Abstract
The integrated stress response (ISR) enables cells to cope with a variety of insults, but its specific contribution to downstream cellular outputs remains unclear. Using a synthetic tool, we selectively activate the ISR without co-activation of parallel pathways and define the resulting cellular state with multi-omics profiling. We identify time- and dose-dependent gene expression modules, with ATF4 driving only a small but sensitive subgroup that includes amino acid metabolic enzymes. This ATF4 response affects cellular bioenergetics, rerouting carbon utilization towards amino acid production and away from the tricarboxylic acid cycle and fatty acid synthesis. We also find an ATF4-independent reorganization of the lipidome that promotes DGAT-dependent triglyceride synthesis and accumulation of lipid droplets. While DGAT1 is the main driver of lipid droplet biogenesis, DGAT2 plays an essential role in buffering stress and maintaining cell survival. Together, we demonstrate the sufficiency of the ISR in promoting a previously unappreciated metabolic state.
Similar content being viewed by others
Introduction
In response to environmental challenges as diverse as viral infection, mitochondrial dysfunction, amino acid deprivation, or the accumulation of misfolded proteins, eukaryotic cells engage a common signaling mechanism called the Integrated Stress Response (ISR)1,2. This highly conserved pathway limits the rate of protein synthesis while simultaneously inducing a stress-responsive gene expression program (Fig. 1a). Four kinases (PERK, HRI, GCN2, and PKR) detect a variety of insults and trigger the ISR through a shared mechanism, the phosphorylation of the alpha subunit of eukaryotic translation initiation factor 2 (eIF2)3. eIF2 is a central participant in mRNA translation, bringing the methionyl-tRNA ternary complex (eIF2-GTP-Met tRNAi) to the ribosome to initiate protein synthesis. Upon initiation of translation, GTP is hydrolyzed. eIF2 is then recharged with GTP to enable a new round of initiation by its own dedicated guanine nucleotide exchange factor (GEF), eIF2B4,5. When eIF2ɑ is phosphorylated on serine 51 by any of the four eIF2 kinases, it binds to eIF2B through a different interface, inhibiting its enzymatic activity6,7. This decrease in GEF activity results in reduced mRNA translation and constitutes a major brake on protein synthesis in stressed cells.
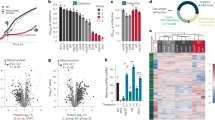
a Schematic illustrating the inputs and outputs of the ISR. The ISR incorporates input from various cellular stress sensors to increase the levels of p-eIF2, the central input into the pathway. Its major outputs include a reduction in protein synthesis, the formation of SGs, and the induction of a specialized gene expression program that is in part mediated by the translational induction of the transcription factor ATF4. b Schematic illustrating the pharmacogenetic Dmr-PERK tool. Upon addition of a small molecule dimerizer, Dmr-PERK phosphorylates eIF2 and activates the ISR. c AlphaLISA measurement of the ratio of p-eIF2/eIF2 in Dmr-PERK U2OS cells following 2 h of dimerizer treatment at the indicated concentrations. Error bars show mean ± SD of three technical replicates. d Quantification of OP-Puromycin incorporation (left axis) and G3BP1-positive SGs (right axis) co-labeled in Dmr-PERK cells following 4 h of dimerizer treatment at the indicated concentration. Error bars depict mean ± SD of three technical replicates e Heatmap of protein coding gene expression in untreated (UT) Dmr-PERK cells or in cells treated with dimerizer (Dmr, 0.2 nM), thapsigargin (Tg, 100 nM), or arsenite (Ars, 0.05 mM), for the indicated amount of time. Columns show the mean expression of three replicates relative to the mean expression at t = 0 on a log2 scale. f Heatmap of genes described in e that have a significantly different time-dependent response between dimerizer (Dmr) and thapsigargin (Tg) treatments. Selected GO terms with significant enrichment by ORA within the groups of Tg-responsive genes are indicated. g Heatmap of genes described in e that have a significantly different time-dependent response between dimerizer (Dmr) and arsenite (Ars) treatments. Selected GO terms with significant enrichment by ORA within the groups of arsenite-responsive genes are indicated. a, b are created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license https://creativecommons.org/licenses/by-nc-nd/4.0/deed.en. Source data are provided as a Source Data file.
Concomitant with the general inhibition of protein synthesis, the ISR facilitates the translation of a subset of specialized mRNAs. These transcripts carry short upstream open reading frames (uORFs) in their 5’ untranslated region (5’UTR) that inhibit translation of the main open reading frame (ORF) when levels of ternary complex are high but result in translation of the ORF when ternary complex is low. A key transcript regulated in this manner encodes ATF4, a bZIP transcription factor that induces the expression of a wide range of genes, including metabolic genes that promote amino acid accumulation and glutathione synthesis2,8,9,10,11. ATF4 is a constitutively expressed transcript across all cell types12 and thus, its translation is a first line of defense when cells encounter an unfavorable environment in mammalian tissues. Other uORF containing mRNAs include DDIT3, which encodes the transcription factor CHOP (C/EBP-homologous protein) and is thought to promote apoptosis during persistent ISR activation13,14, and GADD34, a regulatory subunit of the phosphatase PP1, which dephosphorylates eIF2 and acts as an important negative feedback loop to derepress translation initiation15,16. The reduction in translation initiation also leads to the formation of stress granules (SG), membraneless organelles that are formed by the cytosolic assembly of untranslated mRNAs and their RNA-binding proteins17,18,19.
