Abstract
Lineage plasticity, the ability of cells to transition to an alternative phenotype as a means for adaptation, is an increasingly recognized mechanism of tumor evolution and a driver of resistance to anticancer therapies. The most extensively described clinical settings impacted by such molecular phenomena include neuroendocrine transformation in androgen receptor-dependent prostate adenocarcinoma, and adenocarcinoma-to-neuroendocrine and adenocarcinoma-to-squamous transdifferentiation in epidermal growth factor receptor-driven lung adenocarcinoma, affecting 10%–20% of patients treated with targeted therapy. Recent analyses of human tumor samples and in vivo models of histological transformation have led to insights into the biology of lineage plasticity, including biomarkers predictive of high risk of transformation. However, no clinically available therapies aimed to prevent or revert plasticity are currently available. In the present review, we will provide a biological and therapeutic overview of the current understanding of common and divergent molecular drivers of neuroendocrine and squamous transdifferentiation in tumors from different origins, including descriptive analysis of previously known and recently described molecular events associated with histological transformation, and propose evidence-based alternative models of transdifferentiation. A clear definition of the commonalities and differences of transforming tumors in different organs and to different histological fates will be important to translate molecular findings to the clinical setting.
Similar content being viewed by others
Introduction
Lineage plasticity illustrates an ability of cells in one phenotypic state to transition to an alternative developmental pathway. This phenomenon is an increasingly recognized mechanism of tumor evolution, a driver of resistance to anticancer therapies, and remains a clinical conundrum.1 The most extensively described clinical setting for such process is histological transformation, defined as the transdifferentiation of tumor cells from one initial histologic subtype to an alternative one (Fig. 1). Lineage plasticity most frequently occurs during treatment with targeted therapy, leading to resistance, and includes neuroendocrine (NE) transformation in androgen receptor (AR)-dependent prostate adenocarcinoma,2 estimated to be found in 25% of prostate cancer cases treated with AR-targeted therapies.3 Histological transformation is also frequently observed in epidermal growth factor receptor (EGFR)-driven lung adenocarcinoma (LUAD) transforming to NE and squamous carcinomas, with each occurring in 9%–15% of EGFR-driven LUAD treated with targeted therapies.1,4,5 However, recent reports suggest that histological transformation may occur as a mechanism of resistance to targeted therapy in many oncogene-driven tumor settings, and even spontaneously in the absence of treatment.6
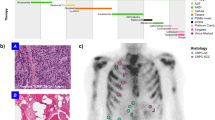
a, b Histological images of the most relevant lung (a) and prostate (b) cancer subtypes associated with histological transformation. Subtypes include lung adenocarcinoma (LUAD), lung adenosquamous (LUAS), squamous cell lung carcinoma (LUSC), small cell lung cancer (SCLC), large cell neuroendocrine cancer (LCNEC), combined SCLC (cSCLC), prostate adenocarcinoma (PRAD), and neuroendocrine prostate cancer (NEPC). Scale bars, 100 µm.
Recent analyses of human tumor samples and in vivo models of histological transformation have led to insights into critical aspects of the biology surrounding lineage plasticity, including biomarkers predictive of high risk of NE transformation.5,7 However, to date, there are no clinically available therapies specifically targeted to prevent or suppress lineage plasticity. Critically, because of substantial overlap in the molecular events across tumor types and histological outcomes, approaches to constrain plasticity may be broadly applicable, making “basket trial” like clinical translation both pragmatic and effective.
A clear definition of the commonalities and differences of transforming tumors in different organs and to different histological fates will be important to inform clinical research, define therapeutic targets, and ultimately, optimize care for these patients. In the present review, we will provide an overview of the current understanding of common and divergent molecular drivers of NE and squamous transdifferentiation in tumors from different origins, from biological and therapeutic perspectives. In addition, we perform a joint analysis of previously known and recently described molecular alterations promoting or driving histological transformation to both squamous and NE tumors and propose evidence-based alternative models of transdifferentiation.
Histological transformations, clinical and molecular context
Besides EGFR-mutant LUADs treated with tyrosine kinase inhibitors (TKIs) and metastatic AR-dependent prostate adenocarcinomas treated with enzalutamide, NE transformation has been described in LUADs with alternative genomic contexts and may represent a general mechanism of acquired resistance to other targeted therapies.1,6,8 NE-transformed small cell carcinomas are typically rapidly progressive and treatment refractory, leading to prognoses similar to or even worse than de novo small cell lung cancer (SCLC).7,9,10 Even if patients with adenocarcinoma at high risk of NE transformation can be identified (i.e., tumors with concurrent TP53/RB1 inactivation), to date, no therapies are available to effectively constrain plasticity and prevent transformation. The identification of drivers of lineage plasticity and pharmacologically targetable effectors of transformation is unmet clinical needs.
Further increasing the complexity of studying lineage plasticity, the documented time between the initiation of targeted therapy and clinical recurrence is variable. Time on therapy is shorter in patients with combined EGFR/TP53/RB1-mutated LUAD bearing TKI-responsive EGFR driver oncogenic alterations and treated with osimertinib.7 In prostate cancer, tumor responsive time on androgen deprivation therapy (ADT) in the era of enzalutamide varies as well, but what is consistent is that virtually all NE prostate cancer (NEPC) subtypes proceed first through a castration-resistant prostate cancer (CRPC) intermediate, with CRPC having brief-to-no objective response to ADT.11 Moreover, while de novo NEPC remains quite rare (~1% of all diagnosed primaries), it is thought to have a similar, recalcitrant clinical course to transformed NEPC.12,13 However, the acquisition of ADT resistance and subsequent development of CRPC do not necessitate transdifferentiation or histological transformation to NEPC.14 Critically, it is important to re-emphasize that most targeted therapy recurrences in prostate or lung cancer are not due to NE transformation.15
Re-biopsy at clinical recurrence is the most reliable method to assess histological transformation, but the decision to do so varies by institution and patient presentation. Such variability may be partially explained by practicality (i.e., metastatic vs local recurrence) and institutional experience with re-biopsy protocols for any patient with lung cancer (receiving targeted therapy or not). While infrequent, re-biopsy-associated complications remain challenges — specifically, pneumothorax.16,17,18 True matched pairs of pre- and post-histological transformation tumor tissues are still relatively rare. When paired biopsies have been characterized, the protein expression of the adenocarcinoma oncogenic driver in the transformed NE component is typically lost, but some expression is retained in some cases.19,20 Of note, similar variations in AR protein production in de novo NEPC and transformed NEPC have been reported.21 While most AR-driven prostate cancers result from amplifications and upregulation of protein expression, the genetic diversity of EGFR-driven LUADs includes single and complex point mutations, deletions, fusions, and amplifications.22,23,24 A recent study demonstrated that putative cases of histological transformation were about twice as frequent in patients harboring EGFR exon 19 deletions as EGFR L858R alterations.25 Such a provocative observation may be most readily supported through parallel datasets in patients of East Asian descent, where LUADs driven by mutant EGFR are more common.25,26
As tumor-informed genomic information may help prioritize the likelihood for a tumor to undergo histological transformation, while on targeted therapy, the clinical dilemma of what to do next remains — in patients with tumors likely to transform, should targeted therapy be stopped once transformation is documented, or should it be combined with NE-directed therapy, such as platinum/etoposide doublet chemotherapy? Indeed, recent efforts to introduce SCLC-directed therapy following treatment with osimertinib in EGFR-mutant LUAD cases with a high risk of histological transformation have demonstrated that chemotherapy alone does not prevent histological transformation.27 In general, detection of NE transformation has historically relied upon available tumor tissue as gold-standard level evidence. Emerging technical advances for the detection of systemic tumor burden utilizing non-invasive approaches, such as circulating tumor DNA (ctDNA),28 methylation patterning of ctDNA28 and radio-immune conjugate-based detection of NE surface markers,29,30 continue to develop and improve sensitivity to detect a spectrum of NE-related cancer subtypes.31 This is especially relevant following the US FDA approval of the delta like ligand 3 (DLL3) bispecific T-cell engager (BiTE) tarlatamab for extensive stage SCLC following progression on platinum-based chemotherapy.32 It remains to be determined whether DLL3 tumor tissue histology staining score (i.e., H-score), protein production, or RNA expression status is correlated with patient responses to tarlatamab. Interestingly, a recent study collecting circulating tumor cells from patients treated with AR signaling inhibitors demonstrated that detection of multiple NE markers — independent of the loss of AR target gene expression — was sufficient for the detection of NEPC. Such studies encourage future non-invasive testing approaches which may perform best when considering multiple targets/analytes in diagnostic “up assays”.33,34
Though most commonly observed in the context of targeted therapy, histological transformation may occur independently of treatment. In lung cancer, ~5% of primary lung tumors present with combined histology, i.e., single tumors showing foci with different histologic subtypes, usually along with areas with intermixed histological phenotypes (Fig. 1).35,36 A number of studies confirm shared driver mutations between the histological subtypes found in combined LUAD/SCLC and lung adeno-squamous (LUAS) tumors,6,37,38 which supports the occurrence of plasticity rather than the development of two concurrent independent tumors. In this line, known LUAD driver oncogenic alterations can be found (1) in SCLC tumors, including KRAS and frequent EGFR ( ~ 5%) mutations, as well as ALK or ROS1 translocations; (2) in squamous cell lung cancers (LUSCs),39,40 where EGFR mutations can be detected in 4%–8% of LUSC patients41,42,43; and also (3) in large cell NE carcinoma (LCNEC) at lower frequencies (Fig. 2a, b). Remarkably, tumors harboring LUAD driver mutations associated with a low/never-smoking profile (EGFR mutations and fusions of ALK, RET or ROS1) are enriched in low/never-smoking patients diagnosed with LUAS, LUSC, combined histology SCLC, LCNEC or SCLC43,44 (Fig. 2c, d), even if these tumor types are strongly associated with a heavy smoking profile. These results suggest the possibility that such LUSC, LCNEC and SCLC tumors may be a result of lineage plasticity in the absence of treatment.
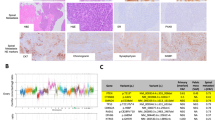
a, b Oncoprint showing LUAD driver mutations, as profiled by MSK-IMPACT next-generation sequencing, and smoking history of Memorial Sloan Kettering Cancer Center (MSK) lung adenosquamous (LUAS, n = 93) and lung squamous carcinoma (LUSC, n = 1379) cohorts (a), as well as MSK combined histology small cell lung cancer (cSCLC, n = 84), large cell neuroendocrine carcinoma (LCNEC, n = 223), and small cell lung cancer (SCLC, n = 772) cohorts (b). cSCLC tumors are defined as tumors containing both SCLC and non-small cell lung cancer (NSCLC Like SCLC tumors) histologic subtypes, where the NSCLC subtype can be LUAD, LUSC, large cell carcinoma (LCC), LCNEC or any other minor NSCLC histologic subtype. c, d Barplots showing the percentage of different patient smoking profiles for patients with LUAS or LUSC (c) or with cSCLC, LCNEC and SCLC (d). Altered groups represent patients with tumors harboring mutations in genes that are known LUAD drivers and are associated with a low smoking profile in the LUAD setting (EGFR, ALK, RET, ROS1). Unaltered groups represent patients with tumors that do not harbor mutations in the previously mentioned genes. Distribution of smoking history groups was compared using χ2 test. *P < 0.05, **P < 0.01, ***P < 0.001. Figures were generated in cBioPortal.org247,248,249 with data from the MSK-IMPACT clinical cohort.250 Histology annotations were obtained from clinical diagnosis, in the style of real word data (RWD). “n” represents the number of patients.
Like SCLC tumors, LCNECs also exhibit a particularly high frequency of KRAS mutations, another driver genomic alteration in LUAD, occurring in > 20% of LCNECs. LCNECs with such LUAD driver mutations have been classified as non-small cell lung cancer (NSCLC)-like LCNEC,45 and it cannot be ruled out that some of these may be histological transdifferentiation cases. Indeed, a minor proportion of LUADs without NE morphology present a certain degree of NE features at diagnosis and may be enriched for EGFR mutations and ALK translocations,46 and similarly, a small percentage of treatment-naive advanced prostate adenocarcinomas exhibit NE differentiation47 (Fig. 3). These observations indicate that targeted therapy may not be essential for histological transformation, and thus, that the convergence of the appropriate molecular alterations in the tumor may lead to spontaneous transformation under no treatment selection pressure.
a–d mRNA expression levels for key NE markers CHGA, INSM1, NCAM1 and SYP in different LUAD (a, b) and PRAD (c, d) cohorts, including The Cancer Genome Atlas251 (TCGA, n = 566) (a, c), OncoSG26 (n = 305) (b) and SU2C/PCF Dream Team47 (n = 444) (d). Each blue dot represents a sample. Plots represent expression values per sample, as pre-processed and normalized in the particular study of interest. Expression values are shown to illustrate interpatient heterogeneity and a subset of tumors exhibiting differentially higher expression. Figures were generated in cBioPortal.org.247,248,249
Poorly differentiated tumors might similarly be a result of plasticity. In these tumors, the features that allow histological classification may be relatively focal, thus requiring examination of several tissue blocks to establish a particular pattern of differentiation.48 Undifferentiated tumors exhibit a stem-like state49 and may represent intermediate dedifferentiated states with the potential to transition to a different histological subtype or may be anchored in a deprogrammed state. Indeed, poorly differentiated solid tumors exhibit expression of a number of molecular factors previously involved in histological transdifferentiation,6,38 including stemness-related transcription factors like SRY-box transcription factor 2 (SOX2) and MYC or activation of the PRC2 epigenetic remodeling complex.49,50,51
Emerging pathways regulating tumor cell plasticity
A tumor’s potential for lineage plasticity is influenced not only by its genomic alterations but also by the underlying developmental and epigenetic landscape of the initiating cell. This foundational state, shaped by the cell of origin, can prime tumor cells for specific responses to oncogenic stress or therapy. However, plasticity is not dictated by developmental identity alone. A growing body of evidence highlights a series of molecular pathways that, in the right molecular context, contribute to phenotypic switching under selective pressure, often overriding or reprogramming lineage constraints. In this section, key signaling and regulatory mechanisms are outlined, including TP53/RB1 co-inactivation, MYC amplification, AKT/mTOR pathway, FGFR signaling, Wnt signaling, epigenetic regulators, immune modulation, inactivation of STK11/KEAP1, and the APOBEC-mediated mutagenesis.
