Abstract
Autism Spectrum Disorder (ASD) is a complex neurodevelopmental condition characterized by social communication deficits and restricted, repetitive behaviors. Growing evidence implicates neuroinflammation-induced blood-brain barrier (BBB) dysfunction as a key pathogenic mechanism in ASD, although the underlying molecular pathways remain poorly understood. This study aimed to identify critical genes linking BBB function and neuroinflammatory activation, with the ultimate goal of evaluating potential therapeutic targets. Through integrative analysis combining differential gene expression profiling with three machine learning algorithms - Least Absolute Shrinkage and Selection Operator (LASSO) regression, Support Vector Machine Recursive Feature Elimination (SVM-RFE), and RandomForest combined with eXtreme Gradient Boosting (XGBoost) - we identified four hub genes, with JUN emerging as a core regulator. JUN demonstrated strong associations with both BBB integrity and microglial activation in ASD pathogenesis. Using a maternal immune activation (MIA) mouse model of ASD, we observed significant downregulation of cortical tight junction proteins ZO-1 and occludin, confirmed through immunofluorescence and qPCR analysis. Bioinformatics analysis revealed a close correlation between JUN and IL-6/MMP-9 signaling in ASD-associated microglial activation. These findings were validated in vivo, with immunofluorescence and qPCR demonstrating elevated IL-6 and MMP-9 expression in ASD mice. Pharmacological intervention using ventricular JNK inhibitor administration effectively downregulated JUN and MMP-9 expression. In vitro studies using IL-6-stimulated BV-2 microglial cells replicated these findings, showing JNK inhibitor-mediated suppression of JUN and MMP-9 upregulation. These results collectively identify the IL-6/JUN/MMP-9 pathway as a specific mediator of barrier dysfunction in ASD, representing a promising target for personalized therapeutic interventions.
Similar content being viewed by others
Background
Autism spectrum disorder (ASD) is a neurodevelopmental disorder with a core diagnosis of repetitive behaviors and social difficulties. According to the 2020 report, 1 in 36 8-year-olds in the United States has been diagnosed with autism [1]. The etiology of ASD remains largely unknown, researchers have increasingly focused on the interactions between genes and the synergies between genetic and environmental factors. In particular, environmental factors such as infections during pregnancy, maternal autoimmune diseases, and other causes of maternal immune activation have been identified as significant risk factors for neurodevelopmental disorders, including ASD, in offspring [2]. Maternal immune activation (MIA) has been extensively validated as a critical factor in the pathogenesis of ASD in both human cohort studies and animal models. In addition, MIA induces the production of several inflammatory mediators (such as IL-6, IL-17,TNF-α…) and increases the permeability of the blood-brain and gut barriers [3].
Intraperitoneal administration of polyinosinic:polycytidylic acid (poly(I:C)) in pregnant mouse models has been demonstrated to elicit significant maternal inflammatory responses, underscoring the remarkable susceptibility of microglial cells to diverse external risk factors during the gestational period [4]. Microglia serve as pivotal regulators in mediating diverse neuroinflammatory processes. These resident immune cells of the central nervous system play a dual role in neuroinflammation: they not only modulate neurogenesis and maintain blood-brain barrier (BBB) integrity, but also orchestrate inflammatory responses through the regulated expression and release of multiple inflammatory mediators, including interleukin-6 (IL-6), interleukin-1β (IL-1β), tumor necrosis factor-α (TNF-α), nitric oxide, and matrix metalloproteinases (MMPs) [3]. microglia also engage in direct neuron-glia interactions to regulate synaptic formation and refinement processes. Through these dynamic interactions, they exert profound influence on neuronal circuit development and functional maturation, ultimately contributing to the modulation of neurobehavioral phenotypes. These microglia-mediated neurodevelopmental alterations may underlie the pathogenesis of various neuropsychiatric disorders characterized by significant behavioral abnormalities [5].
Among these inflammatory factors, interleukin-6 (IL-6) serves as a pivotal pro-inflammatory cytokine that mediates maternal immune activation through multiple pathways. Specifically, IL-6 can induce macrophage polarization and activation in vivo, triggering the subsequent release of secondary cytokines and inflammatory mediators. This cascade of immune responses ultimately disrupts fetal neurodevelopmental processes, potentially leading to long-term neurological abnormalities in offspring [6]. In addition, IL-6 is widely recognized as an inflammatory factor that could increase the permeability of the BBB [7]. However, the precise mechanisms underlying its interactions with specific cytokine networks and signaling pathways remain poorly characterized in the ASD pathogenesis.
Accumulating evidence from neuropathological studies has demonstrated significant alterations in BBB integrity in ASD patients, characterized by marked downregulation of tight junction proteins in both cortical and cerebellar regions, concomitant with elevated expression levels of MMP-9, a key enzyme involved in extracellular matrix degradation [8]. As a zinc-dependent endopeptidase belonging to the matrix metalloproteinase family, MMP-9 serves as a crucial inflammatory mediator in macrophage-mediated immune responses. This multifunctional enzyme plays a pivotal role in neuroinflammatory processes through several distinct mechanisms: (1) modulating immune/inflammatory responses via proteolytic activation and processing of various cytokines and chemokines; (2) facilitating leukocyte transmigration across the BBB through parenchymal infiltration; (3) degrading key tight junction proteins, particularly occludin and zonula occludens-1 (ZO-1), at endothelial cell junctions; (4) inducing structural remodeling of the neurovascular unit, ultimately leading to compromised barrier integrity and increased paracellular permeability [9]. MMP-9 has been mechanistically implicated in the activation of multiple intracellular signaling cascades, particularly the JUN N-terminal kinase (JNK) pathway and Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling axis, in response to diverse pro-inflammatory stimuli, thereby serving as a critical molecular bridge between extracellular proteolytic activity and intracellular signal transduction during neuroinflammatory processes [10]. Although MMP-9 has been found to disrupt barrier function in ASD, its upstream regulators and pathways are not well understood. Combined with recent studies on ASD, we hypothesize that JUN is an important regulator of MMP-9 in ASD.