While these molecular and cellular outputs of the ISR are well documented, it is not known how they are coordinated in response to the strength and duration of input to the pathway. Given that timing and signal intensity can determine whether the ISR promotes cell survival or death, it is critical to define this input-output relationship1. Characterization of the outputs of the ISR is complicated by the fact that it is typically activated as part of a broader cellular response to stressors that engage multiple signaling pathways. For example, the accumulation of unfolded proteins in the ER activates a network of signaling pathways collectively termed the unfolded protein response (UPR). In addition to activating the eIF2 kinase PERK, unfolded proteins induce two other transmembrane ER sensors, IRE1 and ATF6 that trigger their respective signaling programs20,21. Similarly, studies on ISR-dependent metabolic responses typically rely on hypoxia, nutrient stresses or mitochondrial perturbations, which trigger multiple response pathways that have pleiotropic metabolic consequences22,23,24,25,26.
To address these challenges, we generated a cell line expressing a synthetic construct27,28,29 that allowed us to selectively initiate the ISR in a tunable fashion. We harnessed this system to quantitatively explore the transcriptome, metabolome, and lipidome over time and define ISR-specific and -sufficient responses. We found that ATF4, in addition to its well-documented role in promoting amino acid and glutathione synthesis, inhibits carbon utilization by the TCA cycle and promotes the reductive carboxylation of glutamine-derived α-ketoglutarate to support amino acid synthesis. We also found that the ISR was sufficient to drive an ATF4-independent reorganization of cellular lipid content that promotes triglyceride (TG) and cholesterol ester (CE) accumulation in lipid droplets (LDs). Although TG synthesis by DGAT1 was the main contributor to LD formation, we found that DGAT2 was required to buffer ISR activity and maintain cell viability. These responses occurred early and at low levels of pathway input, and thus, we propose that a primary output of the ISR is a metabolic state that acts on amino acid, central carbon and lipid metabolism.
Results
Dmr-PERK provides specific and tunable control of ISR activation
To specifically profile the cellular effects of ISR signaling, we generated a U2OS-derived cell line stably expressing a synthetic construct, dimerizable PERK (Dmr-PERK), consisting of the cytosolic eIF2ɑ kinase domain-containing portion of mouse PERK fused to a chemically inducible DmrB dimerization domain28. Upon addition of ligand AP20187 (dimerizer), the fusion protein dimerizes, leading to its activation and phosphorylation of eIF2ɑ (Fig. 1b). These cells showed a low baseline level of p-eIF2 that increased with dose-dependent addition of dimerizer (Fig. 1c) and initiated canonical downstream signaling events. Inhibition of protein synthesis, as measured by OP-Puromycin incorporation, was dose-dependent and sensitive to Dmr-PERK activation, occurring at even the lowest concentrations of dimerizer (0.04 nM) (Fig. 1d). In contrast, accumulation of SGs, as detected by anti-G3BP1 staining, was only measurable at much higher doses (EC50 of 4.7 nM) (Fig. 1d), consistent with visible G3BP1-positive structures only appearing when inhibition of mRNA translation is significantly reduced18. Translation of uORF-containing transcripts was also sensitive to minimal levels of Dmr-PERK activation, with measurable accumulation of ATF4, GADD34, and CHOP protein at low concentration (0.01 nM), and further increases with higher doses (Supplementary Fig. 1a, b). Protein expression of ATF4 increased within 1 h of Dmr-PERK activation, even at low doses of dimerizer (0.04 nM), likely reflecting the rapid translation of pre-existing mRNA species (Supplementary Fig. 1c). This tunable system therefore allows dose- and time-dependent control of ISR activation.
To evaluate how the transcriptional response elicited by this minimal system compared to that of commonly used pleiotropic ISR-inducing agents, we performed RNA-seq on Dmr-PERK cells treated with a time course of dimerizer (0.2 nM), thapsigargin (100 nM), a SERCA inhibitor that induces ER stress by depleting its Ca++ stores, or sodium arsenite (0.05 mM), which generates ROS and causes oxidative stress. For each compound, we chose a dose that elicited comparable levels of p-eIF2ɑ (Supplementary Fig. 1d). Consistent with more targeted ISR activation, triggering Dmr-PERK induced only a subset of the transcriptional changes elicited by arsenite or thapsigargin (Fig. 1e). This included canonical genes associated with ISR activation8 and the ISR target gene signature of coherently co-expressed genes (ISR GeneCLIC signature), as defined from unbiased, tissue-independent Clustering by Inferred Co-Expression analysis by Wong et al.30,31 (Supplementary Fig. 1e, Supplementary Data 1). Of the 89 genes with a significantly different response to dimerizer as compared to thapsigargin (Fig. 1f), Gene Ontology Biological Process terms associated with the UPR were uniquely enriched by over-representation analysis with thapsigargin treatment, as was expression of known target genes of the ATF6 and IRE1 branches of the UPR32 (Supplementary Fig. 1f), demonstrating that these pathways were not engaged by Dmr-PERK activation. Similarly, 1686 genes were uniquely regulated with arsenite treatment, including genes involved in protein folding, chaperone pathways and non-coding RNA processing (Fig. 1g, Supplementary Data 1). Dimerizer treatment on parental cells that do not carry the Dmr-PERK construct had no significant transcriptional effects over untreated cells (Supplementary Fig. 1g). These results establish that this pharmacogenomic tool elicits a targeted ISR, enabling us to specifically assess downstream cellular events without the contribution of the parallel pathways engaged by pleiotropic stressors.