Co-inactivation of TP53 and RB1
TP53 and RB1 are central regulators of cellular differentiation and plasticity, especially in cancer biology, where their loss fosters tumor progression and therapeutic resistance. TP53, known as the “guardian of the genome”, promotes lineage fidelity by suppressing dedifferentiation and enhancing terminal differentiation. Loss of TP53 function disrupts these processes, increasing cellular plasticity and enabling the acquisition of undifferentiated, stem-like states that drive tumor initiation and progression. RB1, a central cell cycle regulator, enforces lineage-specific differentiation programs through interactions with transcription factors and chromatin remodeling proteins. Disruption of RB1 function permits re-entry into the cell cycle, enhancing plasticity and contributing to tumor heterogeneity. In advanced cancers, such as metastatic CRPC, RB1 loss is associated with phenotypic plasticity and aggressive clinical outcomes. Genomic analyses reveal that RB1 and TP53 deletions are enriched in metastatic CRPC, particularly in low prostate-specific antigen populations, correlating with increased visceral metastasis and shorter survival.52,53 The combined loss of TP53 and RB1 is mechanistically linked to lineage plasticity and antiandrogen resistance. In TP53- and RB1-deficient prostate cancers, tumors evade AR-targeted therapies by transitioning from AR-dependent luminal cells to AR-independent basal-like phenotypes. This phenotypic shift is driven by upregulated SOX2, a reprogramming transcription factor, and can be reversed by restoring TP53 and RB1 function or inhibiting SOX2 expression.51,52,54 Together, TP53 and RB1 maintain differentiation and suppress plasticity; their disruption creates a microenvironment conducive to tumor evolution, with implications for disease aggressiveness and resistance to therapy.55
While TP53 and RB1 loss establishes a permissive state for transformation,56,57 it is insufficient to induce complete transformation into a malignant state. Additional oncogenic alterations, such as KRAS or MYC activation, PTEN loss, or changes in the tumor microenvironment (TME), are required to complete the transition to malignant phenotypes.58,59,60,61,62 In LUAD, concurrent mutations in TP53 and RB1 facilitate histologic transformation to SCLC, but additional mutations and amplifications in PIK3CA, MYC, and NOTCH are also critical for the complete transformation process.58 Similarly, in prostate cancer, loss of TP53 and RB1 leads to lineage plasticity and JAK/STAT activation, giving rise to therapy-resistant, stem-like subclones with suppressed AR and luminal markers. Single-cell RNA sequencing has revealed distinct subpopulations of this kind, exhibiting suppressed expression of luminal lineage markers and increased expression of stem-like and epithelial-to-mesenchymal transition (EMT) genes.63,64,65 Notably, the process of transformation is not typically a clonal sweep but involves multiple subclones with distinct genetic alterations. This intratumoral heterogeneity allows for the selection of subclones that can adapt to therapeutic pressures and contribute to NE transformation.6
Several recent studies on JAK/STAT inflammatory signaling highlight the role of TP53 and RB1 loss in lineage plasticity, showing that organoid models with these deletions enter a smoldering EMT-like state but fail to achieve full NE transition.63,65,66,67 This incomplete transformation suggests that additional microenvironment signals, such as JAK/STAT and FGFR activation, are required. In vivo, genetically-engineered mouse models (GEMMs) reveal further insights; while PtR (PTEN/RB1 loss) models exhibit limited plasticity, PtRP (PTEN/RB1/TP53 loss) models show marked NE phenotype, demonstrating how TP53 loss synergizes with RB1/PTEN deletions, emphasizing the interaction between genetic alterations and systemic factors. Another recent study68 highlights the critical role of in vivo conditions in driving full NEPC transformation using the PARCB (TP53 and RB1 abrogation, AKT overactivation, and MYC and BCL2 overexpression) genetic model. The study shows that primary basal prostate epithelial cells, modified with key oncogenic drivers, underwent partial reprogramming when cultured in an organoid system. Transformation to fully differentiated NEPC required in vivo engraftment to immunodeficient mice, where transplanted cells formed tumors with squamous cell prostate cancer histological features, including high nuclear-to-cytoplasmic ratios, frequent mitoses, and uniform NE differentiation marker expression. This underscores the necessity of non-cell-autonomous signals such as TME signals in driving full transformation. Interestingly, despite successful NEPC transformation in immunodeficient NSG-engrafted xenografts, limited metastasis was observed, suggesting the absence of essential immune or stromal cues critical for tumor dissemination. Collectively, these studies indicate that TP53/RB1 loss is a critical but insufficient driver of NE transformation. Full lineage conversion requires cooperative genetic alterations and signals from the TME to reprogram prostate adenocarcinoma into NEPC.
The role of MYC in plasticity
The amplification of MYC in relapsed or recurrent SCLC is driven by its critical role in tumor plasticity, heterogeneity, and resistance. Studies reveal that MYC is frequently amplified on extrachromosomal DNA (ecDNA), a structure enabling exceptionally high transcriptional activity and rapid genomic heterogeneity through random mitotic segregation. This amplification is often acquired under therapeutic pressure, as seen in patient-derived xenografts (PDXs) of treatment-resistant disease, where MYC drives cross-resistance to DNA-damaging therapies.69,70 The challenge, therefore, may lie in targeting these adaptive mechanisms associated with plasticity in MYC-overexpressing, treatment-resistant cells. The dual involvement of MYC in NE transformation and squamous transdifferentiation38 adds further complexity. In SCLC, MYC can drive NE phenotypes, but under certain contexts, it might also promote squamous characteristics, potentially influencing tumor heterogeneity. In light of these observations, assessment of MYC expression by immunohistochemistry might help identify patients at risk for transformation and relapse.38
AKT/mTOR pathway
Evidence suggests that, in the right molecular context, activation of the PI3K/AKT pathway may act as a driver of lineage plasticity. In the lung setting, AKT/mTOR activation has been associated to transdifferentiation of LUAD to acquire LUSC features. This process is not solely driven by genomic alterations but also involves transcriptional reprogramming and epigenetic changes. Combined overactivation of AKT and MYC in LUAD models synergistically induces squamous markers such as P40, with further augmentation under the selective pressure of EGFR inhibition.38 Similarly, in prostate cancer, the AKT/mTOR pathway has been associated to lineage plasticity, facilitating the transition from AR-dependent adenocarcinoma to AR-independent states like NEPC.68 This transition often follows ADT and is closely linked to resistance mechanisms. The activation of this pathway supports cell survival, interacts with AR signaling, and modulates epigenetic regulators like EZH2 to enable phenotypic shifts.71,72 In both cancer types, the AKT/mTOR pathway functions as a pro-survival signaling mechanism that enhances cell proliferation, supports cellular transitions, and underpins therapy resistance and disease progression.
Epigenetic factors
Epigenetic reprogramming underpins lineage plasticity and therapy resistance in prostate and lung cancers by facilitating transitions to alternative, treatment-resistant phenotypes. This plasticity is driven by dynamic changes in chromatin structure and transcriptional networks, enabling tumor cells to evade therapeutic pressures and adopt new lineages. Key drivers include EZH2, a histone methyltransferase that silences lineage-specific genes through H3K27me3 marks. EZH2 plays a pivotal role in supporting NE differentiation and EMT, both of which contribute to tumor progression and resistance mechanisms.54,73,74,75,76 A recent clinical study showed that combining mevrometostat, an EZH2 inhibitor, with enzalutamide improved progression-free survival and response rates in metastatic CRPC compared to enzalutamide alone.77 FOXA1, a pioneer transcription factor, facilitates chromatin remodeling and regulates AR-dependent transcription. In prostate cancer, FOXA1 is frequently mutated. Studies annotating the landscape of FOXA1 mutations from 3086 human prostate cancers, defined two hotspots in the forkhead domain Wing2 and the highly conserved DNA-contact residue R219. Wing2 mutations are detected in adenocarcinomas at all stages, whereas R219 mutations are enriched in metastatic tumors with NE histology. These mutations in FOXA1 alter its chromatin binding activity, shifting transcriptional programs toward NE states, particularly under ADT.78,79 Beyond genetic alterations, FOXA1 can undergo cistromic reprogramming in NEPC, even in the absence of mutation. In this context, FOXA1 cooperates with transcription factors such as achaete-scute family bHLH transcription factor 1 (ASCL1) and NKX2-1 to activate NE-specific enhancers, supporting an NE phenotype.79 In parallel, FOXA2 promotes NE differentiation by regulating chromatin accessibility and activating pathways such as the KIT pathway.79 FOXA2 facilitates transcriptional reprogramming by cooperating with JUN/AP-1 complexes at lineage-specific enhancers, enabling chromatin remodeling and plasticity. The interaction between FOXA2 and the epigenetic enzyme LSD1 further supports its chromatin binding activity, and inhibition of LSD1 disrupts FOXA2-driven transcriptional programs, thereby reducing lineage plasticity.80
In prostate cancer, the loss of CHD1, a chromatin remodeler, disrupts chromatin integrity, leading to increased transcriptional plasticity and resistance to AR-targeted therapies.81 DNA methylation is another critical epigenetic mechanism driving lineage plasticity. This process is regulated by DNA methyltransferases (DNMT1, DNMT3A, DNMT3B) and TET proteins (TET1, TET2, TET3), which modify the methylation landscape to enable tumor adaptability. A recent study demonstrated that ZNF397 loss triggers a TET2-driven transformation of the 5-hydroxymethylcytosine landscape into a plastic state characterized by AR independence and multilineage potential, further contributing to therapy resistance.82 In lung cancer, alterations in the SWI/SNF complex, including mutations in SMARCA4, destabilize chromatin and facilitate lineage switching, including adenocarcinoma-to-squamous or NE transformation.83 These epigenetic regulators mediate the chromatin remodeling and transcriptional reprogramming necessary for transitions such as adenocarcinoma-to-squamous differentiation and NE transformation. Under therapeutic pressures, tumor cells exploit these transitions to adopt alternative lineages that evade treatment. Together, these findings underscore the central role of epigenetic regulators in shaping lineage plasticity and therapy resistance in prostate and lung cancers. Targeting these mechanisms could provide novel therapeutic opportunities to prevent lineage transitions and improve treatment outcomes.
FGFR signaling
FGFR signaling plays a pivotal role in regulating lineage plasticity and tumor development by influencing cellular trafficking and oncogenic activation. The LIS1/NDE1 complex is crucial for FGFR intracellular trafficking, stability, and recycling, ensuring proper signal strength and localization.84 Dysregulation of FGFR signaling can alter lineage-specific programs and drive phenotypic shifts, with significant implications for cancer lineage plasticity. For instance, in SCLC, FGFR1 oncogenic activation reveals an alternative cell of origin and influences tumor phenotype, promoting low-grade NE lesions in specific contexts while impairing the development of typical SCLC.85 These findings highlight the nuanced roles of FGFR signaling in determining tumor lineage and phenotype. In prostate cancer, Chan et al.65 underscores the importance of FGFR and JAK/STAT signaling as early and essential mediators of plasticity. These pathways drive basal-luminal mixing, EMT, and eventual NE transformation. In Trp53/Rb1-deficient organoids and GEMMs, both pathways are activated and further enhanced under therapeutic pressures, such as androgen deprivation or AR inhibition. Functional experiments have revealed that FGFR activation promotes plasticity-associated phenotypes, including changes in cellular identity and morphology, while JAK/STAT signaling amplifies transcriptional programs critical for the transition to treatment-resistant states. Collectively, these findings position FGFR signaling as a key regulator of cancer lineage plasticity and a potential therapeutic target. Understanding the interplay of FGFR signaling with pathways like JAK/STAT may provide new insights into the mechanisms driving tumor progression and resistance, particularly in cancers characterized by high plasticity and phenotypic heterogeneity.
Immune dysregulation
Lineage plasticity during NE and LUSC transformation actively suppresses the immune response, allowing tumors to evade immune surveillance and thrive in hostile microenvironments. These transformations are characterized by profound transcriptional and epigenetic reprogramming as well as extrinsic microenvironmental changes. A key feature of this transformation is the downregulation of major histocompatibility complex class I (MHC-1) molecules, which impairs antigen presentation and reduces visibility to cytotoxic T lymphocytes. This phenomenon has been observed in NEPC and squamous-transformed leukemia. In these models, promoter hypermethylation was responsible for MHC-1 loss, creating an immunologically “cold” tumor state.6,38 Recent studies also highlight hypoxia as a critical driver of immune dysregulation and lineage plasticity in prostate cancer. Hypoxia stabilizes HIF1a, which cooperates with transcriptional regulators such as ONECUT2 and SMAD3 to promote NE differentiation and downregulation of MHC-1 molecules. By linking transcriptional reprogramming with immune escape, hypoxia-driven signaling creates a permissive environment for NE transformation and therapy resistance.6,38,86 As described above, the ectopic activation of JAK/STAT signaling in tumor epithelial cells,63,65,66,67 a pathway commonly associated with immune and inflammatory responses, plays a pivotal role in these processes. The chronic activation of JAK/STAT not only disrupts normal immune surveillance but also drives the production of inflammatory mediators that reshape the TME. This inflammatory signaling promotes an immunosuppressive milieu that inhibits effective immune cell infiltration and activation. Furthermore, the interplay between JAK/STAT signaling and epigenetic alterations reinforces these immunosuppressive changes, fostering a microenvironment that promotes therapy resistance and tumor progression. These multifaceted changes result in a highly immunosuppressive TME that facilitates tumor growth, enables evasion of immune-mediated destruction, and may contribute to plasticity and resistance to therapies targeting the AR and other key pathways in prostate cancer.
STK11/KEAP1 inactivation
Genomic alterations of the serine/threonine kinase STK11 (also known as LKB1) and of the adaptor subunit of Cullin-based E3 ubiquitin ligase KEAP1, which are often co-occurring, have been detected in different tumor types and associated with tumorigenesis and poor prognosis.87,88,89,90 KEAP1 acts as a redox sensor that gets inactivated under stress, leading to the upregulation of NRF2, a transcription factor triggering transcriptomic reprogramming in response to stress.91 Interestingly, KEAP1 inactivation has been extensively involved in stemness in different settings, including hematopoiesis92 and lung, head and neck, and breast tumors, among others.40,92,93
STK11 is physiologically implicated in energy homeostasis as an upstream kinase of the AMP-activated protein kinase (AMPK) pathway. This tumor suppressor is frequently mutated in human cancers such as lung, cervical, ovarian, skin, pancreas, kidney, and gastrointestinal tumors.94,95 STK11 inactivation has been associated with loss of differentiation and increased stem-like features in lung, gallbladder, and glioblastoma tumors.96,97,98 In the lung cancer setting, where the gene has been most extensively studied, the dedifferentiation induced by STK11 inactivation is mediated by upregulation of stemness factors such as Nanog, inhibition or downregulation of Notch signaling,98 and downregulation of salt-inducible kinase (SIK) family members and the AT2 lineage-defining factor C/EBPα,99 as reported in different Kras-driven LUAD GEMMs. Indeed, LKB1 has been shown to play a role in histological heterogeneity in a Kras-driven LUAD GEMM, where Lkb1 inactivation led to the formation of not only LUADs, but also LUSC and large cell carcinomas.100 Lkb1 inactivation induced chromatin alterations, including the loss of H3K27me3 and gain of H3K27ac and H3K4me3 particularly at squamous lineage genes, including Sox2, ΔNp63, and Ngfr.101 Importantly, even if LKB1 mutations are common in human LUSC ( ~19%), they also occur frequently in LUADs ( ~34%),100 suggesting that LKB1 inactivation may be an enabler of plasticity and histological transformation, rather than a driver in this process. In line with these results, different cells of origin may display different efficiencies of squamous differentiation, where bronchioalveolar stem cells and club cells may be the most prone to transformation to a squamous phenotype.102 Of note, the adeno-to-squamous transition has been thoroughly characterized in Kras/Lkb1-mutant GEMMs, being advanced most significantly by Hongbin Ji’s group.100,103,104,105
Remarkably, inactivating mutations of both STK11 and KEAP1 are common in LCNEC, where they frequently co-occur alone or in combination with KRAS mutations (Fig. 4a, b). LCNECs are considered intermediate entities between NSCLC and SCLC, harboring frequent LUAD-associated genomic alterations, but exhibiting NE features.45,106 Interestingly, recent molecular characterizations of these rare tumors converge into a classification driven by genomic events, which distinguishes SCLC-like LCNEC, enriched for RB1 mutations, and NSCLC-like LCNEC, enriched for STK11 and KEAP1 mutations. The fact that mutations in RB1 and STK11 are mutually exclusive in these tumors (Fig. 4c), and the role of both in maintaining LUAD differentiation, further supports that both tumor suppressors might be master regulators of plasticity, and that LCNECs, or a subset of them, may potentially be cases of histological transformation.
a Frequency of driver mutations in STK11 in the different lung cancer histologic subtypes, large cell neuroendocrine carcinoma (LCNEC), lung adenocarcinoma (LUAD), lung adenosquamous (LUAS), combined small cell lung cancer (cSCLC), squamous cell lung carcinoma (LUSC) and SCLC. b Oncoprint showing mutations in genes of interest in the LCNEC Memorial Sloan Kettering Cancer Center (MSK) cohort. c Co-occurrence or mutual exclusion of mutations in the LCNEC MSK cohort, quantifying how strongly the presence or absence of alterations in A are associated with the presence or absence of alterations in B in the selected samples. d Oncoprint showing mutations in genes related to the Notch signaling pathway in the lung cancer MSK cohort, divided by histological subtypes. e Frequency of genomic alterations in genes related to the Notch pathway (see d) in the different lung cancer histological subtypes from the MSK cohort. Odds Ratio = (Neither × Both)/(A Not B × B Not A). p-values are derived from two-sided Fisher Exact Test and q-values were derived from a Benjamini-Hochberg false discovery rate correction procedure. Log2ratio > 0: tendency towards co-occurrence; log2ratio ≤ 0: tendency towards mutual exclusivity. q-value < 0.05: significant association. Figures were generated in cBioPortal.org247,248,249 with data from the MSK-IMPACT clinical cohort.250 Histology annotations were obtained from clinical diagnosis, in the style of RWD. MSK LUAD (n = 9789), LUAS (n = 93), LUSC (n = 1379), cSCLC (n = 84), LCNEC (n = 223), and SCLC (n = 772) cohorts are depicted. “n” represents the number of patients.
