Abstract
Background
During mammalian gestation, fetal circadian rhythms are thought to be mainly controlled by maternal signals. In humans, the initiation and activity of central and peripheral circadian clocks is largely unknown. This study aimed to elucidate the developmental clock properties in human umbilical vein endothelial cells (HUVECs).
Methods
HUVECs were obtained from (a) preterm infants, subgrouped according to birth weight or gestational age classification, and (b) term infants (in total: n = 60). In vitro clock activity was determined by using live bioluminescence recording of a luciferase reporter gene under circadian control over 120 h. In addition, core clock and clock-associated gene expression were quantified using NanoString technology.
Results
Peripheral clock activity was detected, regardless of prematurity and birth weight classification. The mean period, amplitude, and phase of circadian oscillations were not significantly associated with gestational age or birth weight classification.
Conclusions
Peripheral clock activity can be demonstrated in HUVECs from both preterm and term infants without significant developmental differences in the period, amplitude, and phase of oscillations. This model may be useful to identify perturbation factors of proper development and entrainment of neonatal circadian clock activity.
Impact
-
We established a model system for analyzing the peripheral clock in preterm and term HUVECs.
-
In HUVECs, the peripheral clock exhibits functional in vitro activity independent of gestational age or birth weight categories.
-
In this model system, neither significant developmental differences exist in the period, amplitude, and phase, nor in the expression of circadian core clock and clock-associated genes.
-
Entrainment and proper function of the circadian clock deserve attention in neonatal intensive care.
Similar content being viewed by others
Introduction
The function and impact of the circadian clock in human health and disease have been extensively investigated in recent years, mainly in adults.1 The circadian clock system is essential for the efficiency of metabolic, physiological, and behavioral processes and provides the body with resources in a time-of-day-adapted manner. Disruption of the circadian clock, e.g., by shift work, artificial light, travel across time zones, or social jetlag, has been associated with a variety of diseases, e.g., sleep disorders, cancer, psychiatric and cardiovascular diseases, chronic systemic inflammation, and impaired immune response.2 Disruption of sleep and circadian rhythms is also considered a risk factor for neurodevelopmental disorders and poor outcomes after (neonatal) intensive care.3,4,5
In mammals, the circadian system is organized in a hierarchic fashion, with the suprachiasmatic nucleus (SCN) of the anterior hypothalamus serving as the central pacemaker clock which synchronizes clocks in peripheral tissues through neuronal, neuroendocrine, and behavioral pathways (reviewed in ref. 6). Nevertheless, peripheral clocks have also been shown to be autonomous self-sustained oscillators, hinting at the existence of cell-specific synchronizers.7,8,9 The onset of central and peripheral circadian clock activities is considered a gradual process during mammalian ontogeny, but the exact time point of emergence is still under debate since it varies depending on the species and the experimental approach for detection.10,11 Current data indicate that the activity of the central clock begins in the last trimester of gestation, for example in rats not earlier than developmental stage E19, and in primates not before the third trimester.12,13 Correlation between maternal and fetal rhythms suggests a transplacental entrainment of fetal rhythms by maternal time cues such as hormones, nutrients, body temperature, or physical activity.14,15,16 It is unknown whether and to which extent the activity of peripheral circadian clocks varies through human gestation and upon pregnancy-associated disorders or antenatal treatment. From a perspective of neurodevelopmental outcome, this could be highly relevant, in particular in preterm infants as they are not only separated from transplacentally mediated time cues but also exposed to non-physiological circadian factors such as artificial lighting, noise, and feeding regimens during neonatal intensive care.17,18
While the expression of molecular core clock regulators, such as CLOCK and PER1 mRNA, has been detected in cultured term human neonatal fibroblasts and keratinocytes,19 to the best of our knowledge, peripheral circadian clock gene activity has never been demonstrated in human neonates. In this explorative study, we have studied the onset and activity of the molecular clock in preterm and term neonates at the time of birth by characterizing oscillations in human vein endothelial cells (HUVECs) obtained from their umbilical cords.
Materials and methods
Subjects and samples
Subjects for the NeoCIRCLE (Assessing the Neonatal Circadian Clock and Entrainment Factors) study were recruited at the Department of Obstetrics after obtaining written informed consent (institutional review board approval no. EA2/171/22). Patients’ demographic data are summarized in Table 1 and Supplementary Data 2, respectively. Infants with congenital malformations, syndrome, or perinatal asphyxia were excluded.