Transcriptional targets of the ISR show distinct responses to the strength and duration pathway input
ISR signaling helps cells adapt to changes in their environment but severe and/or chronic stress can trigger cell death pathways and impair physiological processes1,28,30,33. To determine how the transcriptional outputs of the ISR vary with time (duration) and input strength (stress level), we performed RNA-seq on cells treated for 1, 2, 4, 8, 16, or 24 h with one of three doses of dimerizer. We chose a low dose of dimerizer (0.01 nM) where protein synthesis inhibition would be minimal (<15%) but ATF4 protein expression was still induced, an intermediate dose (0.2 nM) that caused a partial (50%) reduction in protein synthesis, and a high dose (3 nM) that led to the strongest (75%) reduction in protein synthesis and accumulation of SGs (Fig. 1d, Supplementary Fig. 1a). To analyze the gene expression time course data, we first normalized the expression of each gene to its average expression at t = 0, filtered for genes that showed a log2 fold change of 1.5 or greater at any time point, and then identified genes with a statistically significant time-dependent response to any dose of dimerizer treatment. Genes with a significant time-dependent response were grouped using hierarchical clustering, revealing four classes of genes with distinct dynamic behaviors (Fig. 2a, b, Supplementary Data 2). A representative response pattern for each class is illustrated in Fig. 2c.
a Heatmap of protein coding genes with time-dependent expression changes in Dmr-PERK cells treated with the indicated concentration of dimerizer for the indicated amount of time. Columns show the mean expression of three technical replicates relative to the mean expression at t = 0 on a log2 scale. Classes describe groups of genes clustered by their dose- and time-dependent responses b Selected GO terms with significant enrichment by ORA of the genes within each class. There were no significant enriched pathways among Class 3 genes, but the PERK-mediated unfolded protein response term identified transcripts exhibiting unique behavior. c Plot of log2 transcripts per million (TPM) over time at the indicated doses of dimerizer for genes representative of the classes described in a. GOT1 is a representative Class 1 gene, IL4I1 is a representative Class 2 gene, IL1RL1 is a representative Class 3 gene, and PCDH18 is a representative Class 4 gene.
The first class (Class 1) was characterized by genes with a rapid (2–4 h) and sustained upregulation at all 3 doses of dimerizer (Fig. 2b, c). Significantly enriched GO pathways included genes related to non-coding RNA processing (ncRNA metabolic process, ncRNA processing), ribosome biogenesis (ribosome biogenesis, rRNA metabolic process), and tRNA metabolism (tRNA modification, tRNA metabolic process). The ISR target genes in amino acid metabolism8,30 were also among the significantly induced GO pathways including “amino acid import” (ATP1A2, SLC47A1, SLC7A2, SLC1A6, SLC7A5, SLC6A14, SLC3A2, SLC7A1, SLC25A38, SLC6A9, SLC1A4, SLC1A5, SLC7A11) and “carboxylic acid biosynthetic process” which included enzymes involved in synthesis of non-essential amino acids including serine (PSAT1, PSPH, PHGDH), asparagine (ASNS), cysteine (CBS, CTH, CBSL), proline (PYCR1), and alanine (GPT2) and transaminases (GOT1, GPT2, BCAT1) (Supplementary Data 2). Class 1 comprises the largest number of genes that are upregulated by this minimal ISR-inducing system and represents the earliest and most sensitive ISR gene-expression outputs. Class I gene expression was inhibited when Dmr-PERK was activated in the presence of the ISR inhibitor ISRIB, which allosterically antagonizes phospho-eIF2ɑ and restores translation initiation34,35,36, confirming that this transcriptional response is a consequence of reduced ternary complex (Supplementary Fig. 2a).
A second and smaller class (Class 2) of upregulated genes showed a pulse-like behavior. Gene expression in this cluster peaked between 4 and 8 h but returned to baseline by 24 h (Fig. 2b, c). The gene sets overrepresented in this class related to immune signaling, including interleukin-4 mediated signaling (IL2RG, JAK3, IL4R) and TNF signaling pathways (VCAM1, NFKB1, CLDN1, SPHK1, TRAF3, TNFRSF9, EDARADD, BIRC3, TNFRSF21, TRAF1, GBP3), and response to osmotic stress (CLDN1, ICOSLG, ABCB1, RELB, EGFR, RCSD1) (Supplementary Data 2). These acute responders were also upregulated at low levels of pathway input, but their rapid downregulation suggests that their prolonged expression may not be adaptive.
In contrast to Classes 1 and 2, genes in a third class (Class 3) were induced only at higher doses of dimerizer (Fig. 2b, c). While there were no significantly enriched pathways among this class, we found transcripts encoding two major effectors of the ISR: PPP1R15A, encoding for GADD34, and DDIT3, encoding CHOP (Supplementary Fig. 2b, c, Supplementary Data 2). Although these transcripts are only induced when protein synthesis is strongly reduced, both PPP1R15A and DDIT3 contain a uORF that allows the rapid translation of their baseline pre-existing mRNA, even at low levels of Dmr-PERK activation2,37.
Finally, the fourth and largest class (Class 4) consisted of genes that were downregulated over time proportionally to dimerizer dose (Fig. 2b, c). The timing of the downregulation of Class 4 targets varied but was generally initiated at later time points compared to the upregulation of genes in Class 1. Associated GO terms included those related to cell adhesion and extracellular matrix pathways, as well as several pathways related to developmental signaling and differentiation and may reflect the slowing of cell growth processes with prolonged ISR activation. Together, this analysis revealed that the gene expression changes downstream of ISR activation exhibit distinct profiles, both in terms of their dynamics and in response to the strength of the input. These modules suggest that multiple control mechanisms coordinate the transcriptional network downstream of the ISR and that the dynamics of these outputs is important in defining downstream cellular responses.
ATF4-dependent transcriptional targets are early responders and enriched for metabolic regulators
Transcriptional profiling revealed an ISR gene-expression program that reflects the duration and strength of stress input. Because ATF4 is a central ISR transcription factor that quickly accumulates even at very low levels of pathway input (Supplementary Fig. 1a) we assessed the contribution of ATF4 to the transcriptional response elicited by Dmr-PERK. We used CRISPR to knock out the ATF4 gene from Dmr-PERK cells (ATF4 KO) and isolated homozygous mutant clones (Supplemental Fig. 3a). As expected, dimerizer induction of p-eIF2ɑ and inhibition of puromycin incorporation, which are upstream events, was not compromised in these cells (Supplementary Fig. 3b, c). We also observed the basal gene expression changes reported in other ATF4-null cell types8,38 (Supplementary Data 3) and rescued the resulting impaired cell growth by supplementation with nonessential amino acids and β-mercaptoethanol8. Accordingly, all comparisons between WT and ATF4 KO cells were performed in supplemented medium.