Wnt pathway
The Wnt/β-catenin pathway plays critical roles in embryonic development and adult tissue homeostasis107 and has been reported to have pro-oncogenic roles in different tumor types, including gastrointestinal carcinomas, leukemia, melanoma, and breast cancers.108 One of the most studied roles of this signaling pathway includes the maintenance of stemness through a variety of mechanisms, including TERT overexpression109 and induction of dedifferentiation.110 Multi-omic analyses of lung tumor clinical specimens undergoing histological transformation revealed upregulation of genes involved in the Wnt signaling pathway, as well as upregulation of β-catenin at the protein level, occurring convergently in both LUSC and NE transformations.6,38 A recent study integrating ATAC-sequencing and RNA-sequencing data from patient-derived organoids (PDOs), PDXs, and cell lines identified four subtypes of CRPC and predicted key transcription factors of each. Among these, the so-called “CRPC-WNT” subtype was characterized as AR-negative/low, Wnt-dependent, and driven by TCF/LEF transcription factors. Even if the “CRPC-WNT” and the NE-like “CRPC-NE” subtypes represented distinct molecular subsets,111 components of the Wnt signaling pathway have been implicated in promoting NE-like features and lineage plasticity in the prostate. AR inhibition leads to Wnt signaling upregulation through the downregulation of Wnt ligand secretion mediator (WLS), a transcriptional regulator and major driver of the NE phenotype.112 In line with this, other effectors of the Wnt pathway, such as TCF4, FOXB2-Wnt7B and Wnt11, have been reported to facilitate NE transformation in the prostate, by inducing a transcriptional program favoring neuronal differentiation.113,114,115
Notch pathway
The notch pathway controls cell fate decisions critical for proper organ development during embryogenesis and homeostasis in adult organisms. This pathway relies on cell surface receptors and ligands for activation that leads to receptor cleavage. The cleaved receptor then acts as a transcriptional regulator and controls differentiation.116 Notch signaling may have both oncogenic and tumor suppressor effects depending on the molecular context and has been reported to be incompatible with the NE phenotype. Specifically, Notch signaling is a strong suppressor of NE differentiation via upregulation of the transcription factors HES1 and REST, which suppress expression of the master NE regulator ASCL1 and other NE genes in prostate cancer and pancreatic carcinoma.116 In line with these results, Notch signaling downregulation was observed at early stages of NE transformation in the lung, specifically in pre-transformation LUAD, as compared to control, never-transformed LUAD clinical specimens.6 Similarly, in the prostate setting, it has been reported that hypoxia-driven Notch signaling inactivation is able to induce NE differentiation.117 Consistently, genomic alterations in Notch family genes are common in NE tumors (Fig. 4d, e).
Remarkably, even if Notch signaling is incompatible with the NE phenotype, non-NE SCLC subtypes exhibit activation of Notch signaling,118 whose induction has been described in the transition of NE to non-NE SCLC observed during disease progression or development of chemotherapy resistance.119,120 These observations suggest that Notch signaling may have a role in SCLC biology after NE transformation. Indeed, different works have described intratumoral heterogeneity within SCLC tumors, containing both NE and non-NE populations. In these, expression of Notch ligands in the NE compartment activates Notch receptors in the non-NE compartment, and the interaction of both populations may represent a selective advantage in terms of disease progression and metastasis.121,122 However, a recent study suggests that conditional expression of a single inhibitory Notch ligand during SCLC development does not necessarily anchor SCLC to a pure NE state, where the authors observed similar heterogeneity of NE and non-NE tumor cell types.123 Such heterogeneity is reminiscent of the occurrence of persistent, even if minor, adenocarcinoma components in some transformed NE tumors,6 where similarly NE and non-NE cells may potentially be cooperating with pro-oncogenic consequences.
Although Notch signaling appears to be at odds with NE tumors, RNA sequencing analyses on clinical specimens undergoing adenocarcinoma-to-squamous transformation indicate a transcriptomic profile compatible with Notch signaling activation.38 Certainly, Notch signaling has been associated with poor prognosis and stemness in NSCLC.124 Nonetheless, Notch signaling activation may have differential effects in different molecular or histological contexts, with oncogenic effects in LUAD but potential tumor suppressive effects in LUSC as well as in other squamous tumors.124 Interestingly, we observed increased frequency of mutations in members of the Notch signaling pathways in LUAS tumors relative to LUAD, and further enrichment of mutations in these genes in LUSC tumors (Fig. 4d, e). Remarkably, mutations in genes related to Notch signaling are very common ( ~70%) in other squamous tumors, such as head and neck cancer,125 further associating the dysregulation of this pathway with squamous differentiation. The role of Notch signaling in histological transformation remains undefined and in need of additional study.
APOBEC-mediated hypermutation
In addition to single genetic alterations, large-scale genomic changes and mutational processes, such as APOBEC-mediated hypermutation and whole-genome doubling, are critical drivers of lineage plasticity and therapy resistance. APOBEC enzymes, a family of cytidine deaminases, introduce mutations that can lead to widespread genomic instability, contributing to tumor heterogeneity and adaptive evolution. Enrichment of APOBEC hypermutation signatures has been observed in cohorts of patients with SCLC tumors undergoing phenotypic transformation, highlighting its role in facilitating tumor plasticity under therapeutic pressure.19 Recent findings have elucidated the mechanisms underlying APOBEC-driven mutagenesis, with the loss of SYNCRIP, an RNA-binding protein that inhibits APOBEC3B activity, identified as a key contributor.126 The loss of SYNCRIP unleashes unchecked APOBEC3B activity, leading to extensive mutational processes that target critical regulatory genes. Among these are FOXA1, EP300, and STAT3 — genes central to the maintenance of transcriptional networks. Mutations in these genes disrupt normal regulatory functions, enabling transcriptional reprogramming that fosters lineage transitions and confers resistance to therapy.126 Moreover, APOBEC-mediated mutagenesis is thought to generate genetic diversity that equips tumors to adapt to changing selective pressures, including those imposed by therapeutic interventions. This mutational process can amplify genomic alterations that promote epigenetic reprogramming and phenotypic plasticity, further reinforcing a feedback loop that drives therapy resistance and tumor progression. Understanding the interplay between APOBEC activity and lineage plasticity may reveal novel therapeutic vulnerabilities to overcome resistance mechanisms.
Influence of cell of origin on lineage plasticity
Evidence suggests that the cell of origin plays a significant role in lineage plasticity during cancer transformation by introducing a bias toward specific histological fates. However, this influence is not absolute, as molecular and environmental factors often dominate the determination of the final histological state, allowing flexibility in lineage transitions.127 This interplay between intrinsic lineage programming and extrinsic molecular cues underscores the complexity of tumor plasticity. In prostate cancer, tumors predominantly display a luminal phenotype. Previously, basal cells were identified as the cell of origin for prostate cancer,128,129 but increasing evidence implicates luminal cells, particularly luminal progenitor cells (LPs), as a preferred cell of origin. LPs with stem-like properties appear critical for both tumor initiation and progression.130 A lineage tracing study has identified a rare population of castration-resistant Nkx3-1-expressing luminal cells that function as bipotent stem cells during regeneration and serve as efficient targets for oncogenic transformation, rapidly forming carcinoma following Pten deletion.131 Functional studies using human prostate epithelial cells transplanted into immunodeficient mice, as well as organoid-based transformation assays further support this paradigm. In these models, basal and luminal cells isolated from primary benign human prostate tissue were transduced with defined oncogenes (e.g., MYC, AKT, ERG), and then either xenografted into NSG mice or cultured in 3D organoid systems. These studies revealed that while basal cells can also initiate prostate cancer when transformed, they often give rise to tumors with more poorly differentiated phenotypes, whereas luminal-derived tumors retain glandular morphology and strong AR activity.128,129 This indicates that the epithelial cell of origin shapes not only tumorigenic potential but also differentiation state. Additionally, the transcriptional and epigenetic landscape of LPs renders them particularly susceptible to lineage plasticity and NE reprogramming under therapeutic pressures such as ADT. Studies investigating the cell of origin for prostate cancer using tissue regeneration models and GEMMs have yielded different results, highlighting the influence of model context. These differences indicate the critical role of microenvironmental components in shaping the susceptibility of specific epithelial lineages to transformation.132 These findings underscore that both intrinsic lineage identity and the microenvironmental context of transformation critically influence plasticity trajectories and therapeutic resistance in prostate cancer.
In lung cancer, similar principles apply, with different progenitor cells showing varying propensities for transformation into specific histological subtypes.119,133 For example, AT2 cells frequently give rise to adenocarcinomas, while basal cells are more likely to transform into squamous cell carcinoma. Additionally, club cells have been implicated in contributing to SCLC. These cell-specific biases reflect intrinsic lineage determinants that shape tumor histology in response to oncogenic or therapeutic pressures. Despite these similarities, notable differences exist between the behavior of NE cells in the lung and prostate. In the lung, NE cells differentiate at specific anatomical sites, such as branch points, and exhibit a unique migratory behavior known as “slithering”, which enables them to form neuroepithelial bodies.134 In contrast, NEPC cells emerge more broadly under stress conditions, such as ADT, and arise from luminal progenitors without requiring notable migration. This difference in NE cell behavior reflects tissue-specific adaptations in response to environmental and molecular cues. These observations suggest that while the cell of origin establishes a foundation for lineage plasticity, the eventual histological outcome is shaped by a complex interplay between intrinsic lineage programming and external molecular influences. Understanding these mechanisms is crucial for identifying vulnerabilities that can be targeted to limit lineage plasticity and overcome therapy resistance.
Preclinical models to interrogate histological transformation and lineage plasticity
Several important questions surrounding mechanisms and timelines in histologic transformation remain unanswered but can be directly addressed using currently available models. First, it is unclear whether inhibition of oncogenic drivers in tissue types other than the lung or prostate will display similar frequencies of histologic dedifferentiation as mechanisms of acquired resistance to said targeted therapies. For example, it is clear that loss of Rb1 and Trp53 alone is insufficient to permit the transition or adoption of an NE fate in an LUAD tumor when the oncogenic driver, such as Kras(G12D), is still active. While not the primary intention of the study, recent work utilizing an Rb1 allele that could be inactivated and then restored had shown that triple mutant Kras, Trp53, Rb1 (KPR) mice develop aggressive, metastatic LUAD with features of dedifferentiation, which is often not observed in the KP LUAD model until much later in tumor development.135 However, the histologic characterization of these lesions was neither NE nor consistent with SCLC. Whether Kras(G12D) or pan-Ras inhibition encourages NE transformation remains open questions. Recent studies on mechanisms of Kras inhibitor resistance in LUAD have suggested that lineage plasticity towards a more squamous fate (if Lkb1 is deactivated) or perhaps even dedifferentiation towards an alternative alveolar state (i.e., AT1-like) are more common mechanisms of acquired resistance than NE differentiation.104,136,137 In line with the models mentioned above, a role for Rb1 genetic restoration in the maintenance of an NE state would be directly addressable. Similar tools have been instrumental in deciphering a role for Trp53 and Lkb1 inactivation during multiple stages of tumorigenesis and in the maintenance of a transformed state.99,138 Further, whether expanded therapeutic application of mutant-selective and/or pan-Ras inhibitors in pancreatic, colorectal/gastrointestinal, and lung cancers result in NE transformation remains to be seen.139,140,141
To the best of our knowledge, most studies describing model systems to study histologic transformation have failed to observe fully differentiated NE transformation outside of a host animal or patient, and the reason for this remains an open question. For example, isolation of reporter-positive prostate cancer cells from the PtRP mouse model of luminally-derived prostate cancer display progressive downregulation of the AR and eventual dedifferentiation to NEPC but fail to fully commit to an Ascl1+ NE state ex vivo.65 This appears to be similarly true in other systems utilizing engineered human cells, requiring in vivo engraftment into well-perfused sites (i.e., renal capsule) or sub-cutaneous engraftment.68 Indeed, in our own experience, working with organoid-derived cultures from a GEMM that combines the conditional expression of Myc, rtTA3, and tdTomato with the loss of Rb1 and Trp53, along with a doxycycline (Dox)-inducible, oncogenic EGFR transgene (ERPMT model), we observed that LUAD histological transformation to SCLC failed to adopt an NE fate following de-induction of the EGFR oncogenic driver.142
GEMMs offer a critical platform for investigating microenvironmental influences on lineage plasticity. Complementary preclinical models, such as PDOs, PDXs, and co-culture systems, enable investigation of TME interactions in human contexts. Notably, a panel of 22 prostate cancer PDOs generated at MSK, along with four NEPC PDOs (WCM154, WCM155, WCM1262, WCM1078) and their corresponding xenografts, constitute a functionally annotated, lineage-diverse model system,143,144 that serve as an effective platform for dissecting the transcriptional and epigenetic mechanisms underlying lineage plasticity. Co-culture systems using these organoids with immune components can be a useful model system for the study of tumor–immune interactions and the microenvironment. A longitudinal study using the Pten(i)pe–/– mice, which carry a prostate-specific deletion of Pten, and Pten/Hif1a(i)pe−/− mice, with combined Pten and Hif1a deletion, demonstrated that HIF1a is essential for initiating the transcriptional programs associated with lineage plasticity, immune evasion, and NE differentiation in the context of Pten loss.86
Meanwhile, PPR (Pten, TP53, and RB1 loss) and PRP (Pten, RB1, and TP53 loss) models have been used to underscore the pivotal role of combined genetic alterations in driving lineage plasticity. Loss of RB1 and TP53, along with Pten deletion, facilitates the transition of prostate cancer cells from AR-dependent luminal states to basal-like or NE phenotypes. One of the critical mediators of this plasticity is Sox2, a transcription factor known for its role in embryonic stem cell maintenance. Sox2 reactivates embryonic transcriptional programs that enable dedifferentiation, lineage switching, and adaptive tumor evolution. This reprogramming involves the suppression of luminal differentiation markers, thereby facilitating the loss of AR dependency and promoting a basal-like or NE phenotype.51,54 Targeting key regulators, such as Sox2, or the pathways activated by the combined loss of RB1, TP53 and Pten, may offer new therapeutic strategies to overcome resistance and prevent tumor evolution in advanced prostate cancer.
NE transformation exhibits distinct subtype mechanisms in NEPC and SCLC, each driven by unique combinations of genetic and epigenetic alterations. In NEPC, MYCN overexpression and RB1 loss can occur independently or co-occur in patients and have been identified as key drivers of NEPC. To model this, Pb-Cre4+/–;Ptenf/f;Rb1f/f;LSL-MYCN+/+ (PRN) mice were generated, leading to MYCN overexpression in prostate epithelium alongside Pten and Rb1 deletion. Analysis of these models revealed that MYCN and RB1 cooperate to foster the NE differentiation into distinct lineages, including ASCL1+ and POU2F3+ subtypes. The ASCL1+ subtype follows canonical neuronal differentiation pathways, marked by the expression of neuronal genes associated with synaptic functions. In contrast, the POU2F3+ subtype represents a rare tuft cell-like lineage, characterized by MYCN-mediated chromatin reprogramming and a mutual exclusivity with ASCL1 expression. These divergent subtypes highlight the role of MYCN as a master regulator of chromatin dynamics, enabling NEPC cells to adopt diverse NE identities.145 In SCLC, NE transformation mechanisms are similarly complex, with tumors often displaying varying expression levels of lineage-defining markers such as ASCL1, NEUROD1, and POU2F3.146 A distinct subtype with YAP1 expression has been identified, demonstrating low levels of canonical NE markers. Using a series of lineage tracing GEMMs, including CgrpCreER;Trp53f/f;Rb1f/f;Ptenf/f (TKO (triple knockout), CgrpCreER;TKO;Rosa26mTmG, and inducible YAP overexpression models (CgrpCreER;TKO;TetOn-YAPS127A and CgrpCreER;TKO;TetOn-YAP5SA), in combination with Rbpjf/f to modulate Notch signaling, these studies demonstrated that YAP activation promotes a dynamic transition from NE to non-NE states. Mechanistically, YAP interacts with transcriptional regulators such as REST and Notch2, leading to the suppression of neuronal genes like ASCL1 and the induction of non-NE markers, including HES1 and CC10. This NE-to-non-NE plasticity highlights YAP’s role as a key modulator of lineage state transitions, enabling tumor cells to adapt to therapeutic and microenvironmental pressures.147
NE transformation is not solely driven by single factors such as ASCL1 expression or RB1 loss but rather involves a complex interplay of genetic, transcriptional, and epigenetic regulators. For instance, while ASCL1 expression and RB1 loss are necessary, they are insufficient to fully convert AR-positive or CRPC cells into NE phenotypes. Research by Cejas and colleagues identified SRRM4 as a novel and critical driver of NE transformation in prostate cancer. SRRM4 reprograms alternative splicing to activate NE-specific gene expression by modifying the function of REST, a transcriptional repressor of neuronal genes. SRRM4 induces the splicing of REST into a truncated isoform, REST4, which lacks functional repressor domains. This splicing event derepresses neuronal and NE-specific genes, including ASCL1, INSM1 and SYP, thereby facilitating the transition to an NE phenotype.148 This discovery underscores the importance of alternative splicing as a regulatory mechanism in NE plasticity.
GEMMs of SCLC have been instrumental in replicating the core genetic alterations observed in human tumors, such as TP53 and RB1 loss and MYC amplification. These models have provided crucial insights into subtype heterogeneity, tumor progression, and therapy resistance. Additionally, advances in CRISPR/Cas9-based somatic editing and immunocompetent models have enabled the study of tumor–immune interactions, offering new opportunities to dissect therapeutic vulnerabilities in NE cancers. These state-of-the-art models allow for the exploration of how NE plasticity influences immune evasion, microenvironmental adaptation, and lineage-specific dependencies.149
Conceptual models of plasticity leading to histological transformation
Recent molecular analyses in clinical specimens and preclinical models have identified dysregulated pathways during NE and squamous transformation.6,38 Surprisingly, several oncogenic pathways were commonly upregulated in both NE- and squamous-transforming clinical specimens, including pathways related to cell cycle and DNA repair, the PRC2 epigenetic remodeling complex, AKT and Wnt signaling, as well as MYC targets, the latter induced by MYC or MYCN.6,38,68,145 Similarly, several pathways related to the anti-tumor immune response were downregulated in both NE and squamous transformation, suggestive of a strong anti-tumor immune suppression occurring during histological transformation.6,38 Also, as previously mentioned, STK11 inactivation has been reported as a promoter of the squamous and large cell carcinoma phenotypes in GEMMs,100 and in potential relationship with the latter, STK11 inactivation is also hallmark of a subset of LCNECs enriched for mutations of LUAD drivers such as KRAS,45,106 which could potentially be cases of NE transformation. These observations associate STK11 inactivation with plasticity and suggest a role for this molecular event as an enabler of plasticity, which could potentially lead to different histological outcomes. Additionally, there is evidence of oncogenic pathways exclusively dysregulated in LUSC transformation, like Hedgehog signaling, and even pathways showing opposite dysregulation depending on the histological fate of transformation, such as Notch signaling, suppressed and induced in NE116 and LUSC38 transformation, respectively. The apparent convergence and divergence of molecular events in NE and squamous transformation, as above explained, suggest a model of transformation where these convergent molecular alterations may lead to a potential intermediate dedifferentiated state, common for histological transformation independently of the histological outcome. Indeed, this observation is supported by the fact that many of these have been extensively associated to stemness.110,150,151,152 After this intermediate state has been reached, the occurrence of additional molecular events, potentially those found to occur divergently in LUSC and NE transformation, may ultimately decide the histological outcome (Fig. 5, top).
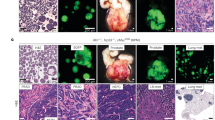
Models of transformation during selective pressure-driven tumor evolution (arrows), such as targeted anti-tumor therapy. Two models are depicted, histological transformation through a dedifferentiated intermediate stem-like state (top) or through a squamous-like intermediate state (bottom). Cell type legend can be found at the bottom of the figure. Question marks account for yet undescribed additional molecular alterations potentially driving transition among different cell states. Figure generated with BioRender.com.
On the other hand, there are data supporting an alternative model where the squamous/basal-like histology may represent the intermediate state of adenocarcinoma-to-NE transition. Even if most combined histology tumors show only two different histological components, the occurrence of tumors exhibiting multiple histologic subtypes during transformation has been documented. Examples of this include baseline prostate adenocarcinomas transdifferentiating into a combined squamous/NE/sarcomatoid tumor,153 or lung tumors with adenocarcinoma, squamous, and NE histological components.154 One potential explanation for these could be the divergent evolution of different adenocarcinoma clones, each transdifferentiating into a different histological subtype, but that seems unlikely. Analogous to what was observed in clinical specimens, preclinical evidence in GEMMs of histological transdifferentiation indicate the co-existence of different histologies. In the NPp53 model, with combined inactivation of Trp53 and Pten in the prostate epithelium, treatment with the androgen blocker abiraterone induced histological transdifferentiation leading to tumors with areas of squamous/basal, sarcomatoid, small cell-like NE, and other non-adenocarcinoma phenotypes.155 In two other GEMMs, one with inactivation of Rb1 and Pten, simultaneously with MYCN transgene overexpression,145 and the other with prostate-specific inactivation of three tumor suppressors Trp53, Rb1 and Pten,65 we observe tumors exhibiting combined histology with AR-positive adenocarcinoma, undifferentiated areas with intermediate adenocarcinoma/NE phenotype, and foci exhibiting squamous differentiation. Importantly, analyses of these models suggest that early steps of plasticity may lead towards a basal-like phenotype characterized by the expression of squamous markers, such as Krt5, Krt14 or Trp63. In line with this, transcriptomic upregulation of basal markers is observed in prostate adenocarcinoma cell lines after the sole inactivation of TP53 and RB1.51 Similar results were obtained in a preclinical model leveraging human prostate epithelial cells with concurrent inactivation of TP53/RB1, as well as overexpression of MYC, BCL2 and a constitutively active AKT isoform.68 Engraftment in vivo of such cells leads to the formation of tumors again exhibiting multiple histologies such as adenocarcinoma, squamous, small cell carcinomas, and mixed phenotypes. Temporal analyses of marker expression in this model revealed early-stage upregulation of basal/squamous cell markers such as TP63, and late-stage upregulation of NE markers such as SYP or NCAM1.156 These results are suggestive of a potential model of transformation where adenocarcinomas, or a subset of them, may transition through a squamous phenotype towards their final NE state (Fig. 5, bottom).
Although it is thought that most molecular changes occurring during histological transformation are governed by epigenomic events,6,38,157 tumors undergoing this process frequently exhibit genomic alterations inducing dysregulation of drivers/promoters of plasticity. For example, in NE transformation, ~90% of tumors exhibit copy number loss or inactivating mutations in RB1 vs only 10% showing wild-type RB1 with protein downregulation, and transforming tumors often exhibit activating mutations/amplifications in gene members of the AKT pathway.6 If histological transformation is the result of the convergent dysregulation of promoter/driver genes or pathways, when such dysregulation is driven by epigenomic events, it might be reversible. Thus, cells might be free to transition between different epigenomic states (Fig. 6, top). However, when the pathway dysregulation occurs as a consequence of a genomic event, which is less likely to be reversible, this may anchor the cells in a particular state (Fig. 6, middle). Both molecular scenarios may coexist, with the occurrence of (1) co-existence and interconversion between histological subtypes (reversible, changing epigenomic states) as part of an equilibrium, that may or may not be temporary; and of (2) genomic alterations anchoring the cells in a given state, such as squamous or undifferentiated phenotypes, thus preventing transition to NE carcinoma, and resulting in a squamous transformation or undifferentiated tumor with loss of adenocarcinoma features.
Cells with purple color tones represent cells which underwent reversible epigenomic changes, while red cells represent cells where irreversible genomic alterations were acquired. The green cell represents a cell state to which a red cell cannot transition, as it has acquired an irreversible genomic alteration that anchors the cell in the green state. Figure generated with BioRender.com.
Therapeutic targets to constrain plasticity and histological transformation
Two key considerations inform therapeutic approaches to combat tumor plasticity, and we choose to view these as potential questions for consideration, (1) are pre-existing, therapy-resistant subclones with transcriptomic and/or morphological features of an alternative histology present prior to therapeutic intervention and therefore likely to be selected for over time with adequate pressure; or, (2) does therapeutic pressure directly convert some part of the measurable residual disease into a therapy-refractory disease, now presenting with differing histologic features?
While both scenarios are possible, their differences may impact the design of optimal therapeutic approaches, where different drugs may be combined or staggered, depending on variables including the kinetics of resistance and additive toxicities of each approach. If the goal of any treatment strategy is to prolong healthy years of life for the patient, strategies prioritizing the temporal control of disease over maximal depth of response may be appropriate.158 However, during consolidation of primary lesions, several key points about control of residual disease are important to stress. Using EGFR-driven LUAD as an example, despite the impressive responses and clear clinical benefit to the TKI osimertinib, now widely used and FDA-approved in the first-line setting, the median best percentage change in target-lesion size in approval trials was ~54%, thus leaving substantial residual disease to address.159 Next, following any large debulking event, the residual disease is different, now immersed in a fibrotic, necrotic, and often hypoxic TME that may or may not have been present prior to starting therapy.160,161 These differences may impact the exposure, and thus efficacy, for any therapy introduced following an objective response.
Therefore, targeting mechanisms responsible for histological transformation may be broadly grouped into several categories, including (1) epigenetic mechanisms thought to be responsible for mediating the transitions between adenocarcinoma and NE tumor types; (2) conduction of replication stress associated with a change in oncogenic driver programs; (3) upstream signaling mechanisms responsible for the survival and immune evasion of the residual disease; and (4) molecular features of NE or adeno-squamous tumor types that are uniquely present following histological transformation.
Exploiting epigenetic plasticity as a therapeutic approach for NE tumors
Loss of the RB1 tumor suppressor plays a key role in unveiling epigenetic heterogeneity in advanced cancers.162,163,164,165,166 While the direct transcriptional activity derived from RB1 inactivation may not be pharmacologically targetable yet, RB1 inactivation leads to increase of dependency on enzymatic targets susceptible for pharmacological inhibition. These include critical regulators of cancer progression and/or NE transition — namely LSD1167 and EZH2.168,169
Studies in CRPC models have demonstrated that LSD1 inhibition may act through disruption of enhancer networks that are partially facilitated by MYC and BET protein interactions.167,170 More has been demonstrated in the context of SCLC, where deletion of Kdm1a has been shown to suppress de novo SCLC tumorigenesis, partly through the upregulation of Rest, thus directly repressing Ascl1 target gene activity.171 Additional preclinical studies have demonstrated that exceptional responses to LSD1 inhibitors (including ORY-1001 and GSK2879552) may occur through the activation of Notch signaling, leading to direct or indirect suppression of Ascl1.172,173 Additional follow-up studies have found that genes involved in MHC-1 expression are upregulated following LSD1 inhibition, thereby potentiating the activity of immune checkpoint blockade.174,175 This is a critical finding, as patients with SCLC experiencing the greatest benefit from immune checkpoint blockade are those with the weakest mRNA expression of LSD1 and EZH2 or the greatest expression of an antigen presentation machinery pathway, specifically HLA-A, HLA-B, HLA-C, B2M, TAP1, and TAP2.176 Whether these findings are extendable to NEPC remains to be determined.
Chemical inhibition of EZH2 has been demonstrated to suppress the emergence of anti-androgen resistance — both EZH2 and SOX2 have been described as critical “re-programming” factors in NE transformation.51,54 Indeed, there may be substantial overlap in how chemical inhibition of the enzymatic activity of EZH2 or LSD1 may impact epigenetically silenced antigen presentation and oncogenic driver pathway activity. Ongoing trials will help directly address whether EZH2 inhibitors can be safely combined with AR-targeted therapy and delay the emergence of CRPC and/or NEPC (NCT04179864, NCT03460977). Collectively, these studies highlight the importance of addressing two inter-related issues underscoring immune evasion in NE transformation — antigen presentation and the added reinvigoration of immune effector function. On the lung side, EZH2 pharmacological inhibition showed no efficacy in combination with osimertinib in an NE-transformed, EGFR-mutant PDX model,6 although the efficacy of this therapeutic approach at preventing transformation is yet to be investigated in LUAD models undergoing NE transformation. In an EGFR-mutant PDX model of LUSC transformation, exhibiting high sensitivity to osimertinib, the combination of osimertinib with an EZH2 inhibitor led to suppression of LUSC relapse in vivo.38 This combination was also efficacious at reverting resistance to osimertinib in this same PDX model, after LUSC relapse on osimertinib. Such results suggest that EZH2 inhibition may be a promising therapeutic strategy for tumors undergoing LUSC transformation.
Clinical trials combining either LSD1 or EZH2 chemical inhibitors with SCLC-centric cytotoxic chemotherapy may have seemed logical based on preclinical mechanism;177 however, two points of caution should be raised based on current trial results — (1) toxicity may be limiting, and thus preclude the combination of certain cytotoxic chemotherapies with EZH2 catalytic inhibitors,178 and (2) if LSD1 inhibition does suppress an Ascl1-dependent transcriptional program, then this may also suppress the chemotherapy-sensitive NE fraction, limiting activity.
Another epigenetic effector sustaining the NE phenotype in both prostate and lung cancer settings may represent an alternative therapeutic option — the SWI/SNF chromatin remodeling complex, whose catalytic subunit, SMARCA4, is highly expressed in NE tumors.179,180 SWI/SNF complexes interact with different lineage-specific factors in NEPC compared to prostate adenocarcinoma.179 In the lung setting, SMARCA4 genetic or pharmacological inhibition leads to the loss of NE features and transition towards a non-NE, adenocarcinoma-like state with induction and overactivation of HER family receptor tyrosine kinases180 and MAPK signaling, known to be incompatible with the NE phenotype.181 Combined treatment with a SMARCA4 small-molecule inhibitor and the HER receptor inhibitor afatinib exhibited high efficacy in an array of SCLC PDXs. The availability and further development of potent and selective SMARCA4 inhibitors and degraders offers a promising therapeutic strategy for NE tumors.
Targeting replication stress
A hallmark of solid tumor types with the losses of tumor suppressors RB1 and/or TP53 are features of high replication stress, genomic instability, mitotic/proliferative indices, and general transient sensitivity to DNA damaging chemotherapy.62,182,183,184 The list of therapeutic targets exploiting vulnerabilities in replication stress is ever-expanding, but in general, can be grouped into agents that impact DNA fidelity directly (i.e., GC-groove binding agent lurbinectedin185) or indirectly (i.e., DNA damage signaling and repair machinery). Most substantially advanced from the bench to the patient have been the translation of PARP and ATM/ATR inhibitor therapies.186,187,188,189,190 For example, a recent trial combining the ATR inhibitor barzosertib with topotecan demonstrated a significant extension in overall survival in relapsed SCLC but fell short in achieving the primary endpoint of extending progression-free survival.191 While such agents have demonstrated clinical activity in relapsed SCLC, CRPC, and NEPC, the search for biomarkers to help identify patients most likely to benefit as well as mechanistic explanations for exceptional responders is an on-going pursuit. It is important to emphasize that the overall challenge remains in identifying therapies with durable responses, as most patients have been found to progress on study in less than one year. Such limited duration of efficacy falls short of the observed benefits of upfront targeted therapies, like ADT and EGFR TKIs.
In pursuit of identifying mechanisms responsible for cross or “pan” resistance to agents that exploit replication stress, a recent study modeling acquired resistance using patient-derived models of SCLC found that MYC family paralog amplification via ecDNA drives acquired cross-resistance by amplifying replication stress response pathways and altering transcriptional landscapes.70,192 Recent evidence in mouse models from our groups suggests that the pulmonary NE cell can be transformed by the Myc oncogene alone, thus representing a fundamental oncogenic driver event. Furthermore, MYC may have a critical role in driving histological transformation of lung cancers to either a NE or squamous fate.6,38,142 Whether or not NE cells within other tissue, namely the prostate, are directly transformed by any Myc family alone remains to be determined; however, several key studies have clearly demonstrated that Myc family members including cMyc and nMyc are capable of cooperating with truncal, inactivating events in Rb1, Trp53, and/or Pten, to produce aggressive, metastatic NEPC.74,193,194,195 Pharmacological inhibition of MYC family members has proven challenging.196 The original studies demonstrating potent, direct MYC inhibition made use of a peptide (Omomyc) that was rationally designed to compete with the Myc–Max interface, acting as a dominant-negative Myc protein.197 This concept has been improved over the last two decades and has recently entered clinical testing in solid tumors.198,199 Additional preclinical strategies to target the MYC–MAX interface have also been recently introduced and hold promise for preclinical translation.200 Indeed, prior data in SCLC models suggested that Omomyc had potent in vitro activity.201,202 On the other hand, our group has recently identified a therapeutic target, the cell division cycle 7 kinase (CDC7), whose inhibition induces indirect MYC inactivation by its proteasomal degradation, thus hindering NE transformation and significantly extending response to targeted therapy in both lung and prostate tumor models.60 Perhaps these results are not all that surprising, as SCLC is the only solid tumor found to lose the MYC-binding effector MAX as a mechanism of tumor progression, further highlighting a role for MYC in SCLC tumorigenesis.203,204
Considering that NE cancers are bi-modally distinct from adenocarcinomas based on the expression and activity of YAP/TAZ,205 histological transformations can be understood as complex events involving both genetic and epigenetic remodeling over large timescales. Several recent studies have focused on the intermediate phases of tumorigenesis leading up to histological transformation. In prostate cancer models of acquired resistance to AR-targeted therapies, a critical role for JAK/STAT signaling has been identified in promoting an inflamed, stem-like precursor to NEPC.63,65 This same stem-like intermediate has been observed in other studies where in vivo systems were used to chart the transformation of normal luminal and/or basal precursor cell types into NE tumor types.68,156 However, it remains unclear when and how an inflamed or stem-like state may help evade key immune checkpoints in tumor control, leaving this as an area of active investigation. Interestingly, JAK/STAT signaling may also play a role in promoting the adeno-to-squamous transition observed in some LUADs following treatment with targeted therapies, including recently approved KRAS inhibitors.4,104,105,206 Encouragingly, recent activity combining the JAK/STAT inhibitor itacitinib with immune checkpoint blockade has shown promise in advanced LUAD.207 These findings suggest a broader role for JAK/STAT signaling in mediating lineage plasticity across different cancer types. Collectively, the inhibition of JAK/STAT signaling raises the possibility of trapping an intermediate state during histological transformation that is both inflamed and expressing MHC-1 prior to transformation to an NE state. Understanding the temporal relationship between an early inflamed TME of an adenocarcinoma, the introduction of immune checkpoint blockade to exploit this inflammation, and the eventual acquisition of a cold TME of the NE state are active areas of research. Collectively, such an approach may have the potential to limit the full transition to NE phenotypes while simultaneously enhancing immune-mediated tumor control.
Along similar lines of reasoning, targeting of the PI3K/AKT signaling pathways has also been proposed to address the survival of the residual disease and directly target a common genetic alteration found in advanced prostate cancer.208,209,210 Several smaller trials in CRPC patients have explored the combination of either mTOR, PI3K, or AKT inhibitors with chemotherapy or ADT and have not demonstrated benefit, some with toxicities that limited the dosing intervals.211,212,213,214,215 Mutations in the PI3K/AKT/mTOR pathway are frequent in NE transformation,6,216 and overactivation of the pathway has been associated with NE transformation in the lung and prostate68,155 and even with squamous transformation.38 The fact that this pathway may have a promoting role in either squamous or NE transformation, and that its dysregulation occurs in adenocarcinomas with no NE features,217 is suggestive of a potential role for this pathway to contribute to the previously mentioned initial, stem-like state prior to histological transition. In the lung setting, PI3K/AKT/mTOR pharmacological targeting has been tested in preclinical models of transformation. In the NE transformation setting, the pathway inhibitor samotolisib was combined with osimertinib in an EGFR-mutant PDX model of NE transformation.38 Even if the combination slowed tumor growth more than either monotherapy alone in this model, the response observed was modest. Importantly, this particular PDX model exhibited baseline combined histology with both LUAD and NE components, with predominant NE histology, and was not responsive to osimertinib. This observation suggests that such a therapeutic approach might not be effective after transformation has occurred and opens the question of whether this combination therapy may hinder plasticity if administered before transformation. In line with this, analyses of tumors collected after treatment in this model revealed that samotolisib avoided the complete disappearance of the LUAD component observed after osimertinib monotherapy, suggesting that PI3K/AKT/mTOR inhibition may be a promising strategy in tumors at high risk of NE transformation. Targeting of PI3K/AKT/mTOR has also been tested in an EGFR-mutant PDX model of LUSC transformation,38 which exhibited limited sensitivity to osimertinib. In this model, samotolisib resensitized tumors to osimertinib, suggesting that AKT inhibition may be a promising strategy for LUSC-transformed tumors. However, the efficacy of such inhibitors in preventing LUSC transformation is yet to be studied. Unfortunately, the toxicity of current PI3K/AKT/mTOR inhibitors, which would be potentiated if combined with targeted therapy, may be a limitation to bring these drugs into the clinic successfully. Although prior efforts to inhibit this signaling axis have been disappointing, this does not preclude the target as relevant, where perhaps alternative approaches (i.e., AKT degraders218) leading to reduced toxicity will have beneficial activity.
NE-specific vulnerabilities
Historically, one of the best examples of “good target, wrong approach” is the DLL3-targeted therapies. Originally identified as elevated in high-grade NE cancers — including SCLC, LCNEC, and NEPC — DLL3 functions as a Notch inhibitory ligand and was considered “targetable” due to its unique expression in tumors relative to normal tissues.30,219,220,221 However, later-phase translational efforts utilizing antibody–drug conjugates (ADCs) encountered significant setbacks, as poor stability between the monomethyl auristatin E warhead and the DLL3-targeting antibody precluded further development of this approach.222 Despite these early challenges, the field has since evolved, encompassing new approaches such as traditional ADCs, BiTEs, radioimmune conjugates, and others, all focused on DLL3.223 In May 2024, tarlatamab was FDA-approved for extensive-stage SCLC with disease progression after platinum-based chemotherapy.32 With preclinical evidence suggesting that DLL3 is similarly expressed in NEPC and that targeting DLL3 has therapeutic potential,224,225 several ongoing clinical trials aim to broaden the indications for tarlatamab and other DLL3-related therapies (NCT04702737, NCT05652686, NCT05882058).
Recently, our group identified exportin 1, a nuclear exporter, as a promising therapeutic target in NE tumors, including SCLC and NEPC.226,227 Exportin 1 was found to be upregulated during the very early stages of NE transformation, immediately following TP53 and RB1 inactivation. Inhibition of exportin 1 with selinexor significantly extended chemotherapy response in various PDXs of SCLC and NEPC. Selinexor also prevented NE transformation in multiple in vivo models of prostate and lung cancers, greatly enhancing the efficacy of targeted therapies in both settings. Mechanistic studies revealed that exportin 1 inhibition downregulated SOX2.227 These findings, coupled with the clinical availability of selinexor, which is FDA-approved for treatment-refractory hematological malignancies, position exportin 1 as a compelling target to prevent plasticity in these aggressive tumors.
Additional targets implicated in NE transformation have emerged from analyses of de novo SCLC and histologically transformed samples, including B7-H3/CD276, B7-H6, SEZ6, TROP-2, CEACAM5, and others.228,229,230,231,232,233,234 Therapeutic strategies targeting these molecules are diverse, including ADCs, BiTEs, chimeric antigen receptor T cell therapies, and other modalities. However, no clear “winning” approach has yet been identified, fueling continued preclinical development. Furthermore, as multiple NE-targeted therapies gain FDA approval, questions regarding the timing and sequence of these treatments remain unanswered. It is not yet clear how recurrent disease will present (e.g., DLL3 positive or negative, NE or not) or whether specific subtypes of NE tumors, such as ASCL1+ vs NEUROD1+, will preferentially benefit from distinct therapeutic strategies.235
Open questions
Despite progress during the last five years to identify promoters, effectors, and therapies targeting histological transformation, several open questions remain to be addressed. Most urgent in our opinion, is the identification of molecular biomarkers predictive of squamous transformation. Whether certain genetic or epigenetic events lock transdifferentiation of adenocarcinoma into a squamous-like state over an NE one is not well defined and remain areas of active investigation. Accrual of large numbers of genetically annotated samples from patients on institution-specific trials continues to present logistical challenges. Moreover, to date, molecular characterizations of squamous transformation have been limited, with no clear emergent molecular predictors of high risk. In addition, the scarcity of human and mouse preclinical models of squamous transformation represents an important challenge in the validation of potential therapeutic targets to prevent or treat these tumors.
Another open question relates to the molecular differences between de novo and transformed tumors, and whether these may respond differentially to therapy. Molecular analyses of clinical specimens undergoing NE or squamous transformation have revealed that transformed tumors may retain broad-scale molecular features of their previous adenocarcinoma state, and may exhibit greater heterogeneity,5,6,38 potentially making them a different entity from de novo occurring NE and squamous tumors. Particularly in the transformed SCLC setting, it has been suggested that transformed tumors may respond worse to chemotherapy as compared to de novo tumors, with a slightly inferior progression-free survival of 3.4 months vs 5.5 months.236,237 However, these results are yet to be confirmed. Importantly, in the EGFR-mutant LUAD setting, we know that tumors lose expression of EGFR upon NE transformation. However, we still do not know whether that is also the case for tumors undergoing LUSC transformation, or whether in adenocarcinomas undergoing histological transformation with drivers different than EGFR, the adenocarcinoma driver gets downregulated as well.238,239 Understanding the molecular biology of transformed tumors will be key to identifying effective therapeutic targets.
The influence of the TME in the process of histological transformation also remains undefined. Even if slight upregulation of NE or squamous markers can be measured in vitro in adenocarcinoma cell lines and organoid models with the appropriate molecular modifications,38,51 major transformation features are only observed in vivo.65,240 This observation suggests that the TME may have a role in histological transformation. Consistent with this hypothesis, clinical specimens undergoing NE or squamous transformation exhibit a strong downregulation of a few pathways related to immune response,6,38 which may indicate the requirement of a strong suppression of anti-tumor immune response for transformation to occur. As epigenetic reprogramming to a stem-like state is an immunogenic process,241,242,243 this immune suppression may be exerted by tumor cells to protect themselves from the immune system during transformation.
One interesting and potentially key aspect of histological transformation that remains unknown, briefly covered in the present review, is whether there is a strict directionality in transformation. In other words, can histological transformation be reverted? On the NE transformation side, reports indicate that MAPK induction may be incompatible with NE SCLC, where it induces cell cycle arrest and senescence.181 These results indicate that EGFR (or receptor tyrosine kinase) re-expression, which would lead to MAPK signaling, may be challenging in SCLC tumors. Interestingly, recent analyses of clinical specimens and GEMMs have described the existence of plasticity in SCLC. Even if SCLC tumors exhibit homogeneous morphology, different transcriptomic subtypes have been identified with different levels of NE phenotype.146 These subtypes are not static, but plastic, in a way that NE SCLC tumors can transition to a non-NE state with an EMT phenotype and activation of Notch signaling.119 Indeed, as mentioned above, inhibition of the activity of enzymes such as EZH2 or LSD1, thought to be partially responsible for the epigenomic remodeling observed following histological transformation, may induce an NE to non-NE transition.75,174,175 These results indicate that the reversal of the NE phenotype may be possible, but whether additional molecular alterations lead to reprogramming (or “resetting”) all the way back to the original adenocarcinoma phenotype remains unknown.
Finally, a related open question is whether transformation may occur in both directions, with an equilibrium between the adenocarcinoma and the alternative histologic subtypes present in the same tumor. The occurrence of combined histology tumors, particularly adeno-squamous tumors which are relatively frequent representing 2%–4% of lung tumors,244 could indicate that (1) tumors were collected at an intermediate state of transformation, before the initial state (adenocarcinoma?) gets overgrown by the transformed cells; or (2) that there might be an equilibrium of interconversion between cells from both histologic subtypes induced by a selective advantage derived from both cell types being present in a given tumor, such as a potential cellular interaction between both components supporting oncogenicity and progression, as has been reported in other settings.121,122
All data reviewed in the present manuscript highlight the challenges that plasticity poses to the clinical management of tumors able to transition to different states via dysregulation of specific oncogenic pathways (Fig. 7). Both the cell of origin and the driver oncogenic signaling derived from the tumor at a given time may induce contextual molecular constraints in terms of the histological phenotypes that a given tumor may display. However, the inhibition of that particular driver, pharmacologically or otherwise, together with the convergence of the right molecular alterations may overcome those constraints. In this sense, histological subtyping, even if of clinical relevance, may be a rough categorization of what could potentially be a spectrum of phenotypes defined by the activation of a given combination of oncogenic pathways. Indeed, in spite of efforts to design molecular tests to accurately predict tumor histology, including genomic and epigenomic approaches,28,245,246 currently histological subtyping in the clinic relies on microscopic morphological criteria and differential expression of differentiation immunohistochemistry markers. Intermediate or transitioning phenotypes may certainly pose a challenge for the development of such molecular tests. An “all-plastic” model (Fig. 5) has been proposed, positing that tumors may be able to transition from one state to another in response to certain selective pressures (treatments, hypoxia, TME, etc.), unless a state-defining oncogenic pathway occurs in a genomic, irreversible manner (as described in Fig. 6). Additionally, the occurrence of tumors with combined histology, intermediate phenotypes, or undifferentiated/dedifferentiated phenotypes, mentioned in the present review, would be explained by this plastic spectrum model. Nonetheless, additional confirmatory work will be required to support or refute such a model, where the use of cutting-edge single-cell and spatial techniques, as well as the development of potent, reliable lineage tracing methods will be key to studying directionality of transformation and fully characterizing intermediate/admixed histological phenotypes.
Conclusions
Recent multi-omic analysis of clinical specimens and single-cell profiling of preclinical models of plasticity have identified oncogenic pathways dysregulated during histological transformation. Some of these may be promoters of plasticity and some drivers of transdifferentiation towards a specific state (i.e., manifesting as a tumor “histology”). Importantly, intermediate states of transformation have been identified that may be of particular therapeutic relevance — specifically an inflammatory intermediate bearing hallmark of EMT. Early promoters of plasticity driving such states may be key therapeutic targets to prevent or delay histological transformation in patients at high risk. Upcoming studies leveraging state-of-the-art high-resolution, spatial and lineage tracing technologies, will be key to further characterizing the directionality, signaling timing of histological transformation and its potential reversibility, as well as the role of the TME in this plasticity phenomenon.
References
Quintanal-Villalonga, A. et al. Lineage plasticity in cancer, a shared pathway of therapeutic resistance. Nat. Rev. Clin. Oncol. 17, 360–371 (2020).
Beltran, H. et al. Circulating tumor DNA profile recognizes transformation to castration-resistant neuroendocrine prostate cancer. J. Clin. Invest. 130, 1653–1668 (2020).
Wang, H. T. et al. Neuroendocrine Prostate Cancer (NEPC) progressing from conventional prostatic adenocarcinoma, factors associated with time to development of NEPC and survival from NEPC diagnosis-a systematic review and pooled analysis. J. Clin. Oncol. 32, 3383–3390 (2014).
Schoenfeld, A. J. et al. Tumor analyses reveal squamous transformation and off-target alterations as early resistance mechanisms to first-line osimertinib in EGFR-mutant lung cancer. Clin. Cancer Res. 26, 2654–2663 (2020).
Niederst, M. J. et al. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat. Commun. 6, 6377 (2015).
Quintanal-Villalonga, A. et al. Multiomic analysis of lung tumors defines pathways activated in neuroendocrine transformation. Cancer Discov. 11, 3028–3047 (2021).
Offin, M. et al. Concurrent RB1 and TP53 alterations define a subset of EGFR-mutant lung cancers at risk for histologic transformation and inferior clinical outcomes. J. Thorac. Oncol. 14, 1784–1793 (2019).
Rudin, C. M. et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat. Genet. 44, 1111–1116 (2012).
Aggarwal, R. et al. Clinical and genomic characterization of treatment-emergent small-cell neuroendocrine prostate cancer: a multi-institutional prospective study. J. Clin. Oncol. 36, 2492–2503 (2018).
Rudin, C. M., Brambilla, E., Faivre-Finn, C. & Sage, J. Small-cell lung cancer. Nat. Rev. Dis. Prim. 7, 3 (2021).
Akamatsu, S., Inoue, T., Ogawa, O. & Gleave, M. E. Clinical and molecular features of treatment-related neuroendocrine prostate cancer. Int. J. Urol. 25, 345–351 (2018).
Eule, C. J. et al. Outcomes of second-line therapies in patients with metastatic de novo and treatment-emergent neuroendocrine prostate cancer: a multi-institutional study. Clin. Genitourin. Cancer 21, 483–490 (2023).
Aggarwal, R., Zhang, T., Small, E. J. & Armstrong, A. J. Neuroendocrine prostate cancer, subtypes, biology, and clinical outcomes. J. Natl. Compr. Cancer Netw. 12, 719–726 (2014).
Brennen, W. N. et al. Resistance to androgen receptor signaling inhibition does not necessitate development of neuroendocrine prostate cancer. JCI Insight 6, e146827 (2021).
Beltran, H. et al. The role of lineage plasticity in prostate cancer therapy resistance. Clin. Cancer Res. 25, 6916–6924 (2019).
Nosaki, K. et al. Re-biopsy status among non-small cell lung cancer patients in Japan: a retrospective study. Lung Cancer 101, 1–8 (2016).
Tuzi, A. et al. Biopsy and re-biopsy in lung cancer, the oncologist requests and the role of endobronchial ultrasounds transbronchial needle aspiration. J. Thorac. Dis. 9, S405–S409 (2017).
Jekunen, A. P. Role of rebiopsy in relapsed non-small cell lung cancer for directing oncology treatments. J. Oncol. 2015, 809835 (2015).
Lee, J. K. et al. Clonal history and genetic predictors of transformation into small-cell carcinomas from lung adenocarcinomas. J. Clin. Oncol. 35, 3065–3074 (2017).
Zhang, S. L. et al. Expression of EGFR-mutant proteins and genomic evolution in EGFR-mutant transformed small cell lung cancer. J. Thorac. Dis. 15, 4620–4635 (2023).
Kim, D. H. et al. BET bromodomain inhibition blocks an AR-repressed, E2F1-activated treatment-emergent neuroendocrine prostate cancer lineage plasticity program. Clin. Cancer Res. 27, 4923–4936 (2021).
Meador, C. B., Sequist, L. V. & Piotrowska, Z. Targeting EGFR exon 20 insertions in non-small cell lung cancer: recent advances and clinical updates. Cancer Discov. 11, 2145–2157 (2021).
Gainor, J. F. & Shaw, A. T. Emerging paradigms in the development of resistance to tyrosine kinase inhibitors in lung cancer. J. Clin. Oncol. 31, 3987–3996 (2013).
Lemmon, M. A., Schlessinger, J. & Ferguson, K. M. The EGFR family: not so prototypical receptor tyrosine kinases. Cold Spring Harb. Perspect. Biol. 6, a020768 (2014).
Sivakumar, S. et al. Integrative analysis of a large real-world cohort of small cell lung cancer identifies distinct genetic subtypes and insights into histologic transformation. Cancer Discov. 13, 1572–1591 (2023).
Chen, J. et al. Genomic landscape of lung adenocarcinoma in East Asians. Nat. Genet. 52, 177–186 (2020).
Chen, M. F. et al. Osimertinib, platinum, etoposide as initial treatment for patients with EGFR mutant lung cancers with TP53 and RB1 alterations. J. Clin. Oncol. 42, 8565–8565 (2024).
Heeke, S. et al. Tumor- and circulating-free DNA methylation identifies clinically relevant small cell lung cancer subtypes. Cancer Cell 42, 225–237.e225 (2024).
Tendler, S. et al. First-in-human imaging with [(89)Zr]Zr-DFO-SC16.56 anti-DLL3 antibody in patients with high-grade neuroendocrine tumors of the lung and prostate. medRxiv https://doi.org/10.1101/2024.01.10.24301109 (2024).
Sharma, S. K. et al. Noninvasive interrogation of DLL3 expression in metastatic small cell lung cancer. Cancer Res. 77, 3931–3941 (2017).
Wang, Z. et al. Molecular subtypes of neuroendocrine carcinomas: A cross-tissue classification framework based on five transcriptional regulators. Cancer Cell 42, 1106–1125.e8 (2024).
Ahn, M. J. et al. Tarlatamab for patients with previously treated small-cell lung cancer. N. Engl. J. Med. 389, 2063–2075 (2023).
Zhao, S. G. et al. A clinical-grade liquid biomarker detects neuroendocrine differentiation in prostate cancer. J. Clin. Invest. 132, e161858 (2022).
Kaelin, W. G. Jr Common pitfalls in preclinical cancer target validation. Nat. Rev. Cancer 17, 425–440 (2017).
Ruffini, E. et al. Lung tumors with mixed histologic pattern. Clinico-pathologic characteristics and prognostic significance. Eur. J. Cardiothorac. Surg. 22, 701–707 (2002).
Mordant, P. et al. Adenosquamous carcinoma of the lung: surgical management, pathologic characteristics, and prognostic implications. Ann. Thorac. Surg. 95, 1189–1195 (2013).
Tochigi, N., Dacic, S., Nikiforova, M., Cieply, K. M. & Yousem, S. A. Adenosquamous carcinoma of the lung: a microdissection study of KRAS and EGFR mutational and amplification status in a western patient population. Am. J. Clin. Pathol. 135, 783–789 (2011).
Quintanal-Villalonga, A. et al. Comprehensive molecular characterization of lung tumors implicates AKT and MYC signaling in adenocarcinoma to squamous cell transdifferentiation. J. Hematol. Oncol. 14, 170 (2021).
Tarigopula, A., Ramasubban, G., Chandrashekar, V., Govindasami, P. & Chandran, C. EGFR mutations and ROS1 and ALK rearrangements in a large series of non-small cell lung cancer in South India. Cancer Rep. 3, e1288 (2020).
Jin, R. et al. EGFR-mutated squamous cell lung cancer and its association with outcomes. Front. Oncol. 11, 680804 (2021).
Liang, H. et al. Real-world data on EGFR/ALK gene status and first-line targeted therapy rate in newly diagnosed advanced non-small cell lung cancer patients in Northern China: a prospective observational study. Thorac. Cancer 10, 1521–1532 (2019).
Liam, C. K., Leow, H. R. & Pang, Y. K. EGFR mutation testing for squamous cell lung carcinoma. J. Thorac. Oncol. 8, e114 (2013).
Joshi, A. et al. EGFR mutation in squamous cell carcinoma of the lung: does it carry the same connotation as in adenocarcinomas?. Onco Targets Ther. 10, 1859–1863 (2017).
Sun, J. M. et al. Small-cell lung cancer detection in never-smokers: clinical characteristics and multigene mutation profiling using targeted next-generation sequencing. Ann. Oncol. 26, 161–166 (2015).
George, J. et al. Integrative genomic profiling of large-cell neuroendocrine carcinomas reveals distinct subtypes of high-grade neuroendocrine lung tumors. Nat. Commun. 9, 1048 (2018).
Chang, H. C., Chen, K. Y., Chang, Y. L., Shih, J. Y. & Yu, C. J. Lung adenocarcinoma with neuroendocrine differentiation: Molecular markers testing and treatment outcomes. J. Formos. Med. Assoc. 122, 731–737 (2023).
Abida, W. et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc. Natl. Acad. Sci. USA 116, 11428–11436 (2019).
Wallace, W. A. The challenge of classifying poorly differentiated tumours in the lung. Histopathology 54, 28–42 (2009).
Ben-Porath, I. et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 40, 499–507 (2008).
Malta, T. M. et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell 173, 338–354.e15 (2018).
Mu, P. et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 355, 84–88 (2017).
Beltran, H. et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 22, 298–305 (2016).
Aggarwal, R. et al. Clinical and genomic characterization of Low PSA Secretors: a unique subset of metastatic castration resistant prostate cancer. Prostate Cancer Prostatic Dis. 24, 81–87 (2021).
Ku, S. Y. et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 355, 78–83 (2017).
Sainz de Aja, J., Dost, A. F. M. & Kim, C. F. Alveolar progenitor cells and the origin of lung cancer. J. Intern. Med. 289, 629–635 (2021).
Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008).
Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012).
George, J. et al. Comprehensive genomic profiles of small cell lung cancer. Nature 524, 47–53 (2015).
Barbie, D. A. et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112 (2009).
Quintanal-Villalonga, A. et al. CDC7 inhibition impairs neuroendocrine transformation in lung and prostate tumors through MYC degradation. Signal Transduct. Target. Ther. 9, 189 (2024).
Chen, Z. et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436, 725–730 (2005).
Nyquist, M. D. et al. Combined TP53 and RB1 loss promotes prostate cancer resistance to a spectrum of therapeutics and confers vulnerability to replication stress. Cell Rep. 31, 107669 (2020).
Deng, S. et al. Ectopic JAK-STAT activation enables the transition to a stem-like and multilineage state conferring AR-targeted therapy resistance. Nat. Cancer 3, 1071–1087 (2022).
Deng, S. et al. Author correction, Ectopic JAK-STAT activation enables the transition to a stem-like and multilineage state conferring AR-targeted therapy resistance. Nat. Cancer 3, 1271 (2022).
Chan, J. M. et al. Lineage plasticity in prostate cancer depends on JAK/STAT inflammatory signaling. Science 377, 1180–1191 (2022).
Lo, U. G. et al. The driver role of JAK-STAT signalling in cancer stemness capabilities leading to new therapeutic strategies for therapy- and castration-resistant prostate cancer. Clin. Transl. Med. 12, e978 (2022).
Westbrook, T. C. et al. Transcriptional profiling of matched patient biopsies clarifies molecular determinants of enzalutamide-induced lineage plasticity. Nat. Commun. 13, 5345 (2022).
Park, J. W. et al. Reprogramming normal human epithelial tissues to a common, lethal neuroendocrine cancer lineage. Science 362, 91–95 (2018).
Pongor, L. S. et al. Extrachromosomal DNA amplification contributes to small cell lung cancer heterogeneity and is associated with worse outcomes. Cancer Discov. 13, 928–949 (2023).
Pal Choudhuri, S. et al. Acquired cross-resistance in small cell lung cancer due to extrachromosomal DNA amplification of MYC paralogs. Cancer Discov. 14, 804–827 (2024).
Cha, T. L. et al. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science 310, 306–310 (2005).
Xu, K. et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 338, 1465–1469 (2012).
Davies, A. et al. An androgen receptor switch underlies lineage infidelity in treatment-resistant prostate cancer. Nat. Cell Biol. 23, 1023–1034 (2021).
Dardenne, E. et al. N-Myc induces an EZH2-mediated transcriptional program driving neuroendocrine prostate cancer. Cancer Cell 30, 563–577 (2016).
Mahadevan, N. R. et al. Intrinsic immunogenicity of small cell lung carcinoma revealed by its cellular plasticity. Cancer Discov. 11, 1952–1969 (2021).
Zhang, Y. et al. Androgen deprivation promotes neuroendocrine differentiation and angiogenesis through CREB-EZH2-TSP1 pathway in prostate cancers. Nat. Commun. 9, 4080 (2018).
Schweizer, M. T. et al. Mevrometostat (PF-06821497), an enhancer of zeste homolog 2 (EZH2) inhibitor, in combination with enzalutamide in patients with metastatic castration-resistant prostate cancer (mCRPC): A randomized dose-expansion study. J. Clin. Oncol. 43, LBA138–LBA138 (2025).
Adams, E. J. et al. FOXA1 mutations alter pioneering activity, differentiation and prostate cancer phenotypes. Nature 571, 408–412 (2019).
Baca, S. C. et al. Reprogramming of the FOXA1 cistrome in treatment-emergent neuroendocrine prostate cancer. Nat. Commun. 12, 1979 (2021).
Wang, Z. et al. FOXA2 rewires AP-1 for transcriptional reprogramming and lineage plasticity in prostate cancer. Nat. Commun. 15, 4914 (2024).
Zhang, Z. et al. Loss of CHD1 promotes heterogeneous mechanisms of resistance to AR-targeted therapy via chromatin dysregulation. Cancer Cell 37, 584–598.e11 (2020).
Xu, Y. et al. ZNF397 Deficiency triggers TET2-driven lineage plasticity and AR-targeted therapy resistance in prostate cancer. Cancer Discov. 14, 1496–1521 (2024).
Concepcion, C. P. et al. Smarca4 inactivation promotes lineage-specific transformation and early metastatic features in the lung. Cancer Discov. 12, 562–585 (2022).
Liu, L. et al. The LIS1/NDE1 complex is essential for FGF signaling by regulating FGF receptor intracellular trafficking. Cell Rep. 22, 3277–3291 (2018).
Ferone, G. et al. FGFR1 oncogenic activation reveals an alternative cell of origin of SCLC in Rb1/p53 mice. Cell Rep. 30, 3837–3850.e33 (2020).
Abu El Maaty, M. A. et al. Hypoxia-mediated stabilization of HIF1A in prostatic intraepithelial neoplasia promotes cell plasticity and malignant progression. Sci. Adv. 8, eabo2295 (2022).
Wohlhieter, C. A. et al. Concurrent mutations in STK11 and KEAP1 promote ferroptosis protection and SCD1 dependence in lung cancer. Cell Rep. 33, 108444 (2020).
Julian, C. et al. Overall survival in patients with advanced non-small cell lung cancer with KRAS G12C mutation with or without STK11 and/or KEAP1 mutations in a real-world setting. BMC Cancer 23, 352 (2023).
Zhang, P. et al. Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol. Cancer Ther. 9, 336–346 (2010).
Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517, 576–582 (2015).
Yamamoto, M., Kensler, T. W. & Motohashi, H. The KEAP1-NRF2 system: a thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol. Rev. 98, 1169–1203 (2018).
Tsai, J. J. et al. Nrf2 regulates haematopoietic stem cell function. Nat. Cell Biol. 15, 309–316 (2013).
Jeong, Y. et al. Role of KEAP1/NRF2 and TP53 mutations in lung squamous cell carcinoma development and radiation resistance. Cancer Discov. 7, 86–101 (2017).
Korsse, S. E., Peppelenbosch, M. P. & van Veelen, W. Targeting LKB1 signaling in cancer. Biochim. Biophys. Acta 1835, 194–210 (2013).
Vaahtomeri, K. & Makela, T. P. Molecular mechanisms of tumor suppression by LKB1. FEBS Lett. 585, 944–951 (2011).
Caja, L. et al. The protein kinase LKB1 promotes self-renewal and blocks invasiveness in glioblastoma. J. Cell Physiol. 237, 743–762 (2022).
Hu, M. T. et al. Tumor suppressor LKB1 inhibits the progression of gallbladder carcinoma and predicts the prognosis of patients with this malignancy. Int J. Oncol. 53, 1215–1226 (2018).
Tong, X. et al. Nanog maintains stemness of Lkb1-deficient lung adenocarcinoma and prevents gastric differentiation. EMBO Mol. Med 13, e12627 (2021).
Murray, C. W. et al. LKB1 drives stasis and C/EBP-mediated reprogramming to an alveolar type II fate in lung cancer. Nat. Commun. 13, 1090 (2022).
Ji, H. et al. LKB1 modulates lung cancer differentiation and metastasis. Nature 448, 807–810 (2007).
Zhang, H. et al. Lkb1 inactivation drives lung cancer lineage switching governed by polycomb repressive complex 2. Nat. Commun. 8, 14922 (2017).
Pierce, S. E. et al. LKB1 inactivation modulates chromatin accessibility to drive metastatic progression. Nat. Cell Biol. 23, 915–924 (2021).
Xue, Y. et al. TET2-STAT3-CXCL5 nexus promotes neutrophil lipid transfer to fuel lung adeno-to-squamous transition. J. Exp. Med. 221, e20240111 (2024).
Tong, X. et al. Adeno-to-squamous transition drives resistance to KRAS inhibition in LKB1 mutant lung cancer. Cancer Cell 42, 413–428.e7 (2024).
Qin, Z. et al. EML4-ALK fusions drive lung adeno-to-squamous transition through JAK-STAT activation. J. Exp. Med. 221, e20232028 (2024).
Rekhtman, N. et al. Next-generation sequencing of pulmonary large cell neuroendocrine carcinoma reveals small cell carcinoma-like and non-small cell carcinoma-like subsets. Clin. Cancer Res. 22, 3618–3629 (2016).
Liu, J. et al. Wnt/beta-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct. Target. Ther. 7, 3 (2022).
Zhan, T., Rindtorff, N. & Boutros, M. Wnt signaling in cancer. Oncogene 36, 1461–1473 (2017).
Park, J. I. et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature 460, 66–72 (2009).
Schwitalla, S. et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 152, 25–38 (2013).
Tang, F. et al. Chromatin profiles classify castration-resistant prostate cancers suggesting therapeutic targets. Science 376, eabe1505 (2022).
Bland, T. et al. WLS-Wnt signaling promotes neuroendocrine prostate cancer. iScience 24, 101970 (2021).
Lee, G. T. et al. TCF4 induces enzalutamide resistance via neuroendocrine differentiation in prostate cancer. PLoS One 14, e0213488 (2019).
Moparthi, L., Pizzolato, G. & Koch, S. Wnt activator FOXB2 drives the neuroendocrine differentiation of prostate cancer. Proc. Natl. Acad. Sci. USA 116, 22189–22195 (2019).
Uysal-Onganer, P. et al. Wnt-11 promotes neuroendocrine-like differentiation, survival and migration of prostate cancer cells. Mol. Cancer 9, 55 (2010).
Shue, Y. T. et al. A conserved YAP/Notch/REST network controls the neuroendocrine cell fate in the lungs. Nat. Commun. 13, 2690 (2022).
Danza, G. et al. Notch signaling modulates hypoxia-induced neuroendocrine differentiation of human prostate cancer cells. Mol. Cancer Res. 10, 230–238 (2012).
Gay, C. M. et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell 39, 346–360.e7 (2021).
Ireland, A. S. et al. MYC drives temporal evolution of small cell lung cancer subtypes by reprogramming neuroendocrine fate. Cancer Cell 38, 60–78.e12 (2020).
Stewart, C. A. et al. Single-cell analyses reveal increased intratumoral heterogeneity after the onset of therapy resistance in small-cell lung cancer. Nat. Cancer 1, 423–436 (2020).
Calbo, J. et al. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell 19, 244–256 (2011).
Lim, J. S. et al. Intratumoural heterogeneity generated by Notch signalling promotes small-cell lung cancer. Nature 545, 360–364 (2017).
Kim, J. W., Ko, J. H. & Sage, J. DLL3 regulates Notch signaling in small cell lung cancer. iScience 25, 105603 (2022).
Liu, L. et al. An RFC4/Notch1 signaling feedback loop promotes NSCLC metastasis and stemness. Nat. Commun. 12, 2693 (2021).
Loganathan, S. K. et al. Rare driver mutations in head and neck squamous cell carcinomas converge on NOTCH signaling. Science 367, 1264–1269 (2020).
Li, X. et al. Loss of SYNCRIP unleashes APOBEC-driven mutagenesis, tumor heterogeneity, and AR-targeted therapy resistance in prostate cancer. Cancer Cell 41, 1427–1449.e12 (2023).
Rubin, M. A., Bristow, R. G., Thienger, P. D., Dive, C. & Imielinski, M. Impact of lineage plasticity to and from a neuroendocrine phenotype on progression and response in postate and lung cancers. Mol. Cell 80, 562–577 (2020).
Goldstein, A. S., Huang, J., Guo, C., Garraway, I. P. & Witte, O. N. Identification of a cell of origin for human prostate cancer. Science 329, 568–571 (2010).
Park, J. W. et al. Prostate epithelial cell of origin determines cancer differentiation state in an organoid transformation assay. Proc. Natl. Acad. Sci. USA 113, 4482–4487 (2016).
Zhang, D., Zhao, S., Li, X., Kirk, J. S. & Tang, D. G. Prostate luminal progenitorcells in development and cancer. Trends Cancer 4, 769–783 (2018).
Wang, X. et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature 461, 495–500 (2009).
Goldstein, A. S. & Witte, O. N. Does the microenvironment influence the cell types of origin for prostate cancer? Genes Dev. 27, 1539–1544 (2013).
Olsen, R. R. et al. ASCL1 represses a SOX9(+) neural crest stem-like state in small cell lung cancer. Genes Dev. 35, 847–869 (2021).
Kuo, C. S. & Krasnow, M. A. Formation of a neurosensory organ by epithelial cell slithering. Cell 163, 394–405 (2015).
Walter, D. M. et al. RB constrains lineage fidelity and multiple stages of tumour progression and metastasis. Nature 569, 423–427 (2019).
Li, Z. et al. Alveolar differentiation drives resistance to KRAS inhibition in lung adenocarcinoma. Cancer Discov. 14, 308–325 (2024).
Punekar, S. R., Velcheti, V., Neel, B. G. & Wong, K. K. The current state of the art and future trends in RAS-targeted cancer therapies. Nat. Rev. Clin. Oncol. 19, 637–655 (2022).
Ventura, A. et al. Restoration of p53 function leads to tumour regression in vivo. Nature 445, 661–665 (2007).
Wasko, U. N. et al. Tumour-selective activity of RAS-GTP inhibition in pancreatic cancer. Nature 629, 927–936 (2024).
Jiang, J. et al. Translational and therapeutic evaluation of RAS-GTP inhibition by RMC-6236 in RAS-driven cancers. Cancer Discov. 14, 994–1017 (2024).
Holderfield, M. et al. Concurrent inhibition of oncogenic and wild-type RAS-GTP for cancer therapy. Nature 629, 919–926 (2024).
Gardner, E. E. et al. Lineage-specific intolerance to oncogenic drivers restricts histological transformation. Science 383, eadj1415 (2024).
Venkadakrishnan, V. B. et al. Lineage-specific canonical and non-canonical activity of EZH2 in advanced prostate cancer subtypes. Nat. Commun. 15, 6779 (2024).
Puca, L. et al. Patient derived organoids to model rare prostate cancer phenotypes. Nat. Commun. 9, 2404 (2018).
Brady, N. J. et al. Temporal evolution of cellular heterogeneity during the progression to advanced AR-negative prostate cancer. Nat. Commun. 12, 3372 (2021).
Rudin, C. M. et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat. Rev. Cancer 19, 289–297 (2019).
Wu, Q. et al. YAP drives fate conversion and chemoresistance of small cell lung cancer. Sci. Adv. 7, eabg1850 (2021).
Cejas, P. et al. Subtype heterogeneity and epigenetic convergence in neuroendocrine prostate cancer. Nat. Commun. 12, 5775 (2021).
Oser, M. G., MacPherson, D., Oliver, T. G., Sage, J. & Park, K. S. Genetically-engineered mouse models of small cell lung cancer: the next generation. Oncogene 43, 457–469 (2024).
Karami Fath, M. et al. PI3K/Akt/mTOR signaling pathway in cancer stem cells. Pathol. Res Pr. 237, 154010 (2022).
Yoshida, G. J. Emerging roles of Myc in stem cell biology and novel tumor therapies. J. Exp. Clin. Cancer Res. 37, 173 (2018).
Vizan, P., Beringer, M., Ballare, C. & Di Croce, L. Role of PRC2-associated factors in stem cells and disease. FEBS J. 282, 1723–1735 (2015).
Weindorf, S. C. et al. Metastatic castration resistant prostate cancer with squamous cell, small cell, and sarcomatoid elements-a clinicopathologic and genomic sequencing-based discussion. Med. Oncol. 36, 27 (2019).
Zhu, Z. et al. Combined large cell neuroendocrine carcinoma, lung adenocarcinoma, and squamous cell carcinoma: a case report and review of the literature. J. Cardiothorac. Surg. 18, 254 (2023).
Zou, M. et al. Transdifferentiation as a mechanism of treatment resistance in a mouse model of castration-resistant prostate cancer. Cancer Discov. 7, 736–749 (2017).
Chen, C. C. et al. Temporal evolution reveals bifurcated lineages in aggressive neuroendocrine small cell prostate cancer trans-differentiation. Cancer Cell 41, 2066–2082.e9 (2023).
Storck, W. K. et al. The role of epigenetic change in therapy-induced neuroendocrine prostate cancer lineage plasticity. Front. Endocrinol. 13, 926585 (2022).
Gatenby, R. A. & Brown, J. S. The evolution and ecology of resistance in cancer therapy. Cold Spring Harb. Perspect. Med. 10, a040972 (2020).
Soria, J. C. et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N. Engl. J. Med. 378, 113–125 (2018).
Sorin, M. et al. Single-cell spatial landscapes of the lung tumour immune microenvironment. Nature 614, 548–554 (2023).
Wu, F. et al. Single-cell profiling of tumor heterogeneity and the microenvironment in advanced non-small cell lung cancer. Nat. Commun. 12, 2540 (2021).
Yamasaki, L. et al. Loss of E2F-1 reduces tumorigenesis and extends the lifespan of Rb1(+/-)mice. Nat. Genet. 18, 360–364 (1998).
Mandigo, A. C. et al. Novel oncogenic transcription fFactor cooperation in RB-deficient cancer. Cancer Res. 82, 221–234 (2022).
Sun, H. et al. E2f binding-deficient Rb1 protein suppresses prostate tumor progression in vivo. Proc. Natl. Acad. Sci. USA 108, 704–709 (2011).
Chellappan, S. P., Hiebert, S., Mudryj, M., Horowitz, J. M. & Nevins, J. R. The E2F transcription factor is a cellular target for the RB protein. Cell 65, 1053–1061 (1991).
Sellers, W. R. & Kaelin, W. G. Jr Role of the retinoblastoma protein in the pathogenesis of human cancer. J. Clin. Oncol. 15, 3301–3312 (1997).
Han, W. et al. RB1 loss in castration-resistant prostate cancer confers vulnerability to LSD1 inhibition. Oncogene 41, 852–864 (2022).
Bracken, A. P. et al. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 22, 5323–5335 (2003).
Bohrer, L. R., Chen, S., Hallstrom, T. C. & Huang, H. Androgens suppress EZH2 expression via retinoblastoma (RB) and p130-dependent pathways: a potential mechanism of androgen-refractory progression of prostate cancer. Endocrinology 151, 5136–5145 (2010).
Li, M. et al. LSD1 inhibition disrupts super-enhancer-driven oncogenic transcriptional programs in castration-resistant prostate cancer. Cancer Res. 83, 1684–1698 (2023).
Jin, Y. et al. Kdm1a promotes SCLC progression by transcriptionally silencing the tumor suppressor Rest. Biochem. Biophys. Res. Commun. 515, 214–221 (2019).
Mohammad, H. P. et al. A DNA hypomethylation signature predicts antitumor activity of LSD1 inhibitors in SCLC. Cancer Cell 28, 57–69 (2015).
Augert, A. et al. Targeting NOTCH activation in small cell lung cancer through LSD1 inhibition. Sci. Signal. 12, eaau2922 (2019).
Nguyen, E. M. et al. Targeting lysine-specific demethylase 1 rescues major histocompatibility complex class I antigen presentation and overcomes programmed death-ligand 1 blockade resistance in SCLC. J. Thorac. Oncol. 17, 1014–1031 (2022).
Hiatt, J. B. et al. Inhibition of LSD1 with bomedemstat sensitizes small cell lung cancer to immune checkpoint blockade and T-cell killing. Clin. Cancer Res. 28, 4551–4564 (2022).
Rudin, C. M. et al. Clinical benefit from immunotherapy in patients with SCLC is associated with tumor capacity for antigen presentation. J. Thorac. Oncol. 18, 1222–1232 (2023).
Gardner, E. E. et al. Chemosensitive relapse in small cell lung cancer proceeds through an EZH2-SLFN11 axis. Cancer Cell 31, 286–299 (2017).
Choudhury, N. J. et al. A phase I/II study of Valemetostat (DS-3201b), an EZH1/2 inhibitor, in combination with irinotecan in patients with recurrent small-cell lung cancer. Clin. Cancer Res. 30, 3697–3703 (2024).
