibited the proliferation of myeloma cells (Fig. 7D, E). However, the remaining three compounds with high docking scores showed no effect on the proliferation of myeloma cells (Supplementary Fig. 5). Immunoprecipitation assays demonstrated that pyrazofurin can significantly inhibit the interaction between LAMP5 and IRF4 proteins, leading to the degradation of the IRF4 protein (Fig. 7F, G). Western blot experiments also confirmed that a lysosomal-protease inhibitor could reverse the degradation of IRF4 protein induced by pyrazofurin (Fig. 7H). We also intravenously injected vk12598 cells into C57BL/6J mice, and after 2 weeks, treated them with pyrazofurin for two weeks (Fig. 7I). The results indicated that pyrazofurin significantly inhibited the progression of myeloma compared to control group (Fig. 7J). Immunofluorescence results also confirmed that pyrazofurin significantly inhibited the expression of IRF4 protein in myeloma cells (Fig. 7K). Furthermore, xenograft models in NSG mice also demonstrated that pyrazofurin inhibited the growth of ARP-1 or IM-9 cells (Fig. 7L, M). Immunofluorescence staining results also indicated that pyrazofurin can suppress tumor proliferation and the expression of IRF4 protein (Fig. 7N, O). In addition, cell proliferation assays indicated that pyrazofurin significantly inhibited the proliferation of the lymphoma cell lines Jurkat and Jeko-1 (Supplementary Fig. 6a, b), demonstrating its potential as a therapeutic agent in lymphoma treatment. Moreover, pyrazofurin shows no significant cytotoxicity to normal peripheral blood mononuclear cells (Supplementary Fig. 7). These collective findings suggest a broader relevance to the elucidation of the pathophysiological mechanisms underlying myeloma progression, potentially offering novel therapeutic drugs for the management of myeloma and lymphoma.
A Workflow of high-throughput virtual screening (HTVS) for LAMP5 inhibitors. B Docking scores of the top 4 candidates obtained from HTVS. C Chemical structure and in silico docking of pyrazofurin into the active pocket of human LAMP5 protein. D Proliferation of myeloma cells (ARP-1 or IM-9) treated with or without pyrazofurin (Pyr.) (0.1 or 1 μM) over time. PBS buffer treated group served as control. E Antiproliferative activities of pyrazofurin were evaluated on human myeloma cell lines ARP-1 or IM-9. F Co-immunoprecipitation of LAMP5 with IRF4 in ARP-1 or IM-9 cells treated with or without pyrazofurin (1 μM). G Co-immunoprecipitation of LAMP5 with IRF4 in HEK293T cells co-transfected with HA-LAMP5 and IRF4 plasmids. H IRF4 protein levels in myeloma cells treated with 1 μM pyrazofurin and 2.5 μM MG-132 or 2.5 μM leupeptin for 7 h. 6-week-old male C57BL/6 J mice were intravenously injected vk12598 cells (n = 9 mice/group), followed by intraperitoneal administration of pyrazofurin (5 mg/kg bodyweight) three times per week for a duration of two weeks, beginning two weeks after cell injection. Shown are the experimental schematic (I). J ELISA analysis shown the concentrations of IgG2b in mouse sera. K Immunofluorescent staining of I mentioned spleen tumor cells of vk12598 mice with DAPI and antibodies against CD138 or IRF4. Scale bar, 40 μm. L Photographic images of tumors in NSG mice after implantation of ARP-1 or IM-9 cells (n = 3 mice/group), and intraperitoneal administration with or without pyrazofurin (5 mg/kg bodyweight). Shown are time course of tumor volume (M). N, O Immunofluorescent staining of Ki67 or IRF4 expression in tumor tissues. D and E are representative of three independent experiments. F–H are representative of two independent experiments. Data are averages ±SD. J P value was determined by unpaired two-tailed t test; D, M P values were determined using two-way ANOVA.