We compared the transcriptional response of WT and ATF4 KO cells after 2, 4, 8, 16, or 24 h of dimerizer treatment at the intermediate concentration (0.2 nM) that resulted in a partial (50%) inhibition of protein synthesis and elicited induction of all four classes of genes in WT cells (Figs. 1d, 2). Surprisingly, the majority of transcriptional changes still occurred in ATF4 KO cells, including upregulation of ncRNA processing pathway genes in Class 1, and the majority of genes in Classes 2, 3, and 4. Of the 3487 genes that were significantly changed over time, only 94 were significantly altered in ATF4 KO cells. Clustering of these ATF4-dependent genes showed that the majority were upregulated with dimerizer treatment in WT cells (Fig. 3a) and were enriched for GO terms related to amino acid synthesis and import, including “cellular amino acid biosynthetic process”, “serine family amino acid metabolic process,” and “amino acid import” (Fig. 3b and Supplementary Data 3). The majority (75%) of the ATF4-dependent genes were from Class 1, which we confirmed in a second independently derived clone (Supplementary Fig. 3d). Thus, ATF4 is responsible for only a very small subset of the transcriptional changes that ensued upon ISR activation but drives one of the earliest and most sensitive transcriptional modules (8% of all upregulated genes) that is characterized by amino acid metabolism.
a Heatmap of protein coding gene expression changes in WT and ATF4 KO Dmr-PERK cells at the indicated time points following treatment with dimerizer (0.2 nM). Genes shown have a significantly altered time course between the two genotypes. Columns show the mean expression of three technical replicates relative to the mean expression at t = 0 on a log2 scale. Color bar indicates the class of each transcript based on the dose response study in Fig. 2. Gray boxes indicate genes that did not meet statistical significance in the dose response study from Fig. 2. b Selected GO terms with significant enrichment by ORA for the upregulated ATF4-dependent genes. Top 20 GO terms are in Supplementary Data 3.
ATF4 diverts carbon utilization away from energy production and towards amino acid metabolism
Gene-expression profiling showed that ISR activation led to a rapid and sustained upregulation of several metabolic enzymes and transporters (Class 1, Fig. 2), many of which were ATF4 dependent (Fig. 3). To determine how this transcriptional program impacts cellular metabolism, we performed a time course of untargeted metabolomic profiling of polar metabolites by tandem liquid chromatography-mass spectrometry (LC-MS) in both WT and ATF4 KO Dmr-PERK cells. Measurements were performed at 0.5, 4, 8, and 24 h of dimerizer treatment, which allowed us to capture any metabolic changes that occurred rapidly following Dmr-PERK activation, as well as at timepoints that correspond with the transcriptional analysis. To minimize gross metabolic alterations caused by reduced protein synthesis, we chose a low concentration of dimerizer (0.05 nM) that only mildly (<15%) inhibited bulk protein synthesis (Fig. 1d) but still induced ATF4 protein expression (Supplementary Fig. 1c), the transcription of Class 1 genes, and the ATF4-dependent metabolic module (Figs. 2, 3). We identified metabolites that changed significantly over time following Dmr-PERK activation and clustered these to reveal distinct responses between the two genotypes (Fig. 4a and Supplementary Data 4).
a Heatmap of significant metabolites (FDR q < 0.01) as measured by LC-MS at the indicated time points following addition of dimerizer (0.05 nM) to WT or ATF4 KO Dmr-PERK cells. Relative abundances are shown as row mean centered z-scores. b Heatmap of TCA cycle metabolites from a. c Schematic of incorporation of 13C-label from U-13C6-glucose into the TCA cycle and in the synthesis of fatty acids. d Bar plots representing fractional labeling of TCA cycle metabolites from U-13C6-glucose in WT and ATF4 KO cells treated with vehicle (Veh.) or dimerizer (Dmr, 0.05 nM) for 24 h. Error bars depict mean ± SD of three technical replicates. e Schematic of incorporation of 13C-label from U-13C5-glutamine from oxidation (blue) vs. reductive carboxylation (red). Bar plots representing fraction labeling of malate, Asn or Asp from oxidative (M + 4) (f) or reductive (M + 3) (g) metabolism of U-13C5-glutamine in WT and ATF4 KO cells treated with vehicle (Veh.) or dimerizer (Dmr, 0.05 nM) for 24 h. Error bars depict mean ± SD of three technical replicates. Bar plots representing the fraction of palmitate carbons labeled from U-13C5-glutamine (h) or U-13C6-glucose (i) in lipids from WT or ATF4 KO Dmr-PERK cells treated with vehicle (Veh.) or dimerizer (Dmr, 0.05 nM) for 24 h. Error bars depict mean ± SD of three technical replicates. Statistical significance was evaluated by two-way ANOVA followed by Tukey’s HSD test. Source data are provided as a Source Data file.
A first cluster of metabolites showed a rapid (4 h) and sustained increase over time (Fig. 4a, marked in red) and included most amino acids, both essential and non-essential, as well as intermediates in glutathione synthesis. The timing and content of this cluster suggests that it is driven by the upregulation of Class 1 genes, which includes both amino acid transporters and biosynthetic enzymes, including the transsulfuration pathway enzymes (Fig. 2a, b). This transcriptional module was ATF4-dependent and, as anticipated, accumulation of metabolites in this cluster was absent in ATF4 KO cells.