Our previous investigations have revealed a significant correlation between cerebellar JUN activation and neuroinflammatory responses in ASD patients [11]. Recent high-profile studies using single-nucleus sequencing (snATAC-seq) and spatial transcriptomics in the brains of ASD patients have demonstrated a peculiar response state in microglia. Many inflammatory genes have been identified with significant differences in ASD, and JUN is one of the core genes [12]. Functionally, JUN serves as a pivotal mediator in inflammatory signaling pathways through dual mechanisms: responding to diverse stimuli including pro-inflammatory cytokines, oxidative stress, and pathogen-associated molecular patterns, all of which are strongly implicated in ASD pathogenesis, and orchestrating the activation of downstream inflammatory gene networks [13]. The MIA hypothesis of ASD pathogenesis emphasizes the critical role of maternal inflammatory cell activation and subsequent signaling pathway dysregulation, wherein JUN activation may represent a key molecular event. However, the precise regulatory mechanisms underlying JUN’s downstream functions remain to be fully elucidated [12]. Molecular characterization studies have demonstrated that JUN encodes c-Jun, a component of the activator protein-1 (AP-1) transcription factor complex, which directly modulates neuronal survival and apoptosis through binding to promoter/enhancer regions of target genes. Furthermore, JUN significantly contributes to BBB maintenance by regulating the expression of tight junction-associated structural molecules [14]. This transcription factor also governs the expression of multiple pro-inflammatory genes and demonstrates functional interactions with key inflammatory mediators, including MMP-9 [10]. Despite these advances, the functional interplay between JUN and MMP-9 in ASD pathogenesis remains poorly understood, warranting further mechanistic investigations to elucidate their roles in neurodevelopmental disorders.
In this research, JUN was identified as a pivotal candidate gene through comprehensive machine learning approaches, including three distinct algorithms: least absolute shrinkage and selection operator (LASSO), support vector machine recursive feature elimination (SVM-RFE), and randomforest combined with eXtreme gradient boosting (XGBoost) analysis. This computational identification established a significant association between immune activation and JUN-mediated BBB dysfunction. The detailed analytical workflow and experimental validation procedures are systematically presented in Fig. 1. Based on those findings, we propose a novel mechanistic hypothesis wherein IL-6-mediated activation of the microglial JUN/MMP-9 signaling axis in MIA-exposed ASD mouse models contributes to the exacerbation of autism-like behavioral phenotypes through BBB disruption.
Materials and methods
Microarray source
The gene expression data from the microarray studies of ASD were obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). The GSE28521 dataset comprises three distinct brain regions from individuals with and without ASD: temporal cortex (13 TD and 13 ASD), frontal cortex (16 TD and 16 ASD), and cerebellum (10 TD and 10 ASD). The differential genes utilizing GEO2R pairs for two datasets are analyzed separately according to the commonly employed threshold parameters (|logFC|> 0.2 and adjust p < 0.05). The dataset underwent pre-processing via log2 transformation and quantile normalization, as implemented by the R package.
A comparison of ASD temporal cortex differentially expressed genes (BH-corrected p-value < 0.05) between tight junctions and autism genes between microglia activation across tissues was conducted using VennDiagram. (Source: https://bioinformatics.psb.ugent.be/webtools/Venn/). The protein and protein network were constructed using the STRING database and the Cytoscape_v3.9.1 software.
Bioinformatics data sources
The genes encoding tight junctions and those involved in microglial activation were obtained from the GeneCards database (https://www.genecards.org/). The genes associated with autism were obtained from the DisGenet database (https://disgenet.com/). The human protein atlas was obtained from the Protein Atlas database (https://www.proteinatlas.org/). The ASD Single-Cell Gene Expression Portal was obtained from the ASD scGENE Portal (http://solo.bmap.ucla.edu/asdscgene/) [12].
Machine learning
Least Absolute Shrinkage and Selection Operator (LASSO) regression is a popular regularization method for linear models. Based on the traditional linear regression model, Lasso introduces the L1 norm as a regularization term, which effectively solves the multicollinearity problem, promotes feature selection, and helps to improve the predictive power and explanatory ability of the model.
Feature selection technology is a very important part of machine learning. It can help us find the feature subset that best represents the data feature from the original data, so as to improve the performance and generalization ability of machine learning models. Support Vector machine-recursive Feature Elimination (SVM-RFE) is a feature selection method based on Support Vector Machine (SVM), which gradually removes unimportant features and finally gets important feature subsets.
Extreme Gradient Boosting (XGBoost) is an optimized distributed gradient boosting library and a tool for massively parallel boosting trees. It is known for its Regularized Boosting technique, which controls the complexity of the model by adding regular terms to the cost function to prevent overfitting.
LASSO regression is analyzed by the “glmnet” package. SVM-RFE is realized utilizing the “e1071” and “caret” packages. XGBoost is implemented using the “xgboost” package within the R software (version 4.4.1). Venn diagrams were constructed by a website tool (http://www.ehbio.com/test/venn/).
Resampling strategy was used in three machine learning methods: 10-fold cross-validation (CV) of the training dataset to avoid overfitting and perform internal validation.
Mice
The experimental animals used in this study were 6-week-old female mice and 6-week-old male mice C57BL/6 mice provided by Hubei Biont Technology Company. Maternal immune activation (MIA) was conducted by the methodology previously research described [15]. The mice were mated overnight, and the presence of seminal plugs was monitored each morning, with the day of gestation recorded as embryonic day 0.5 (E0.5). On embryonic day 12.5 (E12.5), pregnant mice were administered an intraperitoneal injection of 20 mg/kg of Poly-I:C (#P9582, Sigma). In all studies involving adult offspring, only male mice were used, and their behavior was assessed at six weeks of age. 3 μl of 5 μM JNK inhibitor or equal volume of saline injection was injected into the ventricles of control and poly group mice at 6 weeks, and collect samples for testing 3 days later. Based on preliminary experimental data, a minimum sample size of five animals per group is required to detect statistically significant behavioral differences. In this reseach, six male mice were tested in each group and statistically analyzed. For all experiments, animals were randomly allocated to experimental groups, and investigators were blinded to group assignments throughout the experimental procedures and data analysis.