Isolation and culture of primary HUVECs
Umbilical cord specimens were obtained immediately after birth and stored in Hank’s Balanced Salt Solution (HBSS, Thermo Fisher, Henningsdorf, Germany, #14175053) at 4–8 °C for a median of 2 h (95% confidence interval (CI) 1.1–2.9 h) until isolation. For isolation, the umbilical vein was rinsed with HBSS, instilled with 0.5 mg/mL collagenase A solution (Roche Diagnostics, Mannheim, Germany, #10103586001) in phosphate-buffered saline (PBS, Thermo Fisher, #14190-094), gently massaged and incubated at 37 °C for 15 min immersed in water. After another gentle massage, the HUVECs were rinsed from the umbilical vein drop-by-drop with 40 mL HBSS and collected in a tube filled with 10 mL Medium 199 (Thermo Fisher, #31153026). Isolated cells were pelleted at 788 × g for 10 min, resuspended and cultured in Medium 199 supplemented with 20% heat-inactivated fetal bovine serum (FBS, Sigma–Aldrich, St. Louis, MO, #F7524), 2 mM L-glutamine (Bio&Sell, Feucht, Germany, #BS.K0283) and 20.7 mM NaHCO3 (Bio&Sell, #BS.L1713). In the following, this composition is referred to as “Medium 199 s”. The next day, Medium 199 s was exchanged to remove erythrocyte-containing residues. To attain proliferation, 19.95 µg/ml endothelial growth factor (ECGF, Biozol, Hamburg, Germany, #CYT-026948) and 15 U/mL heparin (Ratiopharm, Ulm, Germany, #X01336A) were directly added to the culture dish. Cultures were kept at 37 °C and 5% CO2 and expanded up to four passages (mean:15.5 d, range: 11–22 d) until bioluminescence recording.
Immunocytological staining of HUVECs
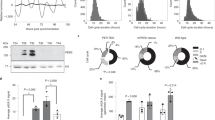
The endothelial origin of the cells was confirmed by immunocytological staining of the exemplary cell pools for the endothelial marker protein platelet endothelial cell adhesion molecule 1 (PECAM1/CD31). HUVEC isolates were grown on fibronectin-coated (Sigma–Aldrich, #F2006-1MG; 0.031 mg/mL) chambered coverglass slides (Thermo Fisher, #177445) and fixed with 10% Roti Histofix (Roth, Karlsruhe, Germany, #A146.6) at a density of 7.5 × 104 cells/cm2. Cells were permeabilized with 0.5% Triton X-100 (Sigma–Aldrich, #T9284) for 30 min and blocked with 5% skimmed milk in PBS for 1 h. The target protein was detected with a polyclonal rabbit anti-PECAM1 antibody (Abcam, Cambridge, UK, #ab28364), diluted 1:50 in antibody diluent (Invitrogen, Thermo Fisher, #00-3118) and incubated at 4 °C. The next day, a secondary Alexa Fluor® 488-coupled goat anti-rabbit IgG antibody (Jackson ImmunoResearch, PA, #111-545-003; 1:200) was added for 1 h at room temperature. Polyclonal rabbit IgG (PeproTech, Hamburg, Germany, #500-00; 1:50) served as a negative control, and cell nuclei were counterstained with Hoechst 33342 (Sigma–Aldrich, #14533) at a final concentration of 0.01 mg/mL. Results were observed under a fluorescent microscope (Keyence BZ-900, Neu-Isenburg, Germany). Images were taken with the built-in camera and processed with BZ-II-Viewer and BZ-II-Analyzer (Keyence). All samples tested (term: 38 + 4, LBW: 34 + 1, ELBW: 26 + 1 weeks gestation) showed >98% PECAM1-positive cells (Supplementary Data 1).
Lentiviral transduction
Human embryonic kidney 293T cells (DSMZ, Braunschweig, Germany), cultivated in Dulbecco’s modified Eagle’s medium/Ham’s F12 (1:1, Corning, Corning, NY, #10-090-CV), supplemented with 10% FBS, were plated on T75 flasks at 5 × 104 cells/cm2. Cells were transiently transfected with 7.5 µg DNA containing 3.375 µg of the lentiviral vector pLenti6.4_mPer2promExon1-Luc (a kind gift of Andrew Liu, first described in ref.20, Fig. 1), 2.625 µg psPAX2 (Addgene #12260), 1.5 µg pMD2.G packaging vectors (Addgene #12259) and 22.5 µl Fugene® 6 Transfection Reagent (Promega, Fitchburg, WI, #E2691) in Medium 199 s. The virus-containing supernatant was collected after 24 and 48 h of transfection, pooled, and either snap-frozen for storage at −80 °C or directly used for transduction of HUVECs. Then, HUVECs were seeded on 60 mm dishes at 5.25 × 104 cells/cm2 (75% confluency) and incubated with 5 mL virus-containing supernatant for 24 h under standard cell culture conditions. After removal of the virus, the samples were selected for stably transduced cells using 6 µg/mL blasticidin S (Fisher Scientific, #10264913) at a cell density of <6.3 × 104 cells/cm2 (approx. 90% confluency) for 3 days. Cells were then either stored in liquid nitrogen (n = 40) or directly underwent bioluminescence recording (n = 20). We did not observe differences in growth, transduction efficiency, or bioluminescence recording quality between freeze-thawed and fresh HUVEC samples, which were evenly distributed amongst the study groups.
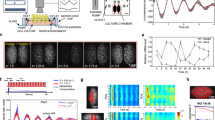
a Steps before the final analysis of recorded time-series data included pre-processing of all signals and ensemble-averaged rhythmical feature extraction of the entire data set of term and preterm samples vs. non-transduced (No-Luc) samples that served as null models (control). A schematic drawing of the lentiviral Per2prom-driven luciferase reporter can be found in the top right corner. AmpR: Ampicillin resistance gene, BlastR: Blasticidin resistance gene. b PyBOAT creates plots of the median and the interquartile range for each oscillatory characteristic over time (wavelet power, period, amplitude) for No-Luc and c for specimens from preterm and term infants. The point of maximal wavelet power (48.817 h post dexamethasone synchronization, red line) was used to extract oscillatory characteristics at a time point with the best data quality with regards to culture condition, duration of cell synchronization, and cell vitality as the ensemble-averaged data showed a decline of wavelet power, amplitude strength and a larger dispersion of period lengths with increased recording time.
Cell synchronization and bioluminescence recordings
HUVECs were seeded into fibronectin-coated 35 mm dishes (Thermo Fisher, #150318) at a near-confluent density of 7 × 104 cells/cm2. After a minimal period of 16 h and prior to bioluminescence recordings, cell rhythms were synchronized following an established protocol.21 In short, 1 µM dexamethasone (Sigma–Aldrich, #D4902-25MG, 1 mM stock in ethanol) was added to the media, and the sample was incubated for 20 min. After washing with PBS, Medium 199 without phenol red (Thermo Fisher, #11043023) was added, supplemented as Medium 199 s with addition of 19.95 µg/ml ECGF and 15 U/mL heparin, 1% penicillin-streptomycin (Bio&Sell, #BS. A 2212), 20 mM HEPES (Bio&Sell, #BS. L 1613) and 0.25 mM D-luciferin (PJK, Kleinbittersdorf, Germany, #102113). The samples were measured in a tailor-made luminometric system consisting of lightproof boxes kept at 36 °C and 5% CO2 in a standard cell culture incubator without humidity, connected to highly sensitive photomultiplier tubes (H7360-02, Hamamatsu Photonics, Hamamatsu, Japan). Live bioluminescence was recorded in at least technical duplicates for 5–7 days. Cell cultures were microscopically inspected for contamination and vitality at the end of each recording.
RNA extraction and expression analysis of core clock and clock-associated genes
Different HUVEC samples were seeded at a density of 7 × 104 cells/cm2 on 12-well plates (Corning, #353043) in 8 replicates each, one per time point. All samples were synchronized in parallel with 1 µM dexamethasone for 20 min and washed with PBS before the media was exchanged to fresh Medium 199 s. RNA was extracted from each well with 500 µL TRIzol Reagent (Thermo, #15596018) at 24, 28, 32, 36, 40, 44, 48, and 52 h after synchronization, according to the manufacturer’s instructions.
Expression quantification of 20 clock and clock-associated genes (ARNTL2, BHLHE40, BHLHE41, BMAL1, CIART, CIPC, CLOCK, CRY1, CRY2, CSNK1D, CSNK1E, DBP, NFIL3, NPAS2, NR1D1, NR1D2, PER1, PER2, PER3, RORA) was performed from HUVEC RNAs obtained from different gestational ages as well as different culture times after isolation by NanoString technology as described.22 To evaluate whether the average gene expression levels associated with birth weight classification, the HUVECs were synchronized by dexamethasone treatment (1 µM, 20 min) and sampled as described above. To obtain mean gene expression values, the resulting RNAs were averaged by pooling equal RNA amounts from each time point. For determining the influence of the culture time since isolation, RNA of HUVEC samples was pooled according to maturity and birth weight classification and culture time (6/8/10/15.5 d after preparation). Gene expression in the different groups was tested for normality (all log-normal) and the log10 values were analyzed for statistical differences by one-way ANOVA.
Data analysis
For data analysis, all time-series data were cropped to the recording time frame of 24–120 h. The software pyBOAT was used - a wavelet-based time-frequency analysis software, specifically designed for non-stationary and noisy rhythmical data.23 In brief, the signal was pre-processed by detrending and amplitude envelope removal. The recorded signal was then decomposed into different frequency components across the recording period by applying the wavelet transformation, and its time-frequency representation was visualized as the wavelet power spectrum displaying the main oscillatory component of the signal. Ridge tracking allowed extraction of the rhythm parameters amplitude, period, and phase. The user-defined filter parameter “Cut-Off-Period” was set to 30 h, so that the program removed low-frequency signals commonly induced by non-circadian cellular processes like cell division. Signals were normalized with an amplitude envelope estimated from a window size of 30 h.
To distinguish oscillating from non-oscillating signals, we estimated a wavelet power threshold for the background noise of a null model established from bioluminescence recordings of non-transduced HUVECs (in the following referred to as No-Luc). For this, 14 No-Luc samples underwent the same procedures as described above apart from the viral transduction and selection step and were recorded for 120 h. An ensemble-averaged wavelet analysis for (a) all No-Luc and (b) all term and preterm (samples of interest) recordings was performed, which showed that 48.817 h post dexamethasone synchronization was the point of maximal wavelet power (median of the batch analysis). This timestamp was used to extract oscillatory characteristics at a time point with the best data quality regarding culture condition, duration of cell synchronization, and cell vitality. Samples with a wavelet power <30 (mean + 2 SD of the No-Luc samples) were categorized as non-oscillating (Fig. 1). A mean absolute bioluminescence <6700 (mean + 2 standard deviations of the No-Luc samples) was used as a minimal brightness threshold, and recordings below were excluded from the analysis.
Oscillatory parameters at 48.817 h were first tested for outliers by Grubbs’ test (one value for phase removed from the >2500 g/≥37 wks and 2500–1500 g/36 + 6–32 + 0 wks cohorts, respectively), then tested for normality by Shapiro–Wilk test (all normal or log-normal) and analyzed for statistical differences between the groups with a BW > 2500 g, 2500–1500 g, 1499–1000 g and <1000 g, or between groups of term (≥37 + 0 wks gestation), preterm (32 + 0–36 + 6 wks gestation), very preterm (28 + 0–31 + 6 wks gestation) and extremely preterm infants (<28 + 0 wks gestation), respectively, by one-way ANOVA and correlation analysis with a significance threshold of p < 0.05 (GraphPad Prism 9, GraphPad Software Inc., San Diego).
Results
Study population
From 2022 to 2023, 82 term and preterm neonates were enrolled in the NeoCIRCLE study, equal to 82 HUVEC samples, 22 of which had to be excluded from the final analysis due to: Unsuccessful cell isolation (n = 3), bacterial contamination (n = 10), inefficient lentiviral transduction (n = 3) or technical issues during recording (n = 6), resulting in a total of 60 subjects. The final study cohort was subgrouped according to BW (Fig. 2). Clinical conditions were reflected within the BW-specific groups as expected: Gestational or perinatal disorders and secondary cesarean sections were more common in the subgroups of preterm infants that also exhibited a higher proportion of twin births and frequency of antenatal glucocorticoid (GC) treatment (Table 1). The underlying clinical conditions showed almost equal distributions if the final study cohort was subgrouped according to GA (Supplementary Data 2).
A peripheral clock oscillates in term and preterm HUVECs at the time of birth
To determine whether the functional activity of the clock exists during fetal development and if so, might depend on birth weight (BW) or gestational age (GA), respectively, we obtained HUVECs at birth of infants born at 23 + 1 to 40 + 0 weeks gestation (birth weight 430–4210 g). Cells were in vitro transduced with a luciferase reporter gene construct under circadian control. After restarting the intercellular clocks with a synchronizing dexamethasone pulse, the resulting bioluminescence was live-recorded over a period of five days. When processed by the analysis software pyBOAT, recordings showed oscillations of weak and moderate signal strength at all stages of development (Fig. 3, all 60 recording tracks can be found in Supplementary Data 3).
When comparing the oscillation quality of preterm and term specimens by the oscillatory quality control parameter wavelet power, those were similar in all four BW groups (>2500 g: 233 ± 90, 2500–1500 g: 219 ± 89, 1499–1000 g: 206 ± 100, <1000 g: 188 ± 113, one-way ANOVA: p = 0.647), all exceeding the threshold for background noise of 30 (Fig. 4, dotted line). A mean period of about 23–25 h in all subgroups (>2500 g: 23.7 ± 2.5 h, 2500–1500 g: 23.7 ± 1.8 h, 1499–1000 g: 25.2 ± 2.2 h, <1000 g: 22.7 ± 2.6 h, one-way ANOVA: p = 0.183) indicated that the oscillations were indeed of circadian nature, and did not largely differ between the BW-specific groups, even if the period in the ELBW group tended to be lower than that of the other groups (Fig. 4a). The amplitude as a surrogate of the oscillation strength was similar in all subgroups (>2500 g: 0.40 ± 0.12, 2500–1500 g: 0.39 ± 0.12, 1499–1000 g: 0.35 ± 0.12, <1000 g: 0.34 ± 0.14, one-way ANOVA: p = 0.591, Fig. 4b), but revealed a trend in HUVECs from VLBW infants towards lower amplitude oscillations than in (near-)term infants (>2500 g: 0.40 ± 0.12 vs. <1500 g: 0.34 ± 0.13). Finally, the parameter phase indicates the relative position within one oscillatory cycle and represents the sample’s reaction to the synchronization. This phase was similar in all subgroups (>2500 g: 3.9 ± 0.8 h, 2500–1500 g: 3.8 ± 0.6 h, 1499–1000 g: 3.9 ± 1.4 h, <1000 g: 3.8 ± 0.7 h, one-way ANOVA: p = 0.915, Fig. 4c), showing neither delay nor advance between the oscillation tracks of term and preterm samples. Of note, the results were similar if the samples were subgrouped by GA instead of BW (Fig. 5). A correlation analysis of the circadian parameters within the BW and GA groups, respectively, revealed a (technically expected) significant correlation between amplitude and wavelet power in all groups. Of note, there also was a significant correlation between period and amplitude as well as period and wavelet power, respectively, in the group with a BW < 1000 g. All correlation results can be found in Supplementary Data 4.
Plots show the median and 95% CI of infants with a birth weight >2500 g (black square, n = 26) vs. 2500–1500 g (crossed circle, n = 18) vs. 1499–1000 g (open circle, n = 7) vs. <1000 g (black circle, n = 9) for the oscillatory characteristics period (a), amplitude (b), phase (c), and the oscillatory quality control parameter wavelet power (d) at the time point of maximal wavelet power (48.817 h after dexamethasone synchronization). The dashed line in (d) represents the minimal wavelet power threshold of 30 to be considered a “true” oscillation. Wavelet power values of all samples were above this threshold classifying them as “real” oscillations.
Plots show the median and 95% CI of infants with gestational age at birth ≥37 wks (black square, n = 26) vs. 32 + 0–36 + 6 wks (crossed circle, n = 18) vs. 28 + 0–31 + 6 wks (open circle, n = 7) vs. <28 wks (black circle, n = 9) for the oscillatory characteristics period (a), amplitude (b), phase (c), and the quality control parameter wavelet power (d) at the time point of maximal wavelet power (48.817 h after dexamethasone synchronization). The dashed line in (d) represents the minimal wavelet power threshold of 30 to be considered a “true” oscillation. Wavelet power values of all samples were above this threshold classifying them as “real” oscillations.
To test whether there were subtle differences in the expression of core clock and clock-associated genes, we analyzed the expression of 18 out of a panel of 20 genes (BHLHE41 and PER1 expression were below the detection threshold) in synchronized HUVEC cultures of different BW groups pooled from a 28 h time course each (Fig. 6). From a statistical point of view, there were no significant differences, but some genes displayed a trend towards lower expression in preterm samples (Fig. 6, shaded in gray in Supplementary Data 5): the core clock genes PER2, CRY1, NR1D1, NR1D2, and the clock-associated genes ARNTL2 (BMAL1 paralogue) and NPAS2 (CLOCK paralogue).
Mean expression of 8 core clock (a) and 10 clock-associated genes (b) normalized to four control genes (GAPDH, PPIA, HPRT1, PSMB2) in HUVEC samples from infants with a BW > 2500 g (n = 6), 2500–1500 g (n = 4), 1499–1000 g (n = 3) and <1000 g (n = 4). Cells were synchronized and sampled over a 24–52 h time period, then averaged by pooling equal RNA amounts from each time point. The mean gene expression was not statistically different between subgroups classified according to birth weight. Dotted line combine data points of the same gene product. The complete data set can be found in Supplementary Data 5.
Taken together, we found that HUVECs had the capacity of basal circadian oscillations at the time of birth, irrespective of whether the infants were born prematurely or at term, but with a non-significant trend towards lower amplitudes and periods in VLBW and especially ELBW infants.
Discussion
In this study, we show for the first time circadian oscillations of the peripheral clock in primary HUVECs isolated from the umbilical cord of preterm and term infants (Figs. 3–5). Thus, in future studies, our model system may allow a detailed description of factors that could modify the activity of the endogenous peripheral clock during development (e.g. ischemia, metabolic or inflammatory factors).
We were also able to portray clock properties in HUVECs of term vs. preterm infants, and show that endogenous, cell-autonomous (PER2-mediated) clock activity already existed in vitro and that oscillation quality did not differ significantly between the BW- or GA-classified subgroups (Figs. 4 and 5). Neither maturity (and BW classification) nor culture time since isolation significantly affects the expression of the core clock and clock-associated genes (Fig. 6, Supplementary Data 5 and 6). However, period and amplitude and the average expression of some of the core clock and clock-associated genes tended to be lower in the groups with a BW < 1500 g. This might indicate that there are indeed subtle differences in the circadian parameters, but that statistical discriminability is limited by the relatively small sample number. Consistent with this observed trend in the circadian profile of HUVECs, a very recent publication showed circadian rhythm development of the neonatal heart rate in 66 preterm infants (29.8 ± 3.7 wks gestation) over the course of postnatal development.24
Experimental data using peripheral tissues of fetal mice (E18) showed rhythmicity in clock gene expression in culture, but not in vivo.25 Thus, one limitation of our model system could be that the GC dexamethasone was used for cell synchronization prior to bioluminescence recordings. Peripheral tissues and cells lose clock synchrony over time due to a lack of intercellular coupling which results in low signal-to-noise ratios and dampened rhythms.9 Therefore, we have chosen the most commonly used synchronization method of a dexamethasone pulse. The alternative synchronization method, cold shock, did not synchronize the HUVECs as well. Serum shock was excluded as we did not want to present the HUVECs with additional fetal plasma components with specific cytokine and metabolite compositions as potential perturbation factors for the developing clock.26 In addition to this in vitro synchronization pulse of dexamethasone, antenatal exposition of premature infants to synthetical GCs is common in the clinical routine - all ELBW, VLBW, and 70% of the LBW infants of our study population received betamethasone for antenatal GC treatment to improve neonatal outcome (Table 1).27 Maternal GCs are attributed to a synchronizing function in the growing fetus, and it is hypothesized that they are a key signal for the development and maturation of the fetal circadian system in utero.28,29 Thus, we cannot exclude that the observed (PER2-mediated) HUVEC rhythmicity was (partially) induced by the common antenatal GC treatment due to impending prematurity, but this would have only influenced the relative comparison with the most mature group as all others were similarly exposed to antenatal GCs.
Our analysis of circadian clock activity revealed that the mean period, amplitude, and phase of circadian oscillations in HUVECs were not significantly associated with BW or GA classification (Figs. 4 and 5), even if period and amplitude and some core clock and clock-associated genes tended to be lower in more preterm groups (Figs. 4–6). Regulation of core clock and clock-associated genes could be mediated by epigenetic modifications. It has been shown that in placental tissue and umbilical cord leukocytes, disorders such as early-onset preeclampsia rather than gestational age are associated with such CpG modifications.30 These combined findings suggest that the developing circadian system of premature infants will react differently to external (e.g., oxidative, metabolic, inflammatory) stressors present in neonatal intensive care. This is a novel and important finding, not adequately considered in current clinical practice. Yet, only very few studies have investigated potential rhythm patterns in very and extremely preterm neonates (26–32 weeks gestation), mostly focusing on vital parameters like body temperature, heart rate, and activity or motility during the early neonatal period. No strong evidence for circadian patterns in these parameters was found, and ultradian rhythms, probably influenced by interventions and feeding, appeared to mask putative circadian rhythms.31,32,33 Bauer et al. found diurnal variability in oxygen consumption (VO2, defined as a 5% fluctuation over baseline values) in 18 of 22 preterm subjects born at 27–31 weeks’ gestation, studied at 3–4 weeks’ postnatal age, suggesting the presence of central endogenous circadian activity in vivo.34 Although our HUVEC in vitro model may not fully reflect the developing internal clock in vivo, we were able to characterize the activity of the molecular clock independent from external factors that may induce ultradian rhythms or disrupt rhythmicity through clinical conditions like mechanical ventilation, sedation, or inflammation.
During intensive care, the circadian system of adults is often dysregulated, as e.g. clock gene expression was found down-regulated in septic and critically ill patients,22,35, and melatonin secretion was desynchronized in mechanically ventilated patients.36 At the same time, the circadian system seems to influence the course and outcome of critical conditions, such as ventilator-induced lung injury37 or the development of delirium.38 During the last decade, the concept of promoting circadian rhythms, in the meaning of “chronofitness”,4 to improve weight gain, comfort, and neurodevelopment of preterm and term neonates in intensive care has gained increasing attention.3,5 By detecting functional in vitro activity of the peripheral clock in HUVECs independent from gestational age, our data support further efforts to protect and entrain circadian rhythms even in the smallest and most premature infants. Which of the perturbation factors in the NICU might have the highest influence on the developing circadian system of preterm and term infants should be addressed in further in vitro studies using our HUVEC model system.
Conclusions
We have established human umbilical vein endothelial cells as a model system for detecting peripheral clock activity by bioluminescence recording in primary cells of preterm and term neonates. In contrast to previous indirect clues from other species, there were no significant developmental differences in the period, amplitude, or phase of the peripheral clock in human endothelial cells which indicates that circadian activity exists independently from maternal signals and deserves attention during intensive care of very preterm neonates.
Data availability
The data used to support the findings of this study are available from the corresponding author upon request.
References
Logan, R. W. & McClung, C. A. Rhythms of life: circadian disruption and brain disorders across the lifespan. Nat. Rev. Neurosci. 20, 49–65 (2019).
Allada, R. & Bass, J. Circadian mechanisms in medicine. N. Engl. J. Med. 384, 550–561 (2021).
Felten, M. et al. Circadian rhythm disruption in critically ill patients. Acta Physiol. 238, e13962 (2023).
McKenna, H., van der Horst, G. T. J., Reiss, I. & Martin, D. Clinical chronobiology: a timely consideration in critical care medicine. Crit. Care 22, 124 (2018).
Lewis, P. et al. A systematic review of chronobiology for neonatal care units: what we know and what we should consider. Sleep. Med. Rev. 73, 101872 (2023).
Dibner, C., Schibler, U. & Albrecht, U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 72, 517–549 (2010).
Yoo, S. H. et al. Period2::Luciferase real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl Acad. Sci. USA 101, 5339–5346 (2004).
Sinturel, F. et al. Circadian hepatocyte clocks keep synchrony in the absence of a master pacemaker in the suprachiasmatic nucleus or other extrahepatic clocks. Genes Dev. 35, 329–334 (2021).
Finger, A. M. & Kramer, A. Peripheral clocks tick independently of their master. Genes Dev. 35, 304–306 (2021).
Umemura, Y. & Yagita, K. Development of the circadian core machinery in mammals. J. Mol. Biol. 432, 3611–3617 (2020).
Greiner, P., Houdek, P., Sládek, M. & Sumová, A. Early rhythmicity in the fetal suprachiasmatic nuclei in response to maternal signals detected by omics approach. PLoS Biol. 20, e3001637 (2022).
Kováciková, Z. et al. Expression of clock and clock-driven genes in the rat suprachiasmatic nucleus during late fetal and early postnatal development. J. Biol. Rhythms 21, 140–148 (2006).
Serón-Ferré, M. et al. Perinatal neuroendocrine regulation. Development of the circadian time-keeping system. Mol. Cell. Endocrinol. 186, 169–173 (2002).
Lunshof, S. et al. Fetal and maternal diurnal rhythms during the third trimester of normal pregnancy: Outcomes of computerized analysis of continuous twenty-four-hour fetal heart rate recordings. Am. J. Obstet. Gynecol. 178, 247–254 (1998).
Kintraia, P. I., Zarnadze, M. G., Kintraia, N. P. & Kashakashvili, I. G. Development of daily rhythmicity in heart rate and locomotor activity in the human fetus. J. Circadian Rhythms 3, 5 (2005).
Bates, K. & Herzog, E. D. Maternal-fetal circadian communication during pregnancy. Front. Endocrinol. 11, 198 (2020).
Rogers, E. E. & Hintz, S. R. Early neurodevelopmental outcomes of extremely preterm infants. Semin Perinatol. 40, 497–509 (2016).
Jarjour, I. T. Neurodevelopmental outcome after extreme prematurity: a review of the literature. Pediatr. Neurol. 52, 143–152 (2015).
Zanello, S. B., Jackson, D. M. & Holick, M. F. Expression of the circadian clock genes clock and period1 in human skin. J. Invest. Dermatol. 115, 757–760 (2000).
Liu, A. C. et al. Redundant function of rev-erbalpha and beta and non-essential role for bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 4, e1000023 (2008).
Finger, A. M. et al. Intercellular coupling between peripheral circadian oscillators by tgf-β signaling. Sci. Adv. 7, eabg5174 (2021).