Discussion
To enhance the survival prospects for myeloma patients, a plethora of research has been dedicated to deciphering the molecular intricacies that fuel this malignant condition. The current armamentarium of treatments, including radiotherapy, chemotherapy, surgical interventions, immunotherapy, and stem cell transplantation, has modestly improved patient longevity. However, the grim reality of a poor prognosis persists. This enduring challenge is a testament to our incomplete understanding of the molecular orchestration that propels myeloma and paucity of targeted therapeutics. It is imperative to delve deeper into the molecular biology of myeloma genesis and progression with the aim of developing more efficacious, targeted drugs for synergistic treatment strategies. Our research has pinpointed LAMP5 as a critical actor in myeloma cells, where its overexpression is linked to enhanced cell proliferation. Although previous studies suggested that LAMP5 promotes myeloma progression by activating p38 MAPK [19], the mechanism underlying LAMP5-mediated p38 activation remained unresolved. In contrast to prior findings, we uncovered a novel molecular dialogue in which LAMP5 engages with IRF4, shielding it from the autophagy-lysosome pathway degradation machinery. Furthermore, LAMP5 orchestrates an enhanced alliance between IRF4 and the nuclear import facilitator KPNA2, thus ensuring IRF4 nuclear ingress and circumventing its sequestration in the cytoplasm, which would otherwise lead to degradation. Within the nucleus, IRF4 acts as a transcriptional conductor for the c-MYC gene, which feeds back positively to stimulate LAMP5 expression, establishing a self-perpetuating loop that escalates myeloma progression. Our high-throughput drug screening has serendipitously identified pyrazofurin, a compound that disrupts the LAMP5-IRF4 interaction, culminating in IRF4 degradation and consequential inhibition of myeloma progression. This discovery not only sheds new light on the molecular underpinnings of myeloma progression but also presents a promising therapeutic approach. The implications of our findings are profound, suggesting that targeting the LAMP5-IRF4-KPNA2 complex, may disrupt the vicious cycle that drives myeloma progression. This research represents a significant step towards the development of a new class of targeted therapies, offering a glimmer of hope for patients with this devastating disease. As we advance our understanding of the molecular landscape of myeloma, the prospect of more personalized and effective treatment strategies has become increasingly tangible.
IRF4 is a multifaceted transcription factor that plays a critical role in the development of immune cells and the immune response [21]. In myeloma, IRF4 assumes a pivotal position, dictating a plethora of genetic programs that are central to neoplastic cell growth, survival, and metabolism [21, 30]. Despite the commonality of elevated IRF4 levels in myeloma cells, it is often ascribed to genetic alterations that foster its expression. While the transcription of IRF4 can be modulated by its own regulatory mechanisms [21] or by factors such as IKZF1/3 and NF-κB [31, 32], research on the maintenance of IRF4 protein homeostasis in myeloma cells remain unclear. Post-translational modifications are recognized as key regulatory mechanisms of protein function [33]. Notably, acetyl-CoA synthetase 2 (ACSS2) has been reported to interact with IRF4, enhancing its stability and transcriptional activity through lysine acetylation. This process involves ACSS2-mediated acetylation, which impairs lysosome-dependent degradation of IRF4 [34]. Our study delves into the intricate interplay between IRF4 and LAMP5, uncovering a novel axis that preserves IRF4 stability. We’ve discovered that LAMP5, by interacting with IRF4, shields it from proteolytic degradation through the autophagy-lysosome pathway. This finding reveals a molecular mechanism distinct from previous reports that IRF4 avoids degradation through acetylation modifications. This finding adds a new dimension to our understanding of IRF4 regulation in myeloma.