Behavior tests
Open-field test (OFT)
Prior to the commencement of the test, the mouse in question should be placed in the designated testing room for a minimum of 1 h to allow for acclimatization to the laboratory environment. Subsequently, the behavioral recording software should be opened, the dimensions of the experimental center, corner, and surrounding areas should be set, and these areas should be labeled as center, corner, and side. The mice should then be placed for the experiment. The mice will then be placed in an open field for a 5 min test. Subsequently, the data pertaining to the locomotion trajectory of the mice in the open field and the total time spent in each compartment were collated.
Marble buried experiment
Prepare a clean 48 cm × 26 cm × 20 cm mice cage. Put 5 cm of corn cob litter in the cage and flatten it. The mice were left in the cage for 30 min to get used to it. During the experiment, we put glass beads on the bedding in a row of five. Then we put the mouse in the cage and let it move for half an hour. Finally, we record the number. The standard for the bead burial experiment is to require the litter to bury at least two-thirds of the glass beads.
Three-chamber social interaction test
The experiment has three parts, with an hour between each part. In the first part, the mice are placed in the middle of the three boxes and allowed to acclimatise for 5 min. In the second part, a similar-aged, same-sex mouse (S1) is placed in the left cage, and the right cage is empty. The experimental mouse is placed in the middle of the three boxes for 5 min. In the third part, the S1 mouse is placed in the right cage, and then a S2 mouse is added to the left cage. Test the mice for 5 min. The contact time of mice with empty and strange cages was recorded in three experiments [16].
Immunofluorescence
Initially, mice are euthanized, followed by the perfusion of saline into the left ventricle, and subsequently with 4% paraformaldehyde. Next, the mouse brain is removed and stored in phosphate-buffered paraformaldehyde for three days at 4 °C. Afterward, the brain is dehydrated in a 30% sucrose solution until it reaches a sinking state. Finally, the brain tissue is frozen and sectioned into 20 μm slices using a cryomicrotome. After fixation with 4% PFA in PBS, the brain slice was incubated with anti-JUN(24909-1-AP, proteintech), anti-MMP-9(10375-2-ap, PTG), anti-IBA1(10904-1-AP, proteintech), anti-occludin(27260-1-AP, PTG),anti-ZO-1(ab221547, Abcam) overnight. The slices were washed with PBS and incubated with Alexa 488 or Alexa 596 conjugated secondary antibodies (Invitrogen) for 90 min. To label the nucleus, cells were also incubated with DAPI (Invitrogen) for 5 min. A laser-scanning confocal microscope (LSM 800; Zeiss). The data were analyzed by ImageJ Software. Three mice were tested in each group and statistically analyzed.
RNA quantification
RNA was extracted from tissues or cells using the RNeasy Mini Kit. The extracted RNA was then converted to cDNA using M-MLV reverse transcriptase (Invitrogen). RT-qPCR was done with an SYBR Green kit on a LightCycler system. The data were normalized to GAPDH levels and fold changes were calculated using the 2^(−ΔΔCt) method. Primer pairs used were as follows: IL-6 5′-GCCTTCTTGGGACTGATGCT-3′, 5′-GCCACTCCTTCTGTGACTCC-3′; MMP-9 5′-TTCTACGGCCACTACTGTGC-3′, 5′-CTCGGTACTGGAAGACGTCG-3′; JUN 5′ACGACCTTCTACGACGATGC-3′, 5′GGGTCGGTGTAGTGGTGATG-3′
Cell
BV-2 was bought from Wuhan Prosel. BV-2 were maintained in DMEM (11995065, Gibco, NY, USA) medium containing 10% fetal bovine serum (10099141, Gibco). BV-2 cells were seeded into 6-well plates and treated with 10 ng/ml IL-6(PeproTech, Waltham, MA, USA, Cat#: 216-16) or 5 μM/ml JNK-IN-7 inhibitor (MedChemExpress, USA, Cat#: HY-15617) for 24 h. Four samples were tested in each group and statistically analyzed.
Statistical analysis
Depending on the dataset, significance is detected by either t-test or one-way analysis. Significant differences were defined as P < 0.05. The images and analyses shown are from at least three experiments. Pearson correlation coefficients and graphs were created using GraphPad Prism software.
Results
Machine learning to identify hub genes in ASD
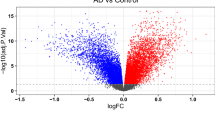
|log2 fold change (FC)|>0.2 and p < 0.05 were selected as the criteria.Transcriptomic analysis of the temporal cortex (TC) region in the GSE28521 dataset revealed 146 differentially expressed genes (DEGs) between ASD and typical development (TD) groups, as visualized through volcano plot analysis using GEO2R software (Fig. 2A). Through integration with the Genecard database, which contains 5476 tight junction-associated genes, we identified 39 overlapping genes at the intersection of ASD-related DEGs and tight junction function (Fig. 2B). Protein-protein interaction (PPI) network analysis, constructed using Cytoscape software, identified IGFBP5, JUN, and RPS6 as central hub genes within the network topology (Fig. 2C).
A Volcano plots of DEGs in GSE28521, |log2 fold change (FC)|>0.2 and p < 0.05 as selection criteria. B 39 intersection genes of DEGs and Tight junction related genes. C Protein-protein internet(PPI) of 39 intersection genes. D LASSO regression algorithm selected 6 genes. E 17 hub genes were screened out by the XGB method. F 10 genes were extracted by using the SVM-RFE algorithm. G Venn network to intersect 3 gene subsets selected by machine learning. H The expression of 4 hub genes in ASD and TD (n = 13). Significant differences are indicated by **P < 0.01, ***P < 0.01, compared with ASD and TD group. I ROC curve of 4 hub gene.