Amino acids in this cluster accumulated in an ATF4-dependent manner and were low in ATF4 KO cells, even at baseline (Supplementary Fig. 4a). A notable exception to the trend of increasing amino acid concentrations was aspartate, whose lack of accumulation is consistent with ATF4 transcriptional induction of asparagine synthetase (ASNS), which converts aspartate to asparagine39 (Fig. 3b). Because several enzymes in amino acid synthesis pathways were identified as ATF4-dependent Class 1 genes, including serine synthesis enzymes (Fig. 3b), we considered whether upregulation of these pathways may be contributing to the cellular accumulation of amino acids. To assess pathway activity, we cultured cells in media containing isotopically labeled glucose (U-13C6-glucose) and quantified the steady state incorporation of heavy carbon into metabolic intermediates by LC-MS after 24 h of dimerizer (0.05 nM) treatment (Supplementary Fig. 4b). Despite a culture media replete with serine, glucose labeling of serine was increased with Dmr-PERK activation in WT cells but remained nearly undetectable in ATF4 KO cells (Supplementary Fig. 4c).
ISR induction also led to an ATF4-dependent increase in the abundance of γ-glutamylcysteine in this cluster (Supplementary Fig. 4d). This is indicative of enhanced synthesis of glutathione, the most abundant antioxidant in cells, and is consistent with the regulation of the transsulfuration pathway enzymes CBS and CTH by ATF4 (Supplementary Data 3)8,25. We determined whether glutathione synthesis was also upregulated by Dmr-PERK activation by culturing cells with isotopically labeled glutamine (13C5-glutamine), a primary precursor to glutathione (Supplementary Fig. 4e). We found that, indeed, metabolic flux from glutamate to glutathione was increased ~50% (FDmr/FVeh = 1.46) with ISR activation in WT cells but was unaffected in ATF4 KO cells (Supplementary Fig. 4f). Together, these results demonstrate that the ISR is sufficient to drive an early ATF4-dependent increase in amino acid and glutathione synthesis, a result in agreement with the gene expression analysis and with previous studies using pleiotropic stressors8,26.
In parallel to the accumulation of amino acids, a second cluster of metabolites rapidly (4 h) decreased upon ISR activation (Fig. 4a, marked in green). This class was composed primarily of TCA cycle intermediates (Fig. 4b), suggesting a possible slowing of the TCA cycle with ISR induction. Surprisingly, this cluster was also ATF4-dependent, even though we did not detect any corresponding transcriptional changes. To test whether depletion of these metabolites was due to reduced oxidation of glucose, we measured U-13C6-glucose labeling of TCA cycle intermediates following Dmr-PERK activation (Fig. 4c) and observed a reduced fraction of α-ketoglutarate, malate, citrate and acetyl-CoA labeling in WT but not ATF4 KO cells (Fig. 4d). Furthermore, glucose labeling of malate and α-ketoglutarate was higher in ATF4 KO cells than WT cells, even without ISR activation (Fig. 4d, white bars), suggesting that, like what we observed with amino acid synthesis, the ATF4-dependent mechanisms that contribute to the slowing of the TCA cycle are also active basally.
In addition to glucose, cultured cells rely heavily on glutamine to fuel the TCA cycle40. This was reflected in the relatively low fraction of malate and α-ketoglutarate (<2%) that was labeled by glucose in WT cells (Fig. 4d). We assessed whether ISR activation also inhibited glutamine oxidation by measuring 13C5-glutamine labeling of oxidative TCA cycle intermediates, which can be identified by their M + 4 mass (Fig. 4e, blue schematic). As expected, the fractional labeling of TCA cycle metabolites from glutamine was higher than from glucose (Fig. 4f). However, labeling of malate derived from glutamine oxidation (M + 4) was also decreased significantly more in WT cells after dimerizer treatment than in ATF4 KO (Fig. 4f). Thus, activation of the ISR reduces oxidation of both glucose and glutamine by the TCA cycle in an ATF4-dependent manner.
This slowing of the TCA cycle would be expected to impact mitochondrial respiration by limiting the amount of NADH available for oxidative phosphorylation. We used a Seahorse bioanalyzer to assess oxygen consumption rate (OCR), a measurement of oxidative phosphorylation, after 24 h of dimerizer treatment, consistent with the timing of the tracing experiments. Basal OCR was decreased with Dmr-PERK induction in both WT and ATF4 KO cells, which is consistent with the decreased energy demand caused by comparable inhibition of protein synthesis in both genotypes (Supplementary Fig. 4g, h). However, the decrease in basal OCR was more pronounced in WT cells, as was the reduction in mitochondrial spare respiratory capacity (Supplementary Fig. 4g, i), consistent with ATF4-dependent TCA cycle inhibition. Addition of ISRIB to WT cells had no effect on OCR in the absence of dimerizer but prevented the decrease in basal OCR and spare respiratory capacity caused by Dmr-PERK activation (Supplementary Fig. 4j, k), indicating that, as expected, slowing of the TCA cycle and of oxidative phosphorylation are a consequence of reduced eIF2B activity downstream of eIF2 phosphorylation.
When oxidative metabolism in the mitochondria is impaired, α-ketoglutarate derived from glutamine can be reductively carboxylated to citrate to support amino acid and fatty acid biosynthesis (Fig. 4e, red schematic)41,42. To assess whether this pathway was engaged with Dmr-PERK activation, we traced 13C5-glutamine labeling of reductive carboxylation products, identified by their M + 3 mass. Malate, aspartate and asparagine labeling through this pathway all increased after Dmr-PERK activation (Fig. 4g), indicating that reductive carboxylation of glutamine was mobilized to support the synthesis of these amino acids. Consistent with the TCA cycle measurements, this was genotype dependent as glutamine labeling of reductive carboxylation products was constitutively low in ATF4 KO cells (Fig. 4g).
Reductive carboxylation-derived citrate can be used to produce cytosolic acetyl-CoA for de novo synthesis of fatty acids, which we assessed by measuring incorporation of U-13C5-glutamine into the acyl chains lipids. We saw that glutamine incorporation into palmitate (C16:0) was unchanged with Dmr-PERK activation in either genotype (Fig. 4h), indicating that fatty acid synthesis via reductive carboxylation of alpha-ketoglutarate was maintained upon ISR induction. However, we observed a significant ATF4-dependent reduction in glucose-derived palmitate with dimerizer treatment (Fig. 4i). This amounted to an overall ATF4-dependent decrease in cellular de novo lipid synthesis, as measured by reduced deuterium labeling of palmitate in cells cultured in heavy water (2H2O) (Supplementary Fig. 4l). Taken together, these results indicate that the ISR induces an ATF4-dependent slowing of the TCA cycle and inhibition of de novo fatty acid synthesis, while maintaining amino acid synthesis derived from glucose or reductive carboxylation of glutamine.
To test whether this metabolic response occurs in other cellular contexts and modes of ISR induction, we used primary embryonic fibroblasts (MEFs) derived from mice that carry a homozygous point mutation (N208Y) in the alpha subunit of eIF2B43. This causes a dramatic reduction of eIF2Bα levels and activation of the ISR without phosphorylation of eIF2 (Supplementary Fig. 5a). This constitutive ISR is attenuated by maintaining MEFs in the presence of the ISRIB analog 2BAct30. Withdrawal of 2BAct leads to upregulation of ATF4 (Supplementary Fig. 5a) and increased ISR target gene expression (Supplementary Fig. 5b) in N208Y MEFs and, to a lesser extent, in WT MEFs. Untargeted metabolomics profiling was performed on N208Y cells maintained in the presence or absence of 2BAct for 24 h. Overall, the change in metabolite levels in N208Y MEFs upon 2BAct withdrawal positively correlated (r = 0.575) with the changes observed with Dmr-PERK activation in U2OS cells (Supplementary Fig. 5c). Further similarities between the metabolic responses of N208Y MEFs and Dmr-PERK cells to ISR activation were observed with 13C6-glucose and 13C5-glutamine tracing. As with Dmr-PERK activation, oxidation of glucose by the TCA cycle was significantly reduced in N208Y MEFs in the absence of 2BAct (Supplementary Fig. 5d–f). 2BAct withdrawal also decreased glucose labeling of TCA cycle intermediates in WT MEFs (Supplementary Fig. 5 e, f), consistent with the basal ISR detected in these cells. We also measured a significant increase in the synthesis of glutathione (Supplementary Fig. 5g) and asparagine (Supplementary Fig. 5h) from glutamine in N208Y MEFs upon 2BAct withdrawal. Intriguingly, N208Y MEFs also synthesize more proline from glutamine (Supplementary Fig. 5i), an effect that was not observed with ISR activation in the Dmr-PERK cells. Fibroblasts have been shown to synthesize high levels of proline to support collagen synthesis44, so this discrepancy may be due to differences in cell lineage. Together, the concordance of metabolic phenotypes in these two very different cell systems suggests that the observed reduction in glucose oxidation in the TCA cycle, as well as the increase in synthesis of glutathione and non-essential amino acids are generalizable responses to ISR induction.
The ISR inhibits glycolysis and lipid synthesis pathways independently of ATF4
In addition to the ATF4-dependent changes described above, we identified two clusters of metabolites that either increased (Fig. 4a, marked yellow) or decreased (marked purple) in both genotypes during ISR induction. Early glycolytic intermediates and pentose phosphate pathway metabolites accumulated with prolonged activation of the ISR in both genotypes while lactate levels were significantly reduced (Supplementary Fig. 6a), suggesting that ISR induction leads to an ATF4-independent decrease in the rate of glycolysis, consistent with Seahorse measurements showing reduced extracellular acidification rates (ECAR), a measurement of lactate production by glycolysis, independent of genotype (Supplementary Fig. 4m). In agreement with the lack of ATF4 effect on glycolysis, there was no defect in basal ECAR in ATF4 KO cells. Interestingly, the ATP/ADP ratio increased with prolonged ISR activation in both genotypes (Supplementary Fig. 6b, c). This suggests that, although activation of the ISR slowed both oxidative phosphorylation and glycolysis, less ATP is being consumed in these cells, possibly as a result of slowed protein synthesis and other anabolic processes. Finally, several polar metabolites of phospholipid metabolism (glycerol-3-phosphate, choline and phosphoethanolamine) accumulated with ISR activation (Supplementary Fig. 6d), suggesting that, in addition to ATF4-dependent inhibition of de novo fatty acid synthesis, the ISR also alters lipid metabolism pathways through ATF4-independent mechanisms.
ISR activation modifies cellular lipid composition and induces lipid droplet formation
The observation that intermediates of phospholipid synthesis were altered with Dmr-PERK activation suggested that the ISR could be modifying cellular lipid composition. To investigate whether this was the case, we measured the lipidome composition at the same time points and dimerizer concentration (0.05 nM) as in the metabolomics analysis. We found that 73% of identified lipid species showed a significant change in abundance over time (Supplementary Data 5). Clustering analysis identified two clear trends which had contrasting dynamic behaviors (Fig. 5a, Supplementary Fig. 7a–c). The largest cluster showed a time-dependent decrease in abundance with Dmr-PERK activation and was composed mostly of diacylglycerol (DG) and phospholipids (Supplementary Fig. 7a,d). A second cluster, consisting of TG and CE species showed a striking accumulation over time (Supplementary Fig. 7b,e) and was enriched in lipids containing polyunsaturated fatty acyl chains (PUFAs) (Supplementary Fig. 7f). Pathway enrichment analysis further demonstrated a significant accumulation of TG and CE with long chain PUFA acyl chains containing at least three double bonds (20:4, 22:4, 22:5, 24:6) after 24 h of Dmr-PERK activation, whereas unsaturated and monounsaturated fatty acids (16:1, 12:0, 14:1, 14:0, 24:0) were underrepresented (Supplementary Fig. 7g). Although there was strong enrichment in 18:0 acyl chains, this is most likely the result of it being typically found in combination with PUFA side chains in TG species (Supplementary Fig. 7h).
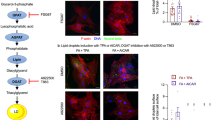
a Heatmap of lipid abundances in Dmr-PERK cells treated for the indicated amount of time with dimerizer (0.05 nM). Columns show three technical replicate measurements for each time point, grouped by lipid class. Measurements are normalized to the mean abundance at t = 0 on a log2 scale. Color bars indicate triglycerides (TG), cholesterol esters (CE), diacylglycerides (DG), glycerophospholipids (GPL), plasmalogens, sphingolipids and other lipid species. b Schematic of the lipid synthesis pathways that supply the constituents of lipid droplets. Enzymes mediating each step are labeled in gray. c BODIPY labeled Dmr-PERK cells treated for 8 h with vehicle (Veh) or dimerizer (Dmr, 1 nM). Scale bar = 5 μm d Quantification of the number of LD per cell in Dmr-PERK cells treated for 8 h with vehicle (Veh.) or the indicated concentration of dimerizer (Dmr). Median with interquartile range is indicated. 240 cells were quantified per condition. Statistical significance was evaluated by one-way ANOVA followed by Dunnett’s multiple comparison test. e Quantification of the number of LD in Dmr-PERK cells at the indicated time points following treatment with the indicated concentration of dimerizer. Error bars show mean ± SD per well across three independent experiments. f, g Bar plots representing the mean number of LD per cell per well in Dmr-PERK cells treated for 8 h with dimerizer (Dmr, 1 nM), ACATi (10 μM), DGAT1/2i (20 μM each) or ACCi (2 μg/mL) as indicated. Error bars show mean ± SD across three independent experiments. Statistical significance was evaluated by one-way ANOVA followed by Dunnett’s multiple comparison test. h Quantification of the percent confluence of Dmr-PERK cells treated with vehicle, dimerizer (Dmr, 0.024 nM), ACATi (10 μM), DGAT1/2i (20 μM each) or ACCi (2 μg/mL), as indicated. Error bars show mean ± SD of two technical replicates and is representative of three independent experiments. Source data are provided as a Source Data file.
Because cells deposit TGs and CEs in LDs, ER-derived organelles composed of a phospholipid monolayer membrane encapsulating a hydrophobic lipid core (Fig. 5b)45,46, we assessed whether Dmr-PERK activation of the ISR was accompanied by the formation of these structures. Dimerizer treatment led to a dose and time-dependent increase in the number of LDs per cell, as visualized with BODIPY 493/503 (Fig. 5c, d), with timing that was consistent with our measurement of neutral lipid accumulation (Fig. 5e). LD formation has been reported to coincide with the formation of SGs in response to arsenite treatment47. Although Dmr-PERK activation induced the formation of both SGs and LDs, they accumulated with different kinetics (Supplementary Fig. 8a). SGs abundance peaked at 4 h and resolved by 8 h, whereas LDs steadily increased over 8 h. Furthermore, we observed no colocalization between SGs and LDs in these conditions (Supplementary Fig. 8b), indicating that these are distinct structures whose ISR-induced formation is not correlated.
Previous studies have shown that insults such as hypoxia, nutrient deprivation and ER stress cause LDs to accumulate48,49. To test whether the ISR contributes to LD formation in response to pleiotropic stressors, we measured LDs across a time course in cells treated with oligomycin or thapsigargin in the presence or absence of ISRIB50. Both stressors induced LD formation with kinetics similar to that of Dmr-PERK activation (Supplementary Fig. 8c). In both cases, addition of ISRIB reduced the number of LDs per cell, indicating that the ISR is necessary for cell stress-induced LD biogenesis (Supplementary Fig. 8c).
DGAT activity is required for ISR-driven lipid droplet formation
To determine the source of these neutral lipids, we tested whether LD formation depended on increased lipid uptake by comparing LD induction in Dmr-PERK cells grown in standard or delipidated media. ISR induction with either dimerizer or thapsigargin treatment still upregulated LDs in delipidated media (Supplementary Fig. 8d), indicating that this process does not depend on extracellular fatty acids and suggesting that cell-intrinsic remodeling underlies ISR-dependent LD formation. This change is independent of ATF4, as ATF4 KO cells were still able to induce LD formation (Supplementary Fig. 8e), a result consistent with the ATF4-independent accumulation of lipid related molecules glycerol-3-phosphate, choline and phosphoethanolamine (Fig. 4a, Supplementary Fig. 6d).
The ISR has been reported to inhibit signaling by the nutrient-sensitive mechanistic target of rapamycin complex 1 (mTORC1)51, a master regulator of cellular metabolism whose inhibition is sufficient to increase cellular LD abundance and is required for LD biogenesis during nutrient deprivation48,52. Although activation of Dmr-PERK induced expression of DDIT4 and SESN2 (Supplementary Data 3), known regulators of mTORC1 signaling51,53,54, we saw no evidence of decreased mTORC1 activity over a time course of Dmr-PERK activation, as measured by either western blot (Supplementary Fig. 8f, g) or using a fluorescent immunoassay to monitor phosphorylation of the mTORC1 target S6K (Supplementary Fig. 8h). This indicates that mTORC1 is not modulated by the ISR in this system and is therefore not required for ISR-driven LD formation.
Finally, we assessed whether transcriptional changes to pathways involved in TG and CE synthesis could account for their accumulation. Consistent with the reduced deuterium labeling of palmitate (Supplementary Fig. 4l), fatty acid synthesis enzymes were unchanged or reduced in expression with dimerizer treatment (Supplementary Fig. 9a), indicating that increased de novo fatty acid synthesis was not responsible for increased TG content. Conversely, several enzymes involved in TG synthesis and LD biogenesis were induced by the ISR (Supplementary Fig. 9a). Most notably, the transcript for the diacylglycerol acyltransferase DGAT2 was elevated over time with dimerizer treatment as a part of the Class 1 responders. DGAT1 and DGAT2 catalyze the final, rate-limiting step in TG synthesis by esterifying DG to yield TG which is then packaged into LDs46,55. DGAT1 showed no change in expression over time with ISR induction but was constitutively expressed at a high level relative to DGAT2 (Supplementary Fig. 9b). Consistent with LD formation being independent of ATF4, the induction of DGAT2 transcript was also observed in ATF4 KO cells (Supplementary Fig. 9c).
To investigate the role of DGATs in mediating the ISR-driven upregulation of LDs, we activated Dmr-PERK in the presence of the pharmacological inhibitors of DGAT1, T863, and DGAT2, PG-06424439 (combined, DGAT1/2i). We compared the effects of DGAT1/2i to inhibiting CE synthesis with the pan-cholesterol acyltransferases (ACAT) inhibitor Avasimibe (ACATi), or to inhibiting the rate limiting step in de novo fatty acid synthesis with the acetyl-CoA carboxylase (ACC) inhibitor TOFA (ACCi). In the absence of dimerizer, cellular LD content at 8 h was decreased by all inhibitors, a result consistent with the role of these pathways in regulating LD biogenesis (Fig. 5f)46,55. However, with the addition of dimerizer, only DGAT1/2i abolished the induction of LDs, while ACATi and ACCi did not prevent the relative increase in cellular LD content by Dmr-PERK activation (Fig. 5f, g). Together, these findings demonstrate that ISR-driven LD formation depends critically and specifically on TG synthesis by DGAT activity through an ATF4-independent mechanism.
Inhibition of LD formation upon activation of the ISR impairs cell survival
LDs are increasingly recognized as serving a protective role during cell stress by providing a source of fatty acids for energy production, regulating membrane composition, and sequestering deleterious proteins and lipid species45,48,49. We therefore considered whether LD induction protected against cell death during a prolonged ISR1. We performed time-lapse microscopy on Dmr-PERK cells and measured their confluence over 96 h of ISR activation in the presence or absence of lipid synthesis inhibitors. Dimerizer treatment alone had a dose-dependent effect on cell growth and viability, with extensive cell death occurring at the highest doses (15 nM, 3 nM) and only moderate effects on confluency measured at the lowest dose (0.024 nM) (Supplementary Fig. 9d). Despite their effects on basal LD content (Fig. 5f), ACATi, DGAT1/2i or ACCi alone had no effect on cell confluence (Supplementary Fig. 9e). However, when a sublethal dimerizer treatment was combined with DGAT1/2i, cell viability was severely compromised, an effect not observed with ACATi or ACCi co-treatment (Fig. 5h). Collectively, our findings indicate that the ISR leads to a reorganization of cellular lipid content, favoring DGAT-dependent TG production and storage in LDs, as part of a protective response to prolonged stress signaling.
DGAT1 and 2 have distinct functions in ISR-induced LD formation and cell survival
Although DGAT1 and DGAT2 catalyze the same reaction, they are evolutionarily unrelated proteins that have overlapping but distinct functions55. To test the relative contribution of DGAT1 and DGAT2 to ISR-induced LD formation and cell viability, we activated Dmr-PERK in the presence of each DGAT inhibitor individually. Following 8 h of dimerizer treatment, LD formation was fully prevented by DGAT1i but not by DGAT2i which, instead, further increased LD content (Fig. 6a, b), indicating that TG synthesis by DGAT1 is sufficient to drive LD formation after Dmr-PERK activation. When DGAT1i was combined with a sublethal dose of dimerizer (0.024 nM), there was no effect on cell confluency (Fig. 6c, Supplementary Fig. 10a), despite its ability to prevent LD formation. However, inhibition of DGAT2 was sufficient to reduce cell viability (Fig. 6c) and induce apoptotic caspase activation (Fig. 6d, Supplementary Fig. 10b) to the same extent as DGAT1/2i. Similarly, siRNA knockdown of DGAT1, but not DGAT2, blunted LD formation when Dmr-PERK was activated (Supplementary Fig. 10c, d), whereas only knockdown of DGAT2 was sufficient to impair cell growth during prolonged treatment with a low dose of dimerizer (0.12 nM) (Supplementary Fig. 10e, f). Therefore, although ISR-driven LD formation is DGAT1-dependent, cell viability depends on the enzymatic activity of DGAT2.
a, b Bar plots representing the mean number of LD per cell per well in Dmr-PERK cells treated for 8 h with dimerizer (Dmr, 1 nM), DGAT1i (20 μM) or DGAT2i (20 μM) as indicated. Error bars show mean ± SD across three independent experiments. Statistical significance was evaluated by one-way ANOVA followed by Dunnett’s multiple comparison test. c Quantification of the percent confluence of Dmr-PERK cells treated with vehicle, dimerizer (Dmr, 0.024 nM), DGAT1i (20 μM) or DGAT2i (20 μM), as indicated. Error bars show mean ± SD of two replicates and is representative of three independent experiments. d Western blot for cleaved caspