Abstract
Nuclear ubiquitous casein and cyclin-dependent kinase substrate 1 (NUCKS1) functions as an oncogene in colorectal cancer (CRC), promotes the progression of CRC, and is associated with poor prognosis in patients. Studies have found that NUCKS1 promotes tumor cell metastasis, yet its role in CRC invasion and metastasis remains unclear. Our findings revealed higher NUCKS1 expression in metastatic CRC compared to non-metastatic samples. Upregulation of NUCKS1 expression promoted the migration and invasion of CRC cells, while knockdown of NUCKS1 significantly inhibited the migration and invasion of CRC cells. Mechanistically, NUCKS1 was initially found to upregulate HDAC2 expression by inhibiting the lysosomal pathway, activating AKT, and thus promoting CRC invasion and metastasis. Moreover, HDAC2 inhibitor Santacruzamate A or AKT inhibitor LY294002 rescued the migration and invasion of CRC cells caused by NUCKS1 overexpression. In vivo, by injecting CRC cells into the tail vein of a nude mouse model, we found that overexpression of NUCKS1-induced lung and liver metastasis was suppressed by HDAC2 knockdown or intraperitoneal administration of the HDAC2 inhibitor Santacruzamate A. Meanwhile, AKT inhibitor LY294002 significantly inhibited lung and liver metastasis caused by overexpression of HDAC2. The expression levels of NUCKS1, HDAC2, and phosphorylated AKT were significantly positively correlated in human CRC tissues. These findings suggest that NUCKS1 contributes to CRC invasion and metastasis by stabilizing HDAC2 and activating AKT, highlighting NUCKS1 and HDAC2 as potential therapeutic targets for CRC.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is the third most common cancer worldwide and the second leading cause of cancer death [1, 2]. CRC is characterized by its heterogeneous nature and early asymptomatic presentation. As the disease progresses to a certain extent, it manifests through various symptoms such as changes in stool shape and bowel habits, bloody or purulent stools, painful urination, weight loss, and fatigue [3]. Although early screening programs can reduce the incidence of CRC, early-stage CRC often remains undetected due to the lack of symptoms, leading to missed diagnoses. Surgical resection is the primary treatment for CRC, yet 30–40% of postoperative patients still experience tumor metastasis [4]. Hence, understanding the mechanisms of CRC metastasis, identifying related proteins and pathways, and developing effective therapeutic drugs are crucial for improving patient outcomes. Additionally, it provides a basis for research and scientific evidence for establishing biomarkers for early diagnosis of CRC. It also offers feasible and effective strategies for the clinical treatment of advanced metastatic CRC.
Nuclear ubiquitous casein and cyclin-dependent kinase substrate 1 (NUCKS1) is a member of the high mobility group family, highly expressed in various tumor cell types and tissues [5, 6]. NUCKS1 functions as an oncogene in several cancers, including CRC [7], cervical squamous cell carcinoma [8], breast cancer [9], gastric cancer [10], and lung cancer [11]. Additionally, NUCKS1 acts as a transcription factor to promote the growth and metastasis of osteosarcoma cells [12] and regulates tumor cells’ entry into the S phase [13]. Previous research demonstrated that NUCKS1 is highly expressed in CRC and promotes its proliferation [14]. However, its role in CRC invasion and metastasis remains unclear.
Zn2+-dependent histone deacetylases (HDACs) are classified into classes I to IV, with histone deacetylase 2 (HDAC2), a class I enzyme, located in the nucleus [15]. HDAC2 is crucial in tumorigenesis, promoting cancer cell proliferation by regulating signaling pathways, apoptosis, and cell cycle progression [16,17,18]. Moreover, HDAC2 inhibitor Trichostatin A leads to weakened cell invasion and migration ability [19]. HDAC2 is highly expressed in CRC and mediates the proliferation of CRC in nude mice [20]. The promoting effect of claudin-1 on invasion and metastasis of colon cancer depends on HDAC2, and HDAC inhibitors can inhibit claudin-1 induced invasion of colon cancer cells [21]. In CRC, HDAC2 is highly expressed and promotes epithelial-mesenchymal transition (EMT) by binding HDAC1 and EZH2 [22], playing a significant role in CRC development [23]. Thus, HDAC2 emerges as a potential therapeutic target for CRC treatment. At present, over 300 HDAC inhibitors are under clinical research, with over 10 approved drugs. HDAC2 inhibitors exert anti-tumor effects by inhibiting HDAC activity, regulating histone acetylation status, promoting transcription and expression of anti-tumor transcription factors, and modulating related signaling pathways. The primary indications for these inhibitors include tumors, neurological diseases, metabolic diseases, autoimmune diseases, and cardiovascular diseases [24].
This study revealed that NUCKS1 promotes CRC invasion and metastasis in vitro and in vivo. NUCKS1 influences CRC invasion and metastasis by stabilizing HDAC2 and enhancing AKT phosphorylation. Importantly, NUCKS1 interacts with HDAC2, regulating its protein levels through the lysosomal pathway. Additionally, HDAC2 inhibitors may serve as effective clinical treatments for patients with CRC exhibiting high NUCKS1 expression.
Results
NUCKS1 promotes the invasion and metastasis of CRC
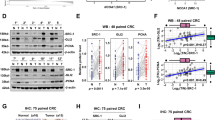
Expression of NUCKS1 in metastatic and non-metastatic CRC was analyzed using the human cancer metastasis database (HCMDB). Databases GSE71222 and GSE14095 indicated that NUCKS1 is highly expressed in metastatic CRC (Fig. 1a, b). To evaluate the function of NUCKS1 in CRC, stable NUCKS1-silenced HCT116 cells and NUCKS1-overexpressing SW480 and DLD1 cells were constructed (Fig. 1c, d). Transwell assays revealed that NUCKS1 silencing significantly reduced the migration and invasion abilities of tumor cells compared to controls (Fig. 1e). Conversely, NUCKS1 overexpression markedly promoted the migration and invasion of SW480 and DLD1 cells (Fig. 1f). Wound healing assays further demonstrated that NUCKS1 silencing decreased migration in HCT116 cells (Fig. 1g), whereas NUCKS1 overexpression enhanced migration in SW480 and DLD1 cells (Fig. 1h). Additionally, FITC phalloidin staining showed that NUCKS1 silencing reduced the distribution of central actin microfilaments and their connection with dense peripheral bands in HCT116 cells (Fig. 1i). Control SW480 and DLD1 cells exhibited dense peripheral bands, and NUCKS1 overexpression increased peripheral actin staining (Fig. 1j). Furthermore, Western blotting also confirmed that knockdown of NUCKS1 in HCT116 cells reduced the expression of vimentin while elevating that of E-cadherin (Supplementary Fig. S1a). This was consistent with changes in RNA levels (Supplementary Fig. S1b). When NUCKS1 was overexpressed, the protein and RNA levels of vimentin in CRC cells were significantly increased, while the level of E-cadherin was significantly decreased (Supplementary Fig. S1c, d, e, f). Overall, these findings suggest that NUCKS1 plays a crucial role in promoting CRC invasion and metastasis.
a, b Expression levels of NUCKS1 in metastatic and non-metastatic CRC analyzed by HCMDB. c, d Detection of NUCKS1 expression in stable cell lines using Western blotting. Numbers #1 and #2 indicate different shRNAs targeting NUCKS1. NUCKS1-OE indicates NUCKS1 overexpression. The number below each band represents the corresponding grayscale value. e, f Impact of NUCKS1 silencing and overexpression on invasion and metastasis of HCT116, SW480, and DLD1 cells detected by transwell assays. Scale bar = 100 μm. g, h Wound healing assay detecting the effect of NUCKS1 silencing and overexpression on CRC migration. Green fluorescence indicates live cells. Scale bar = 400 μm. i, j FITC-phalloidin staining of F-actin in HCT116 cells treated with shRNAs targeting NUCKS1, and in SW480 and DLD1 cells overexpressing NUCKS1. Scale bar = 20 μm. ***p < 0.001, ****p < 0.0001.
NUCKS1 regulates the invasion and metastasis of CRC via the HDAC2/AKT signaling pathway
To elucidate the molecular mechanism by which NUCKS1 promotes CRC invasion and metastasis, label-free quantitative proteomics identified differentially expressed proteins mediated by NUCKS1. Results showed 643 differentially expressed proteins in NUCKS1-knockdown cells compared to controls (Supplementary Table S1). A literature review further screened 183 tumor-related proteins (Supplementary Table S2). GEPIA analysis identified 28 differentially expressed proteins significantly associated with NUCKS1 and highly expressed in CRC (Fig. 2a). Among these, HDAC2 is known to promote EMT in CRC cells [22, 23], and among the proteins associated with CRC metastasis, only HDAC2 showed an increase in protein levels when NUCKS1 was highly expressed (Supplementary Fig. S2a). In addition, NUCKS1 has been found to mediate the PI3K/AKT signaling pathway [14], which was consistent with the analysis of NUCKS1 label-free quantitative proteomics data (Supplementary Fig. S2b). Subsequently, we detected the expression levels of NUCKS1, HDAC2, and phosphorylated AKT proteins in CRC cell lines. The results showed that when NUCKS1 expression decreased, the expression levels of HDAC2 and phosphorylated AKT also decreased accordingly (Supplementary Fig. S2c). Furthermore, NUCKS1 silencing in HCT116 cells reduced HDAC2 and phosphorylated AKT levels, whereas NUCKS1 overexpression increased these levels (Fig. 2b and Supplementary Fig. S2d).
a Heat map showing 28 differentially expressed proteins in NUCKS1-silenced HCT116 cells compared to control cells. b Detection of HDAC2 and phosphorylated AKT expression levels in NUCKS1-silenced and NUCKS1-overexpressing cell lines using Western blotting. c, d Western blotting assay determining HDAC2 and phosphorylated AKT protein expression levels in HCT116 and SW480 cells under various conditions. e, f Transwell assays measuring migration and invasion abilities of HCT116 and SW480 cells under different conditions. Scale bar = 100 μm. g Changes in phosphorylated AKT levels in SW480 cells overexpressing HDAC2 treated with LY294002 inhibitor (10 µM). h Changes in cell invasion and migration in SW480 cells overexpressing HDAC2 treated with LY294002 inhibitor (10 µM). Scale bar = 100 μm. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
To investigate HDAC2’s role in CRC invasion and metastasis, HDAC2-silenced HCT116 cells and HDAC2-overexpressing SW480 and DLD1 cells were constructed. HDAC2 silencing reduced phosphorylated AKT levels in HCT116 cells, while HDAC2 overexpression increased phosphorylated AKT levels (Supplementary Fig. S2e). Transwell assays demonstrated that HDAC2 silencing significantly reduced HCT116 cell invasion and migration, whereas HDAC2 overexpression enhanced the invasion and migration of SW480 and DLD1 cells (Supplementary Fig. S2f). Furthermore, Western blotting and q-PCR confirmed that knockdown of HDAC2 reduced the expression of vimentin, while increasing the expression of E-cadherin, and overexpression of HDAC2 increased the levels of vimentin, while decreasing the level of E-cadherin (Supplementary Fig. S2g, h).
To confirm whether NUCKS1 promotes CRC invasion and metastasis via HDAC2, HDAC2 was overexpressed in NUCKS1-silenced HCT116 cells. This rescued the expression levels of HDAC2 and phosphorylated AKT (Fig. 2c). Conversely, knocking out HDAC2 in NUCKS1-overexpressing SW480 and DLD1 cells significantly reduced HDAC2 and phosphorylated AKT levels (Fig. 2d and Supplementary Fig. S3a). Transwell assays showed that the suppression of migration and invasion by shNUCKS1 in HCT116 cells was partially reversed by HDAC2 overexpression (Fig. 2e). Similarly, HDAC2 silencing reduced the invasion and migration of NUCKS1-overexpressing SW480 and DLD1 cells (Fig. 2f and Supplementary Fig. S3b). In addition, in CRC cells overexpressing HDAC2, treatment with AKT inhibitor LY294002 resulted in a significant decrease in phosphorylated AKT levels compared to untreated cells (Fig. 2g and Supplementary Fig. S3c), while the invasion and migration of treated CRC cells were significantly reduced compared to untreated cells (Fig. 2h and Supplementary Fig. S3d). These results indicate that NUCKS1 induces CRC cell invasion and metastasis by regulating HDAC2/AKT.
HDAC2 inhibitor Santacruzamate A reduces the invasion and metastasis caused by NUCKS1 overexpression
Santacruzamate A, a selective and potent HDAC2 inhibitor, was employed to further investigate the role of NUCKS1 in promoting CRC invasion and metastasis through HDAC2 [25, 26]. In NUCKS1-overexpressing SW480 and DLD1 cells, Santacruzamate A significantly inhibited invasion and metastasis abilities, as shown by transwell assays (Fig. 3a, b). Additionally, wound healing assays demonstrated increased migration in NUCKS1-overexpressing cells compared to controls, which was inhibited by Santacruzamate A (Fig. 3c, d). F-actin analysis further confirmed that Santacruzamate A reduced the distribution of central actin microfilaments in NUCKS1-overexpressing cells compared to untreated cells (Fig. 3e, f). While Santacruzamate A inhibited HDAC2 protein expression, it did not affect NUCKS1 protein levels (Supplementary Fig. S4a). In NUCKS1-silenced HCT116 cells, treatment with Santacruzamate A further decreased migration and invasion abilities (Supplementary Fig. S4b). These findings suggest that NUCKS1 promotes CRC invasion and metastasis by regulating HDAC2, whereas HDAC2 does not regulate NUCKS1 protein expression.
a, b Transwell assays detecting the effects of HDAC2 inhibitor Santacruzamate A (50 μM) on invasion and migration of NUCKS1-overexpressing SW480 and DLD1 cells. Scale bar = 100 μm. c, d Wound healing assays showing the effect of Santacruzamate A on the migration of NUCKS1-overexpressing SW480 and DLD1 cells. Green fluorescence indicates live cells. Scale bar = 400 μm. e, f FITC-phalloidin staining of F-actin in NUCKS1-overexpressing SW480 and DLD1 cells treated with Santacruzamate A. ***p < 0.001 and ****p < 0.0001.
NUCKS1 interacts with and stabilizes HDAC2
To confirm the interaction between NUCKS1 and HDAC2, a correlation analysis of NUCKS1 and HDAC2 mRNA levels was performed using GEPIA, revealing a significant positive correlation (Supplementary Fig. S5a). Co-IP and Western blotting were utilized to validate the interaction, with total protein immunoprecipitated using an anti-HDAC2 antibody. NUCKS1 was co-immunoprecipitated in HCT116 and SW480 cells (Fig. 4a). Additionally, stable HEK 293 T cells overexpressing flag-tagged NUCKS1 demonstrated binding of HDAC2 to exogenous NUCKS1 via Co-IP and Western blotting (Fig. 4b), indicating their interaction. Immunofluorescence assays showed colocalization of NUCKS1 and HDAC2 in the nucleus (Fig. 4c, d and Supplementary Fig. S5b), consistent with previous reports on NUCKS1 [27] and HDAC2 [28] localization. Mean fluorescence intensity (MFI) analysis indicated that NUCKS1 silencing in HCT116 cells significantly reduced the MFI of NUCKS1 and HDAC2, while NUCKS1 overexpression in SW480 and DLD1 cells increased their MFI compared to controls (Fig. 4e, f and Supplementary Fig. S5c). HDAC2 mRNA levels remained unchanged after NUCKS1 silencing or overexpression (Fig. 4g). Protein synthesis inhibitor cycloheximide (CHX) was used to assess HDAC2 stability, revealing greater stability in control cells compared to NUCKS1-silenced HCT116 cells (Fig. 4h). These findings suggest that NUCKS1 interacts with and stabilizes HDAC2.
a, b Detection of endogenous or exogenous NUCKS1 binding to HDAC2 using Co-IP and Western blotting. c, d Immunofluorescence showing the colocalization of NUCKS1 and HDAC2 following treatment with shRNAs for NUCKS1 or NUCKS1 overexpression. Scale bar = 20 μm. Intensity profiles of NUCKS1 and HDAC2 along the white line are plotted on the right. e, f Statistical results of mean fluorescence intensity (MFI) of NUCKS1 and HDAC2. g Detection of HDAC2 mRNA levels in NUCKS1-silenced HCT116 cells and NUCKS1-overexpressing SW480 cells. h Detection of HDAC2 protein levels in NUCKS1-silenced HCT116 cells and control cells after treatment with the protein synthesis inhibitor cycloheximide (CHX) for 8, 24, and 48 hours). ns represents no statistical difference, **p < 0.01 and ****p < 0.0001.
NUCKS1 inhibits lysosome-dependent degradation of HDAC2
Destruction of NUCKS1 significantly increases HDAC2 degradation. To explore the protein degradation pathways through which NUCKS1 affects HDAC2 levels, the effects of specific inhibitors were investigated. Lysosomal inhibitor chloroquine (CQ) inhibited HDAC2 degradation mediated by NUCKS1 silencing (shNUCKS1) (Fig. 5a), whereas the proteasome inhibitor MG132 had no effect (Fig. 5b). These findings suggest that NUCKS1 regulates HDAC2 in a lysosome-dependent manner. Previous studies indicated that shNUCKS1 promotes autophagy [14], while NUCKS1 suppresses autophagy [10]. Therefore, the expression and distribution of HDAC2 to lysosomes were examined. LC3B and LAMP2, markers for autophagy and lysosomes [29, 30], respectively, were analyzed. Knockdown of NUCKS1 significantly reduced the mean fluorescence intensity (MFI) of HDAC2, while increasing LC3B and LAMP2 distribution (Fig. 5c–e). Conversely, NUCKS1 overexpression increased HDAC2 MFI and decreased LC3B and LAMP2 distribution (Fig. 5f–h and Supplementary Fig. S5d–g). These findings consistently demonstrate that NUCKS1 mediates lysosomal degradation of HDAC2.
a, b Western blotting analysis showing the effect of NUCKS1 silencing (shNUCKS1) on HDAC2 expression in the absence and presence of lysosomal inhibitor chloroquine (CQ, 20 µM) and proteasome inhibitor MG132 (5 µM for 4 h). c, d Immunofluorescence showing the colocalization of HDAC2 with LC3B or LAMP2 in shNUCKS1 and control HCT116 cells. Scale bar = 20 μm. e Statistical results of mean fluorescence intensity (MFI) of HDAC2, LC3B, and LAMP2 in shNUCKS1 HCT116 cells. f, g Immunofluorescence showing the colocalization of HDAC2 with LC3B or LAMP2 in NUCKS1-overexpressing SW480 cells. Scale bar = 20 μm. h Statistical results of MFI of HDAC2, LC3B, and LAMP2 in NUCKS1-overexpressing SW480. ns represents no statistical difference, *p < 0.05 and **p < 0.01.
NUCKS1 promotes CRC metastasis in vivo and is significantly correlated with the expression levels of HDAC2 and phosphorylated AKT
To verify the role of NUCKS1 in CRC metastasis in vivo, SW480 cells overexpressing NUCKS1 and control cells were injected into nude mice via the tail vein. Four weeks later, NUCKS1 overexpression significantly promoted liver and lung metastasis compared to controls, whereas HDAC2 knockdown significantly inhibited metastasis induced by NUCKS1 overexpression. Additionally, one week after injecting SW480 cells overexpressing NUCKS1, intraperitoneal administration of the HDAC2 inhibitor Santacruzamate A resulted in reduced liver and lung metastasis compared to the NUCKS1 overexpression group. This reduction is evident in the liver and lung images from four groups of nude mice (Fig. 6a) and the number of liver and lung metastatic lesions (Fig. 6b). Furthermore, HE staining of liver and lung metastases revealed more and larger metastatic nests in mice injected with NUCKS1-overexpressing SW480 cells compared to controls. However, the combination of NUCKS1 overexpression and HDAC2 knockdown or Santacruzamate A treatment significantly reduced the number and size of metastatic nests (Fig. 6c). Subsequently, we examined whether the metastasis caused by overexpression of HDAC2 in vivo could be inhibited by AKT inhibitor LY294002. The results showed that in the group of SW480 cells overexpressing HDAC2, the number of liver and lung metastatic nodules in nude mice was significantly increased compared to the control group. However, when AKT inhibitor LY294002 was administered intraperitoneally to the overexpressing HDAC2 group of nude mice, the number of liver and lung metastatic nodules was significantly reduced (Supplementary Fig. S6a, b). Similarly, compared with the control group, the HDAC2-overexpressing SW480 cell group showed more and larger metastatic nests, while LY294002 inhibited this result (Supplementary Fig. S6c). These data suggest that NUCKS1 significantly promotes CRC liver and lung metastasis in vivo, while HDAC2 knockdown or Santacruzamate A administration can reverse this effect, and AKT inhibitor LY294002 can also inhibit metastasis caused by high HDAC2 expression.
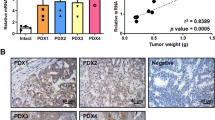
a Representative images of metastatic nodes in the livers and lungs. b Quantification of metastatic nodes. c HE staining of liver and lung metastases. Scale bars = 500 μm (left panel) and 100 μm (right panel). d Immunohistochemical assay for detecting the expression levels of NUCKS1, HDAC2, and p-AKT in 41 pairs of human CRC tissues and adjacent tissues. Magnification: ×100 and ×400, Scale bars = 100 μm (left panel) and 20 μm (right panel). e Statistics of NUCKS1, HDAC2, and p-AKT positive area in human CRC tissues and adjacent tissues. f The correlation analysis of these three proteins was presented. g Schematic diagram of the mechanism by which NUCKS1 promotes CRC invasion and metastasis by regulating the HDAC2/AKT pathway. **p < 0.01, ***p < 0.001, and ****p < 0.0001.
To further demonstrate the relationship between NUCKS1, HDAC2, and p-AKT, we detected the expression levels of the three in 41 pairs of CRC and adjacent tissue samples collected. The immunohistochemical results showed that the expression of NUCKS1 significantly increased in CRC tissues, while the expression of HDAC2 and p-AKT also significantly increased (Fig. 6d, e). Analysis revealed a significant positive correlation between the expression levels of NUCKS1 and HDAC2 and p-AKT (Fig. 6f).
NUCKS1 enhances CRC cell invasion and metastasis by interacting with HDAC2. Overexpression of NUCKS1 elevates HDAC2 levels, whereas the HDAC2 inhibitor Santacruzamate A reduces HDAC2 expression, leading to decreased AKT phosphorylation and suppression of CRC invasion and metastasis. Conversely, low NUCKS1 expression induces lysosomal degradation of HDAC2, thereby inhibiting AKT activation and CRC metastasis. Moreover, AKT inhibitor LY294002 can also inhibit CRC metastasis by suppressing the phosphorylation level of AKT. These findings suggest that Santacruzamate A could be an effective therapeutic agent for patients with CRC exhibiting elevated NUCKS1 expression (Fig. 6g).
Discussion
CRC is a leading cause of cancer-related deaths globally. Despite rising survival rates, metastatic CRC remains deadly, with a 5-year survival rate of approximately 14% [31]. Liver metastasis occurs in over 50% of patients with CRC, making it extremely common [32]. Lung metastasis, particularly in middle and lower rectal cancer, follows, with an incidence rate of ~30% [33]. Exploring the molecular mechanisms of CRC metastasis is crucial for identifying new targets for early diagnosis and treatment.
NUCKS1 has been identified as a biomarker for several cancers [27]. Previous studies indicated that NUCKS1 significantly promotes CRC development [14], but its role in invasion and metastasis was unclear. HCMDB database analysis revealed significantly higher NUCKS1 expression in patients with metastatic CRC compared to non-metastatic ones, suggesting its involvement in CRC metastasis. Further experiments demonstrated that NUCKS1 overexpression enhances CRC cell invasion and metastasis, while its knockdown inhibits these processes. This is consistent with findings in other tumors, such as non-small cell lung cancer [34] and osteosarcoma [12]. Mechanistically, NUCKS1 regulates CRC invasion and metastasis through the HDAC2/AKT pathway, as label-free quantitative proteomics identified 643 differentially regulated proteins, with HDAC2 being particularly noteworthy.
HDAC2 is pivotal in cell proliferation, signaling, and cancer progression, serving as a prognostic marker for various cancers [15]. Upregulation of HDAC2 is noted in metastatic CRC [35]. This study demonstrates that HDAC2 overexpression promotes CRC invasion and metastasis, while HDAC2 knockdown significantly inhibits these processes. Experiments confirmed the interaction between NUCKS1 and HDAC2, showing that HDAC2 degrades via the lysosomal pathway when NUCKS1 is knocked down and is overexpressed when NUCKS1 is highly expressed. Additionally, the HDAC2 inhibitor Santacruzamate A reduces HDAC2 expression in NUCKS1-overexpressing CRC cells. Literature indicates NUCKS1 promotes tumor invasion and migration through AKT activation [36, 37]. HDAC2 facilitates tumor metastasis via pathways like Wnt/β-catenin [38], PI3K/AKT/mTOR [39], and NF-κB [40]. This study reveals that NUCKS1 knockdown leads to decreased HDAC2 expression, inhibiting AKT phosphorylation and CRC metastasis. Moreover, HDAC2 overexpression induced by NUCKS1 is counteracted by Santacruzamate A, which reduces HDAC2 levels and inhibits AKT activity, thereby suppressing CRC metastasis. AKT inhibitor LY294002 also significantly reduced the metastasis of CRC caused by NUCKS1 through the HDAC2/AKT pathway.
Despite the development of 22 HDAC2 inhibitors, successful anti-cancer drugs have not emerged from clinical trials due to issues like lack of specificity, leading to adverse effects. The Zn2+ binding groups in these inhibitors can also bind with other metalloenzymes, causing cytotoxicity and limiting clinical application [41]. Solutions may include screening inhibitors for full-length HDAC2 structures or designing HDAC2-selective chemistries/peptides with anti-cancer activity without off-target effects [15]. Combining HDAC2 inhibitors with immune checkpoint inhibitors has shown promising therapeutic effects in mouse models [42, 43]. Furthermore, the world’s first AKT pathway inhibitor was approved not long ago, and Capivasertib is expected to become a new standard for HR + /HER2 advanced breast cancer targeted endocrine therapy [44]. The combination of AKT inhibitors and HDAC2 inhibitors may also be a feasible strategy for treating metastatic CRC.
Conclusions
In summary, this study reveals a significant correlation between high expression levels of NUCKS1 and HDAC2 and the invasion and metastasis of CRC. NUCKS1 and HDAC2 have the potential to serve as biomarkers for early detection of CRC metastasis. Furthermore, HDAC inhibitors show promise as clinical treatments for metastatic CRC with elevated NUCKS1 expression.
Materials and methods
Patient samples
41 pairs of CRC tissues and adjacent tissues were collected at the Tangdu Hospital of the Air Force Medical University. All specimens were frozen in liquid nitrogen until further analysis.
Cell culture and lentivirus infection
CRC cell lines HCT116, SW480, and DLD1 were obtained from Procell Life Science & Technology Company (Wuhan, China) and verified through Short Tandem Repeat (STR) profiling analysis. HCT116 cells were cultured in McCoy’s 5 A medium (HyClone, Utah, USA), SW480 cells in Leibovitz’s L15 medium (HyClone, Utah, USA), and DLD1 cells in RPMI-1640 medium (HyClone, Utah, USA), each supplemented with 10% fetal bovine serum (Gibco, South America, SA, USA). HEK 293 T cells were maintained in complete DMEM (HyClone, Utah, USA).
For cell treatments, 20 µg/mL cycloheximide (CHX, Glpbio) was added for 8, 24, and 48 h. Cells were incubated with 5 µM MG132 (Selleckchem) for 4 h and treated with 20 µM chloroquine (CQ, MCE) for 8 h. In vitro treatments with Santacruzamate A (Sellechem) were conducted at 50 µM for 6 h. AKT inhibitor LY294002 (MCE) was added to the cells at a concentration of 10 µM for 1 h.
Lentivirus packaging involved co-transfection of HEK 293 T cells (6 × 106 cells/10 cm plate) with psPAX2 (9 µg), pMD2G (3 µg), and target gene plasmid (12 µg) using 76.8 µL HiGene (Applygen, Beijing, China) for 24 h. Lentivirus supernatant was collected after 48 h and used to infect cancer cells for 12 h, followed by selection of positive cells with puromycin (Yeasen, Shanghai, China). The shRNA sequences are detailed in Table 1.
The transwell migration and invasion assays
The experiment utilized 8 μm pore transwell chambers (BD Biosciences). For cell migration assays, cells were seeded into the chamber with 200 μL of serum-free medium, while culture medium containing 10% FBS was added to the lower chamber. For invasion assays, Matrigel (BD Biosciences) diluted 1:20 with serum-free medium was added to the chamber, followed by incubation for 4 h to allow solidification before adding cells. After 24 h, cells were fixed with 95% ethanol for 15 min and stained with 1% crystal violet for 30 min. The chambers were rinsed with tap water, and the cells on the upper surface were removed using a cotton swab. Then, an inverted microscope was used for observation, and ImageJ was used to count the number of migrated or invaded cells in five randomly selected fields of view.
Wound healing assay
Cells were seeded in 6-well plates and allowed to adhere for 24 h. A pipette tip was used to create a scratch in the cell monolayer. Cells were then incubated with cell tracker green CMFDA (Invitrogen) for 15 min in the incubator. After washing with PBS (HyClone, Utah, USA), the cells were observed and photographed under a fluorescence microscope. The medium was replaced with 1% FBS culture medium, and the cells were observed and photographed again after 24 h.
Immunofluorescence staining and confocal microscopy
Cells were seeded in a confocal dish and fixed with 4% paraformaldehyde (Biosharp, Shanghai, China) for 15 min after washing with PBS. Permeabilization was performed with 0.5% Triton X-100 at room temperature for 20 min, followed by blocking for 30 min. Cells were then incubated with primary antibodies overnight at 4 °C. After washing three times with PBS, secondary antibodies were added and incubated for 1 h at room temperature in the dark. After a final wash with PBS, an anti-fade reagent was added, and cells were observed under a confocal microscope, selecting three fields of view for statistical analysis. Mean fluorescence intensity (MFI) was calculated using ImageJ software. Primary antibodies used included NUCKS1 (catalog# 12023-2-AP, Proteintech) at 1:400 dilution; HDAC2 (catalog# 67165-1-Ig, Proteintech) at 1:800 dilution; HDAC2 (catalog# ET1607-78, HUABIO) at 1:100 dilution; LC3B (catalog# 81004-1-RR, Proteintech) at 1:1000 dilution; LAMP2 (catalog# M1603-5, HUABIO) at 1:100 dilution. Secondary antibodies used: anti-rabbit IgG (H + L) Alexa FluorTM 594 (catalog# A-21207, Thermo Fisher) at 1:500 dilution; anti-rabbit IgG (H + L) Alexa FluorTM 488 (catalog# A-21206, Thermo Fisher) at 1:500 dilution; anti-mouse IgG (H + L) Alexa FluorTM 594 (catalog# A-21203, Thermo Fisher) at 1:500 dilution; and anti-mouse IgG (H + L) Alexa FluorTM 488 (catalog# A-21202, Thermo Fisher) at 1:500 dilution.
For FITC-phalloidin staining of F-actin, the TraKind™ F-actin staining kit (Abbkine, Wuhan, China) was used, and cells were observed under a confocal microscope.
Coimmunoprecipitation (Co-IP) and western blotting
Cells were lysed using IP lysis buffer (Applygen, Beijing, China) supplemented with phosphatase and protease inhibitor cocktails (Roche, USA). After 30 min on ice, lysates were centrifuged at 4 °C and 12,000 rpm for 20 min. A 100 µL aliquot of the supernatant was mixed with a loading buffer as input. The remaining supernatant was divided into two parts, with IgG and HDAC2 antibodies added to each. Following overnight incubation at 4 °C, protein A/G magnetic beads (Bimake, Texas, USA) were added and incubated at 4 °C for 4 h. Magnetic beads were collected using a magnetic rack, washed five times with cold PBS, and mixed with loading buffer. Samples were boiled at 100 °C for 10 min, and magnetic beads were removed to obtain protein samples. For HEK 293 T cells overexpressing NUCKS1-flag, anti-flag magnetic beads (Bimake, Texas, USA) were directly added to the protein supernatant, followed by magnetic bead collection and sample preparation.
Western blotting analysis was conducted as previously described [14]. ImageJ software was used to perform grayscale analysis on protein bands, and the experiment was repeated at least three times. Primary antibodies used: NUCKS1 (catalog# 12023-2-AP, Proteintech) at 1:500 dilution; HDAC2 (catalog# ET1607-78, HUABIO) at 1:1000 dilution; CHD4 (catalog# ET1704-53, HUABIO) at 1:1000 dilution; NOLC1 (catalog# HA721113, HUABIO) at 1:1000 dilution; CD166 (catalog# M1012-7, HUABIO) at 1:1000 dilution; LYAR (catalog# ER63885, HUABIO) at 1:1000 dilution; LASP1 (catalog# ER65502, HUABIO) at 1:1000 dilution; ARF6 (catalog# ET1702-91, HUABIO) at 1:1000 dilution; AKT (catalog# 4691, Cell Signaling Technology) at 1:1000 dilution; p-AKT (Ser-473) (catalog# 4060, Cell Signaling Technology) at 1:1000 dilution; E-cadherin (catalog# ET1607-75, HUABIO) at 1:1000 dilution; Vimentin (catalog# HA721174, HUABIO) at 1:1000 dilution; GAPDH (catalog# 60004-1-Ig, Proteintech) at 1:10000 dilution. Secondary antibodies used: anti-rabbit-HRP (catalog# AS014, ABclonal) at 1:4000 dilution; anti-mouse-HRP (catalog# AS003, ABclonal) at 1:4000 dilution.
Quantitative real-time PCR (q-PCR)
Cells were collected and washed twice with PBS, followed by the removal of the supernatant. RNA was extracted from cell pellet using the M5 Universal RNA Mini Kit (Mei5Bio, Beijing, China). Complementary DNA (cDNA) was synthesized using the Evo M-MLV RT Mix Kit (Accurate Biotechnology, Changsha, China). Real-time PCR was performed with the SYBR Green Premix Pro Taq HS qPCR Kit (Accurate Biotechnology, Changsha, China). All experimental procedures were conducted according to the manufacturer’s instructions. Primer sequences are listed in Table 2.
Immunohistochemistry
The CRC patient tissue specimens were fixed in 4% paraformaldehyde for 24 h and then embedded in paraffin. We conducted the experiment according to the instructions of the rabbit SP kit (SP-9001; ZSGB-BIO, Beijing, China). The primary antibodies incubated were as follows: NUCKS1 (catalog# 12023-2-AP, Proteintech) at 1:200 dilution; HDAC2 (catalog# ET1607-78, HUABIO) at 1:2000 dilution; p-AKT (Ser-473) (catalog# 4060, Cell Signaling Technology) at 1:500 dilution. ImageJ software was used to count the positive signals.
Animal experiments
Female nude mice (6–8 weeks old, 18–22 g) were supplied by the Air Force Medical University Animal Center and randomly divided into four groups (n = 6 per group). All animals were maintained in the UTHSC animal facility and the animal studies were conducted in accordance with the National Institutes of Health Animal Use Guidelines and UTHSC IACUC approved protocols. Free software for power analysis online using G * Power, utilizing repeated measures ANOVA with inter subject factors. Each mouse was injected with 1 × 106 CRC cells via the tail vein. The nude mice were randomly divided into 4 groups, with 12 mice injected with the same type of cells and randomly divided into 2 groups (6/group). The monitoring of grouping and tumor metastasis was conducted using a double-blind method. Four weeks later, the mice were euthanized, and lung and liver tissues were collected, embedded in paraffin, and sliced for hematoxylin–eosin (HE) staining using the Hematoxylin and Eosin Staining Kit (Beyotime, Shanghai, China). HDAC2 inhibitor Santacruzamate A (Sellechem) was administered by intraperitoneal injection at a dose of 25 mg/kg once daily for three weeks, with a one-day break each week. And AKT inhibitor LY294002 (MCE) was administered via intraperitoneal injection at a dose of 10 mg/kg, three times a week for four weeks.
Statistical analysis
Quantitative data are presented as mean ± SD and analyzed using GraphPad Prism 8.0 and SPSS Statistics 25.0. Statistical analyses utilized two-tailed Student’s t-test or one-way ANOVA. Significance levels were defined as *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article or its supplementary materials.
References
Siegel RL, Wagle NS, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2023. CA Cancer J Clin. 2023;73:233–54.
Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17–48.
Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–32.
Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, et al. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J Clin. 2017;67:93–9.
Huang P, Cai Y, Zhao B, Cui L. Roles of NUCKS1 in diseases: susceptibility, potential biomarker, and regulatory mechanisms. Biomed Res Int. 2018;2018:7969068.
Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr Opin Genet Dev. 2005;15:496–506.
Zhou Y, Zhang Q, Qiu X, Tian T, Xu Q, Liao B. Hsa_circ_0001550 facilitates colorectal cancer progression through mediating microRNA-4262/nuclear casein kinase and cyclin-dependent kinase substrate 1 cascade. J Clin Lab Anal. 2022;36:e24532.
Gu L, Xia B, Zhong L, Ma Y, Liu L, Yang L, et al. NUCKS1 overexpression is a novel biomarker for recurrence-free survival in cervical squamous cell carcinoma. Tumour Biol. 2014;35:7831–36.
Li L, Wei D, Zhang J, Deng R, Tang J, Su D. miR-641 inhibited cell proliferation and induced apoptosis by targeting NUCKS1/PI3K/AKT signaling pathway in breast cancer. Comput Math Methods Med. 2022;2022:5203839.
Zhao E, Feng L, Bai L, Cui H. NUCKS promotes cell proliferation and suppresses autophagy through the mTOR-Beclin1 pathway in gastric cancer. J Exp Clin Cancer Res. 2020;39:194.
Ma H, Xu J, Zhao R, Qi Y, Ji Y, Ma K. Upregulation of NUCKS1 in Lung Adenocarcinoma is Associated with a Poor Prognosis. Cancer Invest. 2021;39:435–44.
Zheng S, Ji R, He H, Li N, Han C, Han J, et al. NUCKS1, a LINC00629-upregulated gene, facilitated osteosarcoma progression and metastasis by elevating asparagine synthesis. Cell Death Dis. 2023;14:489.
Hume S, Grou CP, Lascaux P, D’Angiolella V, Legrand AJ, Ramadan K, et al. The NUCKS1-SKP2-p21/p27 axis controls S phase entry. Nat Commun. 2021;12:6959.
Zhu LL, Shi JJ, Guo YD, Yang C, Wang RL, Li SS, et al. NUCKS1 promotes the progression of colorectal cancer via activating PI3K/AKT/mTOR signaling pathway. Neoplasma. 2023;70:272–86.
Jo H, Shim K, Kim HU, Jung HS, Jeoung D. HDAC2 as a target for developing anti-cancer drugs. Comput Struct Biotechnol J. 2023;21:2048–57.
Peng XP, Sun ZQ, Kuang PH, Chen JJ. Recent progress on HDAC inhibitors with dual targeting capabilities for cancer treatment. Eur J Med Chem. 2020;208:112831.
Qiao X, Wu XY, Chen ST, Niu MM, Hua HL, Zhang Y. Discovery of novel and potent dual-targeting AXL/HDAC2 inhibitors for colorectal cancer treatment via structure-based pharmacophore modelling, virtual screening, and molecular docking, molecular dynamics simulation studies, and biological evaluation. J Enzym Inhib Med Chem. 2024;39:2295241.
Harms KL, Chen X. Histone deacetylase 2 modulates p53 transcriptional activities through regulation of p53-DNA binding activity. Cancer Res. 2007;67:3145–52.
Wang XX, Xu J, Wang H, Wu L, Yuan WQ, Du J, et al. Trichostatin A, a histone deacetylase inhibitor, reverses epithelial-mesenchymal transition in colorectal cancer SW480 and prostate cancer PC3 cells. Biochem Biophys Res Commun. 2015;456:320–6.
Tang WM, Zhou WJ, Xiang L, Wu XS, Zhang P, Wang J, et al. The p300/YY1/miR-500a-5p/HDAC2 signalling axis regulates cell proliferation in human colorectal cancer. Nat Commun. 2019;10:663.
Krishnan M, Singh AB, Smith JJ, Sharma A, Chen X, Eschrich S, et al. HDAC inhibitors regulate claudin-1 expression in colon cancer cells through modulation of mRNA stability. Oncogene. 2010;29:305–12.
Qi ZP, Yalikong A, Zhang JW, Cai SL, Li B, Di S, et al. HDAC2 promotes the EMT of colorectal cancer cells and via the modular scaffold function of ENSG00000274093.1. J Cell Mol Med. 2021;25:1190–97.
Zhu P, Martin E, Mengwasser J, Schlag P, Janssen KP, Gottlicher M. Induction of HDAC2 expression upon loss of APC in colorectal tumorigenesis. Cancer Cell. 2004;5:455–63.
Khadempar S, Lotfi M, Haghiralsadat F, Saidijam M, Ghasemi N, Afshar S. Lansoprazole as a potent HDAC2 inhibitor for treatment of colorectal cancer: an in-silico analysis and experimental validation. Comput Biol Med. 2023;166:107518.
Zhang T, Wei D, Lu T, Ma D, Yu K, Fang Q, et al. CAY10683 and imatinib have synergistic effects in overcoming imatinib resistance via HDAC2 inhibition in chronic myeloid leukemia. RSC Adv. 2020;10:828–44.
Gromek SM, DeMayo JA, Maxwell AT, West AM, Pavlik CM, Zhao Z, et al. Synthesis and biological evaluation of santacruzamate A analogues for anti-proliferative and immunomodulatory activity. Bioorg Med Chem. 2016;24:5183–96.
Østvold AC, Grundt K, Wiese C. NUCKS1 is a highly modified, chromatin-associated protein involved in a diverse set of biological and pathophysiological processes. Biochem J. 2022;479:1205–20.
Kumar V, Kundu S, Singh A, Singh S. Understanding the role of histone deacetylase and their inhibitors in neurodegenerative disorders: current targets and future perspective. Curr Neuropharmacol. 2022;20:158–78.
Yu X, Wu J, Wu Q, Sun S. Quantitative analysis of autophagy-related protein LC3B by quantum-dot-based molecular imaging. Methods Cell Biol. 2021;165:177–85.
Hubert V, Peschel A, Langer B, Groger M, Rees A, Kain R. LAMP-2 is required for incorporating syntaxin-17 into autophagosomes and for their fusion with lysosomes. Biol Open. 2016;5:1516–29.
Shin AE, Giancotti FG, Rustgi AK. Metastatic colorectal cancer: mechanisms and emerging therapeutics. Trends Pharm Sci. 2023;44:222–36.
Hernandez-Alejandro R, Ruffolo LI, Sasaki K, Tomiyama K, Orloff MS, Pineda-Solis K, et al. Recipient and donor outcomes after living-donor liver transplant for unresectable colorectal liver metastases. JAMA Surg. 2022;157:524–30.
Mitry E, Guiu B, Cosconea S, Jooste V, Faivre J, Bouvier AM. Epidemiology, management and prognosis of colorectal cancer with lung metastases: a 30-year population-based study. Gut. 2010;59:1383–88.
Zhao S, Wang B, Ma Y, Kuang J, Liang J, Yuan Y. NUCKS1 promotes proliferation, invasion and migration of non-small cell lung cancer by upregulating CDK1 expression. Cancer Manag Res. 2020;12:13311–23.
Stypula-Cyrus Y, Damania D, Kunte DP, Cruz MD, Subramanian H, Roy HK, et al. HDAC up-regulation in early colon field carcinogenesis is involved in cell tumorigenicity through regulation of chromatin structure. PLoS One. 2013;8:e64600.
Hu C, Zha Q, Hua P, Xiao L, Pan D. NUCKS promotes the proliferation, migration and invasion of lung cancer cells through Pi3k/Akt signalling pathway. Clin Invest Med. 2021;44:E55–E61.
Huang YK, Kang WM, Ma ZQ, Liu YQ, Zhou L, Yu JC. NUCKS1 promotes gastric cancer cell aggressiveness by upregulating IGF-1R and subsequently activating the PI3K/Akt/mTOR signaling pathway. Carcinogenesis. 2019;40:370–79.
Pei L, Zhao F, Zhang Y. USP43 impairs cisplatin sensitivity in epithelial ovarian cancer through HDAC2-dependent regulation of Wnt/beta-catenin signaling pathway. Apoptosis. 2024;29:210–28.
Tu T, Huang J, Lin M, Gao Z, Wu X, Zhang W, et al. CUDC‑907 reverses pathological phenotype of keloid fibroblasts in vitro and in vivo via dual inhibition of PI3K/Akt/mTOR signaling and HDAC2. Int J Mol Med. 2019;44:1789–1800.
Dong J, Chen J, Li Q, Qiu S. Knockdown of FKBP3 suppresses nasopharyngeal carcinoma cell growth, invasion and migration, deactivated NF-kappaB/IL-6 signaling pathway through inhibiting histone deacetylase 2 expression. Chin J Physiol. 2023;66:85–92.
Sacks FM, Lichtenstein A, Van Horn L, Harris W, Kris-Etherton P, Winston M. Soy protein, isoflavones, and cardiovascular health: an American Heart Association Science advisory for professionals from the nutrition committee. Circulation. 2006;113:1034–44.
Han R, Ling C, Wang Y, Lu L. Enhancing HCC treatment: innovatively combining HDAC2 inhibitor with PD-1/PD-L1 inhibition. Cancer Cell Int. 2023;23:203.
Gao Y, Nihira NT, Bu X, Chu C, Zhang J, Kolodziejczyk A, et al. Acetylation-dependent regulation of PD-L1 nuclear translocation dictates the efficacy of anti-PD-1 immunotherapy. Nat Cell Biol. 2020;22:1064–75.
Turner NC, Oliveira M, Howell SJ, Dalenc F, Cortes J, Gomez Moreno HL, et al. CapivAsertib in Hormone Receptor-positive Advanced Breast Cancer. N Engl J Med. 2023;388:2058–70.
Acknowledgements
The authors thank EditChecks (https://editchecks.com.cn/) for providing professional language polishing services.
Funding
This study was supported by Shaanxi Provincial Natural Science Basic Research Program to Yang Song (2023-JC-YB-782), and the Key Research and Development Projects of Shaanxi Province to Haichuan Su (No.2022ZDLSF03-01) and Shaanxi Provincial Health Research Innovation Team Project (2024TD-04). No disclosures were reported by the other authors.
Author information
Authors and Affiliations
Contributions
Study concept and design: Yang Song, Jun Chen, and Haichuan Su. Performing the experiments and data analysis: Liaoliao Zhu, Ting Zhao, Xiangjing Shen. Animal experiment: Xiangjing Shen and Liang Zhang. Writing-review and editing: Liaoliao Zhu, Junqiang Li, Yang Song, and Jun Chen. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
We confirm that all experiments were performed in accordance with relevant guidelines and regulations. All animal experimental procedures were approved by the Institutional Animal Care and Use Committee of Tangdu Hospital of Air Force Medical University (Number: 20230743). The protocol for using patient tissue specimens has been reviewed and approved by the Ethics Committee of the Tangdu Hospital of Air Force Medical University (No. GKJ-Y-202303-025), and all patients have given written informed consent before being included in this study.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhu, L., Zhao, T., Su, H. et al. NUCKS1 promotes invasion and metastasis of colorectal cancer by stabilizing HDAC2 and activating AKT. Oncogenesis 14, 19 (2025). https://doi.org/10.1038/s41389-025-00562-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41389-025-00562-5