Among the 39 crossover genes of ASD and Tight junction, 6 hub genes were first identified by the least absolute shrinkage and selection operator (LASSO) regression algorithm Fig. 2D). Subsequently, 17 hub genes were screened out by the extreme gradient boosting (XGB) method (Fig. 2E). In addition, 10 genes were extracted by using the support vector machine recursive feature elimination (SVM-RFE) algorithm (Fig. 2F). Ultimately, through the utilization of a Venn diagram, there were four genes able to be identified (Fig. 2G). The remaining genes were insulin-like growth factor binding protein 5(IGFBP5), JUN, flagellar radial spoke protein (RSP2), and secreted protein acidic and cysteine-rich (SPARC). Concurrently, the researchers observed a notable elevation in the expression of all four genes within the TC region in subjects diagnosed with ASD (Fig. 2H). The diagnostic value of the four hub genes was further verified by receiver operating characteristic (ROC) curves. Specifically, RPS2 (AUC = 0.911), JUN (AUC = 0.888), SPARC (AUC = 0.858), and IGFBP5 (AUC = 0.858) demonstrated significant diagnostic value for ASD (Fig. 2I).
JUN is widely expressed in the brain and is mainly derived from microglia
The preceding analysis of DEGs and machine learning in the TC region of ASD patients revealed the existence of four potential hub genes. The single-cell sequencing results of ASD provided further insight into the role of these four hub genes on the BBB. Our analysis revealed particular interest in JUN, which exhibited the highest log2 fold change (LogFC = 0.31), suggesting its potential functional significance in ASD pathogenesis (Fig. 3A). To investigate the regional specificity of JUN dysregulation in ASD, we analyzed RNA-Seq data from the GSE28521 dataset, which demonstrated significantly elevated JUN expression not only in the temporal cortex but also in the prefrontal cortex and cerebellum of ASD individuals compared to TD group (Fig. 3B). To establish the baseline expression pattern of JUN in neurotypical brains, we integrated data from The Human Protein Atlas (HPA) project. Through comprehensive analysis of RNA-Seq expression data from both HPA and FANTOM5 human brain tissue databases, we observed ubiquitous JUN expression across multiple brain regions, with particularly high expression levels in cortical areas (Fig. 3C, D). Furthermore, single-cell RNA sequencing analysis of human brain tissues from the HPA database enabled us to characterize JUN expression patterns at cellular resolution, revealing its presence across diverse neural cell subtypes. The principal marker genes of astrocytes were ALDH1L1, GFAP, and SLC1A3, while the principal marker genes of microglia were AIF1, ITGAM, P2RY12, and TYROBP. The expression of JUN was higher in astrocytes (clusters 35) and microglia (clusters 43) than in other cell subsets in brain tissues. This suggests that JUN is predominantly derived from microglia in part of the brain (Fig. 3E, F).
A The single-cell sequencing results of ASD in four hub genes on the BBB. B The expression of JUN in the frontal cortex (n = 16) and cerebellum (n = 10) in GSE28521. C, D The expression of JUN in the different brain regions of the HPA(C) and FANTOM5(D) human brain RNA-Seq dataset. E, F Expression of JUN in the human brain of single-cell RNA sequencing of HPA dataset. Error bars indicate as means ± SD, *P < 0.05, **P < 0.01, compared with ASD and TD group.
BBB impairment and JUN upregulation in ASD
Behavioral experiments were conducted on six weeks age male offspring mice (poly(I:C) group) born following maternal immune activation induced by poly(I:C) injection in pregnant mice (Fig. 4A). Each group consisted of six male mice tested for behavioral assessments. The results showed that the poly (I: C) group mice exhibited significantly abnormal activity patterns in open spaces (Fig. 4B). Compared with the control group, the mice in the poly (I: C) group showed reduced activity in the center of the open field and significantly increased corner time (Fig. 4C), reflecting anxiety-like behavior in the MIA group within an unfamiliar environment. The marble buried test revealed a statistically significant increase in the number of buried marbles in the poly(I:C) group compared to control animals (Fig. 4D), indicating an increase in stereotyped behavior. The results of the three-chamber social experiment demonstrated that the mice in the MIA group exhibited a statistically significant reduction in social interaction with the two unfamiliar mice, in comparison to the control group (Fig. 4E), indicating that the mice in the MIA group showed a significant decrease in social ability. A comprehensive analysis of the behavioral tests revealed that the mice in the poly(I:C) group exhibited characteristic features consistent with autism phenotypes. Immunofluorescence(IF) staining of the temporal cortex region demonstrated a reduction in the expression of ZO-1 and occludin proteins in ASD mice, indicating damage to the BBB (Fig. 4F). Concurrently, we observed substantial upregulation of JUN expression alongside pronounced microglial activation, as evidenced by increased IBA1 immunoreactivity. Furthermore, a colocalization relationship between JUN and IBA1 was observed (Fig. 4G). This suggests that microglia are activated in ASD mice under MIA conditions, leading to an increase in JUN expression.
A Schematic diagram of modeling a mouse model of ASD with maternal immune activation. Embryonic day 12.5 (E12.5 d). B The movement trajectories in the open field test (OFT) (n = 6). C Time spent in the corner and center of OFT experitment (n = 6). D The number of marbles buried in the Marble buried experiment(n = 6). E Social time in three-chamber social interaction test (n = 6). F Relative protein levels of ZO-1 and occludin in control and poly(I: C) group (n = 6). G Relative protein levels of IBA1 and JUN in control and poly(I: C) group (n = 6). Scale bars, 20 μm. Scale bars, 20 μm. Error bars indicate as means ± SD, *P < 0.05, **P < 0.01, ***P < 0.001 compared with control and poly(I: C) group, unpaired t-test.
Up-regulation of immune activating IL-6/MMP-9 in ASD was positively correlated with JUN
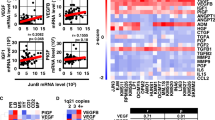
The occurrence of ASD is closely related to immune activation. A total of 326 crossover genes were obtained by using ASD-related genes in the Disgenet database and microglia activation-related genes in the Genecard database (Fig. 5A). The PPI network was constructed using the Cytoscape software, and the nodes with a degree value greater than 80 were selected for display. It can be observed that IL-6 and MMP-9 are among the top 20 genes with the highest closeness in the inner loop and the top 20 genes with the highest betweenness centrality (Fig. 5B, C), which play an important role in the immune activation of ASD and have been closely related to the increase of barrier permeability in previous studies. The construction of the top 10 networks (GenDoma database) of transcription factors with target genes (red) and protein-protein interaction networks (PPI, blue) with MMP-9 as the core reveals a close relationship with JUN (Fig. 5D). Furthermore, an additional finding in the FANTOM5 data set was a modest correlation between the expression levels of JUN and MMP-9 in each brain region (R = 0.7001, P = 0.0053, Fig. 5E). Accordingly, the expression of IL-6 and MMP-9 was quantified in ASD mice, which exhibited a notable elevation in the TC region by IF and real-time quantitative polymerase chain reaction (RT-qPCR) test (Fig. 5F, G).
A 326 intersection genes of autism and microglia activation-related genes. B Top 20 genes with the highest closeness in the inner loop. C Top 20 genes with the highest betweenness centrality in the inner loop. D Top 10 networks of transcription factors with target genes (red) and protein-protein interaction networks (PPI, blue) with MMP-9 in the GenDoma database. E Correlation between the expression levels of JUN and MMP-9 in each brain region of the FANTOM dataset (R = 0.7001, P = 0.0053). F Relative protein levels of MMP-9 in control and poly(I: C) group (n = 6). G Relative mRNA levels of IL-6 and MMP-9 in control and poly(I: C) group (n = 6). Scale bars, 20 μm. Error bars indicate as means ± SD, **P < 0.01, ***P < 0.001 compared with control and poly(I: C) group, unpaired t-test.
IL-6 could activate the JUN/MMP-9 specific in disrupting BBB
In vivo pharmacological intervention studies were conducted through ventricle administration of JNK inhibitors in ASD model mice. Immunofluorescence analysis of cortical tissues collected three days post-injection revealed significant downregulation of both JUN and MMP-9 expression following JNK inhibitor treatment (Fig. 6A).
A Ventricular administration of JNK inhibitor for 3 days significantly altered the protein expression levels of JUN and MMP-9 across four experimental groups (n = 6). B The mRNA expression of JUN and MMP-9 was assessed by RT-qPCR in BV-2 cells after 10 ng/ml IL-6 or 5 μM/ml JNK inhibitor for 24 h (n = 4). Scale bars, 20 μm. Error bars indicate as means ± SD, *P < 0.05, **P < 0.01, ***P < 0.001, One-way ANOVA.
Complementary in vitro experiments utilized the BV-2 microglial cell line, a well-established model for studying neuroinflammatory processes. Cells were stimulated with IL-6 (10 ng/mL) for 24 h to induce microglial activation. RT-qPCR analysis demonstrated significant upregulation of JUN and MMP-9 expression in IL-6-stimulated cells compared to untreated controls. Co-treatment with a JNK inhibitor (10 μmol/L) effectively attenuated this IL-6-induced activation, resulting in significantly reduced expression of both JUN and MMP-9 (Fig. 6B).
These findings collectively demonstrate that IL-6-mediated microglial activation triggers JUN pathway signaling, which subsequently regulates MMP-9 expression. The pharmacological inhibition of JNK signaling effectively suppresses this inflammatory cascade, suggesting a potential therapeutic target for modulating neuroinflammatory processes in ASD.
Discussion
The etiology of ASD is multifaceted, with a combined influence of genetic and environmental factors resulting in alterations to neurons, disruption of the BBB, and modifications to the intestinal flora. We successfully modeled ASD-like mice by MIA, which will lead to the generation of multiple inflammatory responses. On the one hand, at the cytokine level, there is a massive release of factors such as IL-6, IL-17, and TNF-a. On the other hand, at the cellular level, MIA activates microglia in the brain and promotes their amplification of inflammatory responses [5]. These interconnected processes establish a self-perpetuating inflammatory cycle that directly impacts neuronal homeostasis and BBB integrity [17].
Accumulating evidence has identified specific cytokines as critical contributors to ASD pathogenesis. Experimental studies have demonstrated that maternal IL-6 administration during pregnancy can induce ASD-like behavioral and neuropathological phenotypes in offspring, establishing its pivotal role in neurodevelopmental disruption [6]. IL-6 exerts its pathological effects through multiple mechanisms, as a potent inducer of microglial activation, through direct mediation of BBB dysfunction, and by promoting neuroinflammatory cascades that contribute to neuronal damage [18]. This cytokine dysregulation, coupled with aberrant glial cell activation, disrupts neural circuit formation and synaptic plasticity, ultimately leading to altered neuronal function and connectivity.
In recent years the gut-brain axis has served as a focus of attention, and the increased permeability of the BBB in ASD is widely recognized, impairment of barrier function is highly correlated with microglia activation [19]. The integrity of the BBB serves to screen and control the passage of cytokines in immune pathways or metabolites in metabolic pathways into the blood or brain tissues, which leads to the regulation of neurological function [20, 21]. Although, IL-6 and MMP-9 are thought to contribute to BBB damage, however, the specific molecules and pathways involved are still poorly understood. So in this research we conducted a more in-depth exploration by combining bioinformatics analysis.
Machine learning methods to screen the crossover genes of disease differential genes and functional genes have been fully recognized in the diagnosis and mechanism studies of many diseases. For example, the interaction between major depressive disorder and oxidative stress [22], the relationship between ischemic stroke and reticulum (ER) stress [23], etc. Through integrative analysis combining differential gene expression screening and advanced machine learning algorithms, we identified four key tight junction-associated genes in the temporal cortex (TC) of ASD individuals: IGFBP5, JUN, RSPH2, and SPARC. Notably, JUN emerged as a particularly significant candidate based on its dual role in BBB regulation and neuroinflammatory processes. Supporting evidence from cross-disease genomic analyses revealed elevated expression of both JUN and its subunit JUND in ASD patients, mirroring expression patterns observed in certain tumor types. These findings suggest that JUN may play a pivotal role in mediating cellular differentiation processes and barrier function impairment in ASD pathogenesis [24]. Recent single-cell genomics studies have indicated that the pathway for JUN to be upregulated in brain cells of patients with autism may be associated with oxidative phosphorylation and the inflammatory stress response [12]. The activity of c-Jun is regulated by phosphorylation, which is mediated by c-Jun’s N-terminal kinase (JNK) family, including JNK1, JNK2, and JNK3 [25]. JNK1 plays a pivotal role in the regulation of numerous processes associated with neuronal development in the central nervous system (CNS) [26]. JNK2 is specific to the prefrontal cortex and has been demonstrated to induce anxiety-like behavioral and cognitive impairment [27]. JUN may serve as a therapeutic target and biomarker in neurodevelopmental disorders [28]. Activation of the JNK/c-JUN signaling pathway has been mechanistically linked to downregulation of ZO-1 expression and disruption of tight junction integrity, as evidenced by both previous studies and our current findings. This pathway-mediated impairment of barrier function provides a molecular framework for understanding BBB dysfunction in neurodevelopmental disorders, consistent with the observed pathological changes in our ASD model [29].
To determine the cellular origin of JUN, we analyzed data from single-cell sequencing of the brain, which confirmed that JUN is expressed mainly by microglia and astrocytes. The JUN family is widely expressed in the brain, which is consistent with previous findings [30]. IL-6 is an important driver of ASD, and other cytokines such as IL-17 also play important roles. Activation of the IL-17 pathway also causes up-regulation of JUN expression and thus has a deeper impact. IL-1β can upregulate high JUN expression and thus upregulate MMP-9 [31]. Those studies proposed that JUN plays a role in neuromodulation, not only in microglia but also in other immune cytes, which deserves to be further explored.
The role of the other three hub genes, namely IGFBP5, RSP2, and SPARC, also merits further investigation. Insulin-like growth factor-binding protein 5 (IGFBP5) is an inhibitory binding protein of insulin-like growth factor 1 (IGF-1), and its overexpression could cause cognitive deficits [32]. In the cohort study, an elevated level of IGFBP5 has been linked to an accelerated rate of cognitive decline [33]. IGFBP5 have been assumed as an important role in the induction of extracellular matrix production and deposition [34, 35]. Above such results, indicate IGFBP5 may affect the BBB function and cognitive disorder of ASD. Ribosomal Protein S2 (RPS2) is highly related peptide chain elongation and nervous system development [36]. RPS2 has also been found to be a specific molecule in capillary proteomics of AD and may be associated with barrier function and cognition [37]. In Parkinson’s neural stem cell model, Omics Analyses analysis showed that RSP2 may be a hallmark of neurological disease [38]. Although there is still a lack of research on RPS2 in ASD, it may be considered a potential marker for further investigation. SPARC may serve as a potential mediator for the activation of the MMP-9 pathway, which could subsequently result in damage to the BBB following subarachnoid hemorrhage [39]. Combined with the previous research, IGFBP5, RSP2, and SPARC might have a significant impact on neurological function, especially for the diagnosis or mechanism of ASD, which is worthy of further exploration.
In a recent serum study of patients with ASD, a significant association was found between polymorphisms in the MMP-9 gene and its serum concentrations and autism [40]. In a birth cohort study in Denmark, amniotic fluid testing was associated with increased levels of MMP-9 in ASD compared with controls [41]. In other cases, JUN plays a significant role in regulating the expression of MMPs. In a variety of diseases, the JUN/MMP-9 pathway has been demonstrated to regulate the permeability of local structures, thereby inducing tumor metastasis and myocardial metabolism [42]. The impact of JUN/MMP-9 on intestinal permeability in inflammatory astrocytes has also been substantiated [31].
Integrative analysis of microglial activation-associated genes in ASD has identified IL-6 and MMP-9 as central mediators in disease pathogenesis. These findings support a mechanistic model wherein IL-6 serves as the primary initiator of microglial activation, triggering the JUN/MMP-9 signaling cascade that ultimately culminates in barrier dysfunction. To validate this proposed pathway, we conducted complementary in vivo and in vitro experiments that consistently demonstrated pathway upregulation under ASD-relevant conditions.Pharmacological intervention studies revealed that JNK inhibitors effectively attenuated IL-6-induced pathway activation, while JUN inhibition blocked IL-6-mediated effects, establishing JUN as a critical node in this signaling network. These results collectively demonstrate that the IL-6/JUN/MMP-9 axis represents a key regulatory pathway in microglial-mediated barrier dysfunction, providing potential therapeutic targets for modulating neuroinflammatory processes in ASD.
Additional studies need to be further explored, the role of other protease metals in MMPs should also be investigated. MMP-2 and MMP-8 have been shown to have a critical impact on a variety of neurological disorders including ASD, schizophrenia and depression [43, 44]. In current MIA models, male offspring have predominantly served as experimental subjectssubjects [15, 45], reflecting the well-documented male bias in ASD prevalence and phenotypic presentation. This gender disparity in study design stems from several factors including increased male susceptibility to MIA-induced neurodevelopmental alterations, more pronounced behavioral phenotypes in male offspring, and potential protective effects of estrogen in females. However, this male-centric approach has limitations as it potentially overlooks female-specific pathological mechanisms and therapeutic targets. Recent years have witnessed growing recognition of sex as a critical biological variable in neurodevelopmental research. Emerging studies are increasingly incorporating female subjects to provide a more comprehensive understanding of ASD pathogenesis. While the current focus on male mice aligns with human epidemiological data and offers clear phenotypic readouts, future investigations should address several key aspects such as characterization of potential sex-specific molecular pathways, evaluation of estrogen-mediated neuroprotective mechanisms, and determination of whether the JUN/MMP-9 pathway exhibits sex-dependent regulation. Such sex-balanced approaches will be essential for developing more inclusive and effective therapeutic strategies for ASD.
Conclusions
Autism spectrum disorders (ASD) are characterized by substantial phenotypic heterogeneity, with neuroinflammation emerging as a significant environmental contributor to disease pathogenesis. Through comprehensive bioinformatics analysis and machine learning approaches, we have identified JUN as a critical regulator of BBB dysfunction in the ASD cortex. Our experimental findings using both MIA models and microglial cell systems demonstrate that IL-6-mediated activation of the JUN/MMP-9 pathway in microglia represents a key mechanism underlying BBB impairment.
This microglial-mediated barrier dysfunction may amplify the neuropathological effects of maternal immune factors and circulating metabolites on the developing brain through compromised gut-brain axis communication. The current study provides novel insights into BBB alterations in ASD-associated neuroinflammation, highlighting the IL-6/JUN/MMP-9 pathway as a potential therapeutic target for developing effective intervention strategies in autism.
Data availability
The data that support the findings of this study are available from the corresponding authors upon request.
References
Maenner MJ, Warren Z, Williams AR, Amoakohene E, Bakian AV, Bilder DA, et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 years - autism and developmental disabilities monitoring network, 11 sites, United States, 2020. MMWR Surveill Summ. 2023;72:1–14.
Estes ML, McAllister AK. Maternal immune activation: implications for neuropsychiatric disorders. Science. 2016;353:772–7.
Zawadzka A, Cieślik M, Adamczyk A. The role of maternal immune activation in the pathogenesis of autism: a review of the evidence, proposed mechanisms and implications for treatment. Int J Mol Sci. 2021;22:11516.
Yin H, Wang Z, Liu J, Li Y, Liu L, Huang P, et al. Dysregulation of immune and metabolism pathways in maternal immune activation induces an increased risk of autism spectrum disorders. Life Sci. 2023;324:121734.
Hughes HK, Moreno RJ, Ashwood P. Innate immune dysfunction and neuroinflammation in autism spectrum disorder (ASD). Brain Behav Immun. 2023;108:245–54.
Smith SE, Li J, Garbett K, Mirnics K, Patterson PH. Maternal immune activation alters fetal brain development through interleukin-6. J Neurosci. 2007;27:10695–702.
Voirin AC, Perek N, Roche F. Inflammatory stress induced by a combination of cytokines (IL-6, IL-17, TNF-α) leads to a loss of integrity on bEnd.3 endothelial cells in vitro BBB model. Brain Res. 2020;1730:146647.
Fiorentino M, Sapone A, Senger S, Camhi SS, Kadzielski SM, Buie TM, et al. Blood-brain barrier and intestinal epithelial barrier alterations in autism spectrum disorders. Mol Autism. 2016;7:49.
Vafadari B, Salamian A, Kaczmarek L. MMP-9 in translation: from molecule to brain physiology, pathology, and therapy. J Neurochem. 2016;139:91–114.
Liao B, Geng L, Zhang F, Shu L, Wei L, Yeung P, et al. Adipocyte fatty acid-binding protein exacerbates cerebral ischaemia injury by disrupting the blood-brain barrier. Eur Heart J. 2020;41:3169–80.
Li H, Wang X, Hu C, Li H, Xu Z, Lei P, et al. JUN and PDGFRA as crucial candidate genes for childhood autism spectrum disorder. Front Neuroinform. 2022;16:800079.
Wamsley B, Bicks L, Cheng Y, Kawaguchi R, Quintero D, Margolis M, et al. Molecular cascades and cell type-specific signatures in ASD revealed by single-cell genomics. Science. 2024;384:eadh2602.
Wu Y, Li Q, Lv LL, Chen JX, Ying HF, Ruan M, et al. Nobiletin inhibits breast cancer cell migration and invasion by suppressing the IL-6-induced ERK-STAT and JNK-c-JUN pathways. Phytomedicine. 2023;110:154610.
Byrns CN, Perlegos AE, Miller KN, Jin Z, Carranza FR, Manchandra P, et al. Senescent glia link mitochondrial dysfunction and lipid accumulation. Nature. 2024;630:475–83.
Choi GB, Yim YS, Wong H, Kim S, Kim H, Kim SV, et al. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science. 2016;351:933–9.
Willinger Y, Friedland Cohen DR, Turgeman G. Exogenous IL-17A alleviates social behavior deficits and increases neurogenesis in a murine model of autism spectrum disorders. Int J Mol Sci. 2023;25:432.
Suprunowicz M, Tomaszek N, Urbaniak A, Zackiewicz K, Modzelewski S, Waszkiewicz N. Between dysbiosis, maternal immune activation and autism: is there a common pathway. Nutrients. 2024;16:549.
Williams JA, Burgess S, Suckling J, Lalousis PA, Batool F, Griffiths SL, et al. Inflammation and brain structure in schizophrenia and other neuropsychiatric disorders: a mendelian randomization study. JAMA Psychiatry. 2022;79:498–507.
Davoli-Ferreira M, Thomson CA, McCoy KD. Microbiota and microglia interactions in ASD. Front Immunol. 2021;12:676255.
Socała K, Doboszewska U, Szopa A, Serefko A, Włodarczyk M, Zielińska A, et al. The role of microbiota-gut-brain axis in neuropsychiatric and neurological disorders. Pharmacol Res. 2021;172:105840.
Lukens JR, Eyo UB. Microglia and neurodevelopmental disorders. Annu Rev Neurosci. 2022;45:425–45.
Shao X, Wang Y, Geng Z, Liang G, Zhu X, Liu L, et al. Novel therapeutic targets for major depressive disorder related to oxidative stress identified by integrative multi-omics and multi-trait study. Transl Psychiatry. 2024;14:443.
Jiang N, Wang C, Xie B, Xie H, Wu A, Kong X, et al. Identification of endoplasmic reticulum stress genes in human stroke based on bioinformatics and machine learning. Neurobiol Dis. 2024;199:106583.
Yavuz BR, Arici MK, Demirel HC, Tsai CJ, Jang H, Nussinov R, et al. Neurodevelopmental disorders and cancer networks share pathways, but differ in mechanisms, signaling strength, and outcome. NPJ Genom Med. 2023;8:37.
Petrov D, Luque M, Pedrós I, Ettcheto M, Abad S, Pallàs M, et al. Evaluation of the role of JNK1 in the hippocampus in an experimental model of familial Alzheimer’s disease. Mol Neurobiol. 2016;53:6183–93.
Castro-Torres RD, Busquets O, Parcerisas A, Verdaguer E, Olloquequi J, Ettcheto M, et al. Involvement of JNK1 in neuronal polarization during brain development. Cells. 2020;9:1897.
Yang M, Barrios J, Yan J, Zhao W, Yuan S, Dong E, et al. Causal roles of stress kinase JNK2 in DNA methylation and binge alcohol withdrawal-evoked behavioral deficits. Pharmacol Res. 2021;164:105375.
Musi CA, Agrò G, Santarella F, Iervasi E, Borsello T. JNK3 as therapeutic target and biomarker in neurodegenerative and neurodevelopmental brain diseases. Cells. 2020;9:2190.
Qu X, Yang R, Tan C, Chen H, Wang X. Astrocytes-secreted WNT5B disrupts the blood-brain barrier via ROR1/JNK/c-JUN cascade during meningitic Escherichia Coli infection. Mol Neurobiol. 2024;62:661–73.
Nogueiras R, Sabio G. Brain JNK and metabolic disease. Diabetologia. 2021;64:265–74.
Wu CY, Hsieh HL, Sun CC, Yang CM. IL-1beta induces MMP-9 expression via a Ca2+-dependent CaMKII/JNK/c-JUN cascade in rat brain astrocytes. Glia. 2009;57:1775–89.
Rauskolb S, Andreska T, Fries S, von Collenberg CR, Blum R, Monoranu CM, et al. Insulin-like growth factor 5 associates with human Aß plaques and promotes cognitive impairment. Acta Neuropathol Commun. 2022;10:68.
Kim N, Yu L, Dawe R, Petyuk VA, Gaiteri C, De Jager PL, et al. Microstructural changes in the brain mediate the association of AK4, IGFBP5, HSPB2, and ITPK1 with cognitive decline. Neurobiol Aging. 2019;84:17–25.
Yamaguchi Y, Yasuoka H, Stolz DB, Feghali-Bostwick CA. Decreased caveolin-1 levels contribute to fibrosis and deposition of extracellular IGFBP-5. J Cell Mol Med. 2011;15:957–69.
Su Y, Nishimoto T, Feghali-Bostwick C. IGFBP-5 promotes fibrosis independently of its translocation to the nucleus and its interaction with nucleolin and IGF. PLoS ONE. 2015;10:e0130546.
Manchado M, Infante C, Asensio E, Cañavate JP, Douglas SE. Comparative sequence analysis of the complete set of 40S ribosomal proteins in the Senegalese sole (Solea senegalensis Kaup) and Atlantic halibut (Hippoglossus hippoglossus L.) (Teleostei: Pleuronectiformes): phylogeny and tissue- and development-specific expression. BMC Evol Biol. 2007;7:107.
Suzuki M, Tezuka K, Handa T, Sato R, Takeuchi H, Takao M, et al. Upregulation of ribosome complexes at the blood-brain barrier in Alzheimer’s disease patients. J Cereb Blood Flow Metab. 2022;42:2134–50.
Notopoulou S, Gkekas I, Petrakis S. Omics analyses in a neural stem cell model of familial Parkinson’s disease. Adv Exp Med Biol. 2023;1423:149–60.
Okada T, Suzuki H, Travis ZD, Altay O, Tang J, Zhang JH. SPARC aggravates blood-brain barrier disruption via integrin αVβ3/MAPKs/MMP-9 signaling pathway after subarachnoid hemorrhage. Oxid Med Cell Longev. 2021;2021:9739977.
Lord JR, Mashayekhi F, Salehi Z. How matrix metalloproteinase (MMP)-9 (rs3918242) polymorphism affects MMP-9 serum concentration and associates with autism spectrum disorders: a case-control study in Iranian population. Dev Psychopathol. 2022;34:882–8.
Abdallah MW, Pearce BD, Larsen N, Greaves-Lord K, Nørgaard-Pedersen B, Hougaard DM, et al. Amniotic fluid MMP-9 and neurotrophins in autism spectrum disorders: an exploratory study. Autism Res. 2012;5:428–33.
Sun J, Hu JR, Liu CF, Li Y, Wang W, Fu R, et al. ANKRD49 promotes the metastasis of NSCLC via activating JNK-ATF2/c-Jun-MMP-2/9 axis. BMC Cancer. 2023;23:1108.
Pirbhoy PS, Rais M, Lovelace JW, Woodard W, Razak KA, Binder DK, et al. Acute pharmacological inhibition of matrix metalloproteinase-9 activity during development restores perineuronal net formation and normalizes auditory processing in Fmr1 KO mice. J Neurochem. 2020;155:538–58.
Ji Y, Huang W, Chen Y, Zhang X, Wu F, Tang W, et al. Inhibition of MMP-2 and MMP-9 attenuates surgery-induced cognitive impairment in aged mice. Brain Res Bull. 2023;204:110810.
Kim S, Kim H, Yim YS, Ha S, Atarashi K, Tan TG, et al. Maternal gut bacteria promote neurodevelopmental abnormalities in mouse offspring. Nature. 2017;549:528–32.
Acknowledgements
We would like to express our gratitude to all those who took part in this research project, whose contributions have been invaluable.
Author information
Authors and Affiliations
Contributions
CH and HL helped write this paper and collect the data. JC, YL, YZ, HL, and XL helped with analysis, resources, review and editing.YH helped design the experiment and reviewed it. All authors contributed to the article and approved the final version.
Corresponding author
Ethics declarations
Competing interests
This research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethics approval and consent to participate
All animal experiments conducted in this study were performed in strict compliance with the National Institutes of Health (NIH) guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee of Huazhong University of Science and Technology (Approval No. 4025).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hu, C., Li, H., Cui, J. et al. Integrative analysis identifies IL-6/JUN/MMP-9 pathway destroyed blood-brain-barrier in autism mice via machine learning and bioinformatic analysis. Transl Psychiatry 15, 239 (2025). https://doi.org/10.1038/s41398-025-03452-x
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41398-025-03452-x