Lachmann, G. et al. Circadian rhythms in septic shock patients. Ann. Intensive Care. 11, 64 (2021).
Mönke, G., Sorgenfrei, F. A., Schmal, C. & Granada, A. E. Optimal time frequency analysis for biological data - pyboat. bioRxiv 179, 985–986 (2020).
Govindan, R. B. et al. Circadian rhythm development in preterm infants. The role of postnatal versus postmenstrual age. Early Hum. Dev. 196, 106084 (2024).
Dolatshad, H., Cary, A. J. & Davis, F. C. Differential expression of the circadian clock in maternal and embryonic tissues of mice. PLoS ONE 5, e9855 (2010).
Cottrell, E. C., Seckl, J. R., Holmes, M. C. & Wyrwoll, C. S. Foetal and placental 11β-hsd2: a hub for developmental programming. Acta Physiol. 210, 288–295 (2014).
McGoldrick, E., Stewart, F., Parker, R. & Dalziel, S. R. Antenatal corticosteroids for accelerating fetal lung maturation for women at risk of preterm birth. Cochrane Database Syst. Rev. 12, Cd004454 (2020).
Fowden, A. L. & Forhead, A. J. Glucocorticoids as regulatory signals during intrauterine development. Exp. Physiol. 100, 1477–1487 (2015).
Lehmann, M., Haury, K., Oster, H. & Astiz, M. Circadian glucocorticoids throughout development. Front. Neurosci. 17, 1165230 (2023).
van den Berg, C. B. et al. Early- and late-onset preeclampsia and the DNA methylation of circadian clock and clock-controlled genes in placental and newborn tissues. Chronobiol. Int. 34, 921–932 (2017).
Mirmiran, M. & Kok, J. H. Circadian rhythms in early human development. Early Hum. Dev. 26, 121–128 (1991).
D’Souza, S. W. et al. Skin temperature and heart rate rhythms in infants of extreme prematurity. Arch. Dis. Child. 67, 784–788 (1992).
Glotzbach, S. F., Edgar, D. M. & Ariagno, R. L. Biological rhythmicity in preterm infants prior to discharge from neonatal intensive care. Pediatrics 95, 231–237 (1995).
Bauer, J. et al. Circadian variation on oxygen consumption in preterm infants. J. Perinat. Med. 37, 413–417 (2009).
Maas, M. B. et al. Circadian gene expression rhythms during critical illness. Crit. Care Med. 48, e1294–e1299 (2020).
Olofsson, K., Alling, C., Lundberg, D. & Malmros, C. Abolished circadian rhythm of melatonin secretion in sedated and artificially ventilated intensive care patients. Acta Anaesthesiol. Scand. 48, 679–684 (2004).
Felten, M. et al. Ventilator-induced lung injury is modulated by the circadian clock. Am. J. Respir. Crit. Care Med. 207, 1464–1474 (2023).
Li, J. et al. Circadian rhythm disturbance and delirium in ICU patients: a prospective cohort study. BMC Anesthesiol. 23, 203 (2023).
Acknowledgements
We are grateful to Gina Klee for initial support in establishing the HUVEC isolation protocol, to the staff of the Department of Obstetrics, Charité – Universitätsmedizin Berlin for their help with preserving umbilical cord specimens, to Nicole Dinse for technical assistance in quantitative PCR measurements, and to Alex Zuo, who helped automatizing the data extraction by providing programming scripts. We thank all parents of our patients who consented to participate in this study.
Funding
M.Z. received a doctoral scholarship from the Sonnenfeld Stiftung, Berlin. Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
M.Z. recruited subjects, collected samples and data, performed data analyses, and drafted and revised the manuscript. A.K. assisted in study design, provided technical expertise and equipment, helped interpreting data, and reviewed the manuscript for important intellectual content. G.M. assisted with bioinformatic analyses and interpretation of the results and reviewed the manuscript for biostatistics. L.K.S. designed the study and established the required methods, supervised data collection and analyses, interpreted data, and drafted and finalized the manuscript. C.D. conceptualized and designed the study, supervised the analysis, discussed the content, and drafted and finalized the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent for publication
Term and preterm subjects for the NeoCIRCLE study were recruited at the Department of Obstetrics with written informed consent (IRB Approval no. EA2/171/22).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zuo, M., Kramer, A., Mönke, G. et al. Circadian clock activity in human umbilical vein endothelial cells of preterm and term neonates. Pediatr Res 98, 734–742 (2025). https://doi.org/10.1038/s41390-024-03705-3
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s41390-024-03705-3