Moreover, as a transcription factor, IRF4 relies primarily on nuclear translocation to exert its transcriptional regulatory functions. However, there are still very limited reports on how IRF4 is transported into the nucleus. A previous study suggested that SUMOylation of IRF4 mediated by the ubiquitin-conjugating enzyme E2 (Ubc9) enhances its nuclear localization and stability, thereby transcribing IL-4 and arginase 1 (ARG1) to promote the macrophage M2 program [35]. In our study, LAMP5 augmented the affiliation between IRF4 and the nuclear transport facilitator KPNA2, thereby expediting IRF4 nuclear import and circumventing cytosolic sequestration and subsequent lysosomal decay. Previous study has shown that during KPNA2-mediated nuclear transport of cargo proteins, molecular chaperones (e.g., Hsp70) induce conformational changes in cargo proteins by interacting with them, thereby exposing their nuclear localization signal (NLS) sequences and facilitating recognition by KPNA2 [36]. The IRF4 protein contains a conserved bipartite NLS sequence within its N-terminal DNA-binding domain (DBD), spanning amino acids 78–98 [37]. The pull-down assays demonstrated that KPNA2 or LAMP5 directly binds to the DBD domain of IRF4. Additionally, LAMP5 enhances the interaction between KPNA2 and IRF4. Based on our findings, we hypothesize that LAMP5 binding to the DBD domain of IRF4 induces structural rearrangements in IRF4, leading to the exposure of its NLS sequence. This mechanism would enable increased recruitment of KPNA2, thereby enhancing IRF4 nuclear translocation. Importantly, our study suggests that LAMP5 may function in a chaperone-like manner to regulate this process. This nuclear translocation of IRF4 is a critical step, as it promotes the transcription of oncogenic c-MYC, which in turn amplifies LAMP5 transcription, instituting a sinister feedback loop that accelerates myeloma progression. The discovery of pyrazofurin via high-throughput screening is of interest. This small molecule has demonstrated the ability to specifically disrupt the interaction between LAMP5 and IRF4, precipitating IRF4 degradation and thereby curtailing myeloma cell growth. Our findings suggest that pyrazofurin could be a promising therapeutic candidate for myeloma, potentially offering a new avenue of treatment for patients, especially those with refractory disease where existing therapies have failed. Interestingly, pyrazofurin has also shown potential in curbing the proliferation of lymphoma cells, suggesting at its broad applicability in oncology. However, further research is warranted to validate its efficacy and safety in lymphoid malignancies.
The potential of pyrazofurin to interfere with the IRF4-LAMP5 axis represents a novel therapeutic strategy that could complement existing treatments and offer a multifaceted approach to myeloma management. This strategy underscores the importance of understanding the post-translational modifications and protein-protein interactions that dictate the stability and function of oncogenic transcription factors such as IRF4. As research progresses, the prospect of harnessing these mechanisms to develop targeted therapies has become increasingly promising, bringing new hope to the forefront of myeloma treatment.
Data availability
All supporting data have been included in the manuscript and supplemental files. Additional data are available upon reasonable request to the corresponding author. The RNA-seq data generated in this study is available at GEO database (PRJNA1160438).
References
Palumbo A, Anderson K. Multiple myeloma. N Engl J Med. 2011;364:1046–60.
Liu H, He J, Koh SP, Zhong Y, Liu Z, Wang Z et al. Reprogrammed marrow adipocytes contribute to myeloma-induced bone disease. Sci Transl Med. 2019;11:eaau9087.
Liu H, Liu Z, Du J, He J, Lin P, Amini B et al. Thymidine phosphorylase exerts complex effects on bone resorption and formation in myeloma. Sci Transl Med. 2016;8:353ra113.
Cowan AJ, Green DJ, Kwok M, Lee S, Coffey DG, Holmberg LA et al. Diagnosis and management of multiple myeloma: a review. JAMA. 2022;327:464–77.
Boiarsky R, Haradhvala NJ, Alberge JB, Sklavenitis-Pistofidis R, Mouhieddine TH, Zavidij O et al. Single cell characterization of myeloma and its precursor conditions reveals transcriptional signatures of early tumorigenesis. Nat Commun. 2022;13:7040.
Tian E, Zhan F, Walker R, Rasmussen E, Ma Y, Barlogie B et al. The role of the Wnt-signaling antagonist DKK1 in the development of osteolytic lesions in multiple myeloma. N Engl J Med. 2003;349:2483–94.
Kuehl WM, Bergsagel PL. Multiple myeloma: evolving genetic events and host interactions. Nat Rev Cancer. 2002;2:175–87.
Tsuchiya H, Epstein J, Selvanayagam P, Dedman JR, Gallick G, Alexanian R et al. Correlated flow cytometric analysis of H-ras p21 and nuclear DNA in multiple myeloma. Blood. 1988;72:796–800.
Zhang PL, Quiery AT Jr., Blasick TM, Brown RE. Morphoproteomic expression of H-ras (p21ras) correlates with serum monoclonal immunoglobulin reduction in multiple myeloma patients following pamidronate treatment. Ann Clin Lab Sci. 2007;37:34–8.
Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–12.
Roy P, Sarkar UA, Basak S. The NF-kappaB activating pathways in multiple myeloma. Biomedicines. 2018;6:59.
Spaan I, Raymakers RA, van de Stolpe A, Peperzak V. Wnt signaling in multiple myeloma: a central player in disease with therapeutic potential. J Hematol Oncol. 2018;11:67.
Alessandrini F, Pezze L, Ciribilli Y. LAMPs: shedding light on cancer biology. Semin Oncol. 2017;44:239–53.
Deng Y, Bi M, Delerue F, Forrest SL, Chan G, van der Hoven J et al. Loss of LAMP5 interneurons drives neuronal network dysfunction in Alzheimer’s disease. Acta Neuropathol. 2022;144:637–50.
Combes A, Camosseto V, N’Guessan P, Arguello RJ, Mussard J, Caux C et al. BAD-LAMP controls TLR9 trafficking and signalling in human plasmacytoid dendritic cells. Nat Commun. 2017;8:913.
Umeda S, Kanda M, Shimizu D, Nakamura S, Sawaki K, Inokawa Y et al. Lysosomal-associated membrane protein family member 5 promotes the metastatic potential of gastric cancer cells. Gastric Cancer. 2022;25:558–72.
Beird HC, Khan M, Wang F, Alfayez M, Cai T, Zhao L et al. Features of non-activation dendritic state and immune deficiency in blastic plasmacytoid dendritic cell neoplasm (BPDCN). Blood Cancer J. 2019;9:99.
Gracia-Maldonado G, Clark J, Burwinkel M, Greenslade B, Wunderlich M, Salomonis N et al. LAMP-5 is an essential inflammatory-signaling regulator and novel immunotherapy target for mixed lineage leukemia-rearranged acute leukemia. Haematologica. 2022;107:803–15.
Chen Y, Ma T. LAMP5 may promote MM progression by activating p38. Pathol Oncol Res. 2023;29:1611083.
Charan J, Biswas T. How to calculate sample size for different study designs in medical research?. Indian J Psychol Med. 2013;35:121–6.
Shaffer AL, Emre NC, Lamy L, Ngo VN, Wright G, Xiao W et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–31.
Sundararaj S, Seneviratne S, Williams SJ, Enders A, Casarotto MG. Structural determinants of the IRF4/DNA homodimeric complex. Nucleic Acids Res. 2021;49:2255–65.
Li X, Zheng J, Su J, Wang L, Luan L, Wang T et al. Myotubularin 2 interacts with SEC23A and negatively regulates autophagy at ER exit sites in Arabidopsis. Autophagy. 2024;21:141–59.
Zhang P, Xiao Z, Wang S, Zhang M, Wei Y, Hang Q et al. ZRANB1 is an EZH2 deubiquitinase and a potential therapeutic target in breast cancer. Cell Rep. 2018;23:823–37.
Cheng Z, Gan W, Xiang Q, Zhao K, Gao H, Chen Y et al. Impaired degradation of PLCG1 by chaperone-mediated autophagy promotes cellular senescence and intervertebral disc degeneration. Autophagy. 2024;21:352–73.
Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol. 2013;15:713–20.
Alshareeda AT, Negm OH, Green AR, Nolan CC, Tighe P, Albarakati N et al. KPNA2 is a nuclear export protein that contributes to aberrant localisation of key proteins and poor prognosis of breast cancer. Br J Cancer. 2015;112:1929–37.
Agnarelli A, Mitchell S, Caalim G, Wood CD, Milton-Harris L, Chevassut T et al. Dissecting the impact of bromodomain inhibitors on the Interferon Regulatory Factor 4-MYC oncogenic axis in multiple myeloma. Hematol Oncol. 2022;40:417–29.
Wang Y, Chen H, Liu W, Yan H, Zhang Y, Cheung AHK et al. MCM6 is a critical transcriptional target of YAP to promote gastric tumorigenesis and serves as a therapeutic target. Theranostics. 2022;12:6509–26.
Agnarelli A, Chevassut T, Mancini EJ. IRF4 in multiple myeloma-Biology, disease and therapeutic target. Leuk Res. 2018;72:52–8.
Kronke J, Udeshi ND, Narla A, Grauman P, Hurst SN, McConkey M et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–5.
Xie Z, Bi C, Chooi JY, Chan ZL, Mustafa N, Chng WJ. MMSET regulates expression of IRF4 in t(4;14) myeloma and its silencing potentiates the effect of bortezomib. Leukemia. 2015;29:2347–54.
Lee JM, Hammaren HM, Savitski MM, Baek SH. Control of protein stability by post-translational modifications. Nat Commun. 2023;14:201.
Li Z, Liu H, He J, Wang Z, Yin Z, You G et al. Acetyl-CoA synthetase 2: a critical linkage in obesity-induced tumorigenesis in myeloma. Cell Metab. 2021;33:78–93 e77.
Wang F, Sun F, Luo J, Yue T, Chen L, Zhou H et al. Loss of ubiquitin-conjugating enzyme E2 (Ubc9) in macrophages exacerbates multiple low-dose streptozotocin-induced diabetes by attenuating M2 macrophage polarization. Cell Death Dis. 2019;10:892.
Rutledge BS, Choy WY, Duennwald ML. Folding or holding?-Hsp70 and Hsp90 chaperoning of misfolded proteins in neurodegenerative disease. J Biol Chem. 2022;298:101905.
Li W, Ling L, Wang Z, Liang Y, Huang W, Nie P et al. Functional domains and amino acid residues of Japanese eel IRF1, AjIRF1, regulate its nuclear import and IFN/Mx promoter activation. Dev Comp Immunol. 2021;116:103923.
Acknowledgements
We thank the Research Histology and Bone Histomorphometry Core Labs and the Preclinical Imaging Core at the Xiamen University.
Funding
This research was supported by the National Natural Science Foundation of China (82170197 to HL), Fujian Science Fund for Distinguished Young Scholars (2022D015 to HL), Shenzhen Science and Innovation Committee (JCYJ20220530143403007 to HL), Basic Scientific Research Projects of Public Welfare Research Institutes Affiliated to Fujian Province (2024R1036 to HL), Guangdong Basic and Applied Basic Research Foundation (2024A1515010076 to HL), The Science and Technology Innovation Leading Talent Support Program of Henan Province (254000510004 to CW), and Key Scientific Research Funding Program for Higher Education Institutions of Henan Province (24A320060 to CW).
Author information
Authors and Affiliations
Contributions
HL and CW designed all the experiments and wrote the manuscript. ZL, RL, ZHF, RC, DYY, YL and SRL performed the experiments and statistical analysis. ZHF and CW provided the patient samples. All authors have reviewed the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, Z., Liu, R., Fang, Z. et al. LAMP5 modulates IRF4 stability and nuclear transport: a critical mechanism in myeloma progression and therapy. Oncogene 44, 3553–3567 (2025). https://doi.org/10.1038/s41388-025-03513-x
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-025-03513-x