Cyrta, J. et al. Role of specialized composition of SWI/SNF complexes in prostate cancer lineage plasticity. Nat. Commun. 11, 5549 (2020).
Redin, E. et al. SMARCA4 controls state plasticity in small cell lung cancer through regulation of neuroendocrine transcription factors and REST splicing. J. Hematol. Oncol. 17, 58 (2024).
Caeser, R. et al. MAPK pathway activation selectively inhibits ASCL1-driven small cell lung cancer. iScience 24, 103224 (2021).
Tubbs, A. & Nussenzweig, A. Endogenous DNA damage as a source of genomic instability in cancer. Cell 168, 644–656 (2017).
Cybulla, E. & Vindigni, A. Leveraging the replication stress response to optimize cancer therapy. Nat. Rev. Cancer 23, 6–24 (2023).
da Costa, A., Chowdhury, D., Shapiro, G. I., D’Andrea, A. D. & Konstantinopoulos, P. A. Targeting replication stress in cancer therapy. Nat. Rev. Drug Discov. 22, 38–58 (2023).
Bhamidipati, D. & Subbiah, V. Lurbinectedin, a DNA minor groove inhibitor for neuroendocrine neoplasms beyond small cell lung cancer. Oncoscience 10, 22–23 (2023).
Doerr, F. et al. Targeting a non-oncogene addiction to the ATR/CHK1 axis for the treatment of small cell lung cancer. Sci. Rep. 7, 15511 (2017).
Thomas, A. et al. Therapeutic targeting of ATR yields durable regressions in small cell lung cancers with high replication stress. Cancer Cell 39, 566–579.e7 (2021).
Schultz, C. W. et al. ATR inhibition augments the efficacy of lurbinectedin in small-cell lung cancer. EMBO Mol. Med. 15, e17313 (2023).
Pietanza, M. C. et al. Randomized, double-blind, phase II study of Temozolomide in combination with either Veliparib or Placebo in patients with relapsed-sensitive or refractory small-cell lung cancer. J. Clin. Oncol. 36, 2386–2394 (2018).
Byers, L. A. et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov. 2, 798–811 (2012).
Takahashi, N. et al. Berzosertib plus Topotecan vs Topotecan alone in patients with relapsed small cell lung cancer: a randomized clinical trial. JAMA Oncol. 9, 1669–1677 (2023).
Farago, A. F. et al. Combination olaparib and temozolomide in relapsed small-cell lung cancer. Cancer Discov. 9, 1372–1387 (2019).
Rickman, D. S., Schulte, J. H. & Eilers, M. The expanding world of N-MYC-driven tumors. Cancer Discov. 8, 150–163 (2018).
Ellwood-Yen, K. et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell 4, 223–238 (2003).
Iwata, T. et al. MYC overexpression induces prostatic intraepithelial neoplasia and loss of Nkx3.1 in mouse luminal epithelial cells. PLoS One 5, e9427 (2010).
Madden, S. K., de Araujo, A. D., Gerhardt, M., Fairlie, D. P. & Mason, J. M. Taking the Myc out of cancer: toward therapeutic strategies to directly inhibit c-Myc. Mol. Cancer 20, 3 (2021).
Soucek, L. et al. Modelling Myc inhibition as a cancer therapy. Nature 455, 679–683 (2008).
Garralda, E. et al. MYC targeting by OMO-103 in solid tumors: a phase 1 trial. Nat. Med. 30, 762–771 (2024).
Beaulieu, M. E. et al. Intrinsic cell-penetrating activity propels Omomyc from proof of concept to viable anti-MYC therapy. Sci. Transl. Med. 11, eaar5012 (2019).
Speltz, T. E. et al. Targeting MYC with modular synthetic transcriptional repressors derived from bHLH DNA-binding domains. Nat. Biotechnol. 41, 541–551 (2023).
Fiorentino, F. P. et al. Growth suppression by MYC inhibition in small cell lung cancer cells with TP53 and RB1 inactivation. Oncotarget 7, 31014–31028 (2016).
Masso-Valles, D., Beaulieu, M. E. & Soucek, L. MYC, MYCL, and MYCN as therapeutic targets in lung cancer. Expert Opin. Ther. Targets 24, 101–114 (2020).
Romero, O. A. et al. MAX inactivation in small cell lung cancer disrupts MYC-SWI/SNF programs and is synthetic lethal with BRG1. Cancer Discov. 4, 292–303 (2014).
Augert, A. et al. MAX functions as a tumor suppressor and rewires metabolism in small cell lung cancer. Cancer Cell 38, 97–114.e117 (2020).
Pearson, J. D. et al. Binary pan-cancer classes with distinct vulnerabilities defined by pro- or anti-cancer YAP/TEAD activity. Cancer Cell 39, 1115–1134.e2 (2021).
Awad, M. M. et al. Acquired resistance to KRAS(G12C) inhibition in cancer. N. Engl. J. Med. 384, 2382–2393 (2021).
Mathew, D. et al. Combined JAK inhibition and PD-1 immunotherapy for non-small cell lung cancer patients. Science 384, eadf1329 (2024).
Choudhury, A. D. PTEN-PI3K pathway alterations in advanced prostate cancer and clinical implications. Prostate 82, S60–S72 (2022).
Jamaspishvili, T. et al. Clinical implications of PTEN loss in prostate cancer. Nat. Rev. Urol. 15, 222–234 (2018).
Morgan, T. M., Koreckij, T. D. & Corey, E. Targeted therapy for advanced prostate cancer: inhibition of the PI3K/Akt/mTOR pathway. Curr. Cancer Drug Targets 9, 237–249 (2009).
Courtney, K. D. et al. A phase I study of everolimus and docetaxel in patients with castration-resistant prostate cancer. Clin. Genitourin. Cancer 13, 113–123 (2015).
George, D. J. et al. Phase 2 clinical trial of TORC1 inhibition with everolimus in men with metastatic castration-resistant prostate cancer. Urol. Oncol. 38, 79.e15–79.e22 (2020).
Hotte, S. J. et al. A phase II study of PX-866 in patients with recurrent or metastatic castration-resistant prostate cancer: Canadian Cancer Trials Group Study IND205. Clin. Genitourin. Cancer 17, 201–208.e1 (2019).
Nakabayashi, M. et al. Phase II trial of RAD001 and bicalutamide for castration-resistant prostate cancer. BJU Int. 110, 1729–1735 (2012).
Rescigno, P. et al. Capivasertib in combination with enzalutamide for metastatic castration resistant prostate cancer after docetaxel and abiraterone: Results from the randomized phase II RE-AKT trial. Eur. J. Cancer 205, 114103 (2024).
Chen, J. et al. Genomic alterations in neuroendocrine prostate cancer: a systematic review and meta-analysis. BJUI Compass 4, 256–265 (2023).
Grazini, U. et al. Overcoming Osimertinib resistance with AKT inhibition in EGFRm-driven non-small cell lung cancer with PIK3CA/PTEN alterations. Clin. Cancer Res. 30, 4143–4154 (2024).
You, I. et al. Discovery of an AKT degrader with prolonged inhibition of downstream signaling. Cell Chem. Biol. 27, 66–73.e7 (2020).
Saunders, L. R. et al. A DLL3-targeted antibody-drug conjugate eradicates high-grade pulmonary neuroendocrine tumor-initiating cells in vivo. Sci. Transl. Med. 7, 302ra136 (2015).
Puca, L. et al. Delta-like protein 3 expression and therapeutic targeting in neuroendocrine prostate cancer. Sci. Transl. Med. 11, eaav0891 (2019).
Pourquie, O. & Kusumi, K. When body segmentation goes wrong. Clin. Genet. 60, 409–416 (2001).
Blackhall, F. et al. Efficacy and safety of rovalpituzumab tesirine compared with topotecan as second-line therapy in DLL3-High SCLC: results from the phase 3 TAHOE study. J. Thorac. Oncol. 16, 1547–1558 (2021).
Rudin, C. M. et al. Emerging therapies targeting the delta-like ligand 3 (DLL3) in small cell lung cancer. J. Hematol. Oncol. 16, 66 (2023).
Ajkunic, A. et al. Assessment of TROP2, CEACAM5 and DLL3 in metastatic prostate cancer, expression landscape and molecular correlates. NPJ Precis. Oncol. 8, 104 (2024).
Tendler, S. et al. Imaging with [(89)Zr]Zr-DFO-SC16.56 anti-DLL3 antibody in patients with high-grade neuroendocrine tumours of the lung and prostate, a phase 1/2, first-in-human trial. Lancet Oncol. 25, 1015–1024 (2024).
Quintanal-Villalonga, A. et al. Inhibition of XPO1 sensitizes small cell lung cancer to first- and second-line chemotherapy. Cancer Res. 82, 472–483 (2022).
Quintanal-Villalonga, A. et al. Exportin 1 inhibition prevents neuroendocrine transformation through SOX2 down-regulation in lung and prostate cancers. Sci. Transl. Med. 15, eadf7006 (2023).
Zhang, B. et al. Brief report: Comprehensive clinicogenomic profiling of small cell transformation from EGFR-mutant NSCLC informs potential therapeutic targets. JTO Clin. Res. Rep. 5, 100623 (2024).
Feustel, K., Martin, J. & Falchook, G. S. B7-H3 inhibitors in oncology clinical trials: a review. J. Immunother. Precis. Oncol. 7, 53–66 (2024).
Yamato, M. et al. DS-7300a, a DNA topoisomerase I inhibitor, DXd-based antibody-drug conjugate targeting B7-H3, exerts potent antitumor activities in preclinical models. Mol. Cancer Ther. 21, 635–646 (2022).
Kemble, J., Kwon, E. D. & Karnes, R. J. Addressing the need for more therapeutic options in neuroendocrine prostate cancer. Expert Rev. Anticancer Ther. 23, 177–185 (2023).
DeLucia, D. C. et al. Regulation of CEACAM5 and therapeutic efficacy of an anti-CEACAM5-SN38 antibody-drug conjugate in neuroendocrine prostate cancer. Clin. Cancer Res. 27, 759–774 (2021).
Thomas, P. L. et al. Beyond programmed death-ligand 1: B7-H6 emerges as a potential immunotherapy target in SCLC. J. Thorac. Oncol. 16, 1211–1223 (2021).
Wiedemeyer, W. R. et al. ABBV-011, a novel, Calicheamicin-based antibody-drug conjugate, targets SEZ6 to eradicate small cell lung cancer tumors. Mol. Cancer Ther. 21, 986–998 (2022).
Akbulut, D. et al. Differential NEUROD1, ASCL1, and POU2F3 expression defines molecular subsets of bladder small cell/neuroendocrine carcinoma with prognostic implications. Mod. Pathol. 37, 100557 (2024).
Marcoux, N. et al. EGFR-mutant adenocarcinomas that transform to small-cell lung cancer and other neuroendocrine carcinomas: clinical outcomes. J. Clin. Oncol. 37, 278–285 (2019).
Foster, N. R. et al. Tumor response and progression-free survival as potential surrogate endpoints for overall survival in extensive stage small-cell lung cancer, findings on the basis of North Central Cancer Treatment Group trials. Cancer 117, 1262–1271 (2011).
Huang, J. et al. Genomic and transcriptomic analysis of neuroendocrine transformation in ALK-rearranged lung adenocarcinoma after treatments with sequential ALK inhibitors: a brief report. JTO Clin. Res. Rep. 3, 100338 (2022).
Dimou, A., Lo, Y. C., Merrell, K. W., Halling, K. C. & Mansfield, A. S. Small cell transformation in a patient with RET fusion-positive lung adenocarcinoma on pralsetinib. JCO Precis. Oncol. 6, e2200478 (2022).
Lee, J. K. et al. N-Myc drives neuroendocrine prostate cancer initiated from human prostate epithelial cells. Cancer Cell 29, 536–547 (2016).
Wood, K. J., Issa, F. & Hester, J. Understanding stem cell immunogenicity in therapeutic applications. Trends Immunol. 37, 5–16 (2016).
Qiao, Y., Agboola, O. S., Hu, X., Wu, Y. & Lei, L. Tumorigenic and immunogenic properties of induced pluripotent stem cells: a promising cancer vaccine. Stem Cell Rev. Rep. 16, 1049–1061 (2020).
Zhao, T., Zhang, Z. N., Rong, Z. & Xu, Y. Immunogenicity of induced pluripotent stem cells. Nature 474, 212–215 (2011).
Vassella, E. et al. Molecular profiling of lung adenosquamous carcinoma: hybrid or genuine type? Oncotarget 6, 23905–23916 (2015).
Baca, S. C. et al. Liquid biopsy epigenomic profiling for cancer subtyping. Nat. Med. 29, 2737–2741 (2023).
Chakraborty, G., Gupta, K. & Kyprianou, N. Epigenetic mechanisms underlying subtype heterogeneity and tumor recurrence in prostate cancer. Nat. Commun. 14, 567 (2023).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 6, pl1 (2013).
de Bruijn, I. et al. Analysis and visualization of longitudinal genomic and clinical data from the AACR project GENIE biopharma collaborative in cBioPortal. Cancer Res. 83, 3861–3867 (2023).
Cheng, D. T. et al. Memorial sloan kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J. Mol. Diagn. 17, 251–264 (2015).
Liu, J. et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 173, 400–416.e11 (2018).
Acknowledgements
This work was supported or partially supported by the National Cancer Institute/National Institutes of Health (R37CA258730, R01CA288820, R01CA292949 to P.M.), Department of Defense (W81XWH21-1-0520 to P.M., W81XWH2110418 to X.L.), Cancer Prevention Research Institute (CPRIT; RP220473 to P.M.), and Prostate Cancer Foundation (17YOUN12 to P.M.). This study was supported by the NIH T32 CA1600001 (A.Q.V.), by the American Lung Association (A.Q.V.), by Druckenmiller Center for Lung Cancer Research (A.Q.V.). This study was also supported by the Regional Ministry of Health and Consume of Andalucía RC-0004-2020 (S.M.P.) and the Carlos III Health Institute through the projects “PI20/01109 and PI23/01679” (co-funded by European Regional Development Fund/European Social Fund; “A way to make Europe”/“Investing in your future”) (S.M.P.). We acknowledge the use of the PPBC Biobank, Pathology Core Facility, and Integrated Genomics Operation Core, funded by the NCI Cancer Center Support Grant (P30 CA008748), Cycle for Survival, and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology.
Author information
Authors and Affiliations
Contributions
A.Q.V. conceived the manuscript. X.L., E.E.G., P.M. and A.Q.V. collected literature. X.L., E.E.G., S.M.P., C.W., P.M. and A.Q.V. wrote the manuscript. A.Q.V., C.W. and P.M. edited the manuscript, and A.Q.V. prepared the figures.
Corresponding author
Ethics declarations
Competing interests
P.M. served as a scientific consultant to Accutar Biotechnology, Inc. A.Q.V. has received honoraria from AstraZeneca and research funding from Jazz Pharmaceuticals, Foghorn Therapeutics and Duality Biologicals. A.Q.V. is a current employee of AstraZeneca.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, X., Gardner, E.E., Molina-Pinelo, S. et al. Lineage plasticity and histological transformation: tumor histology as a spectrum. Cell Res 35, 803–823 (2025). https://doi.org/10.1038/s41422-025-01180-x
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41422-025-01180-x
This article is cited by
-
KMT2D loss drives adeno-to-squamous transition and sensitizes TKI-resistant lung cancer to AURKA inhibition
Cell Death & Differentiation (2026)
-
Lineage plasticity: a new dilemma in lung cancer treatment
Molecular Biology Reports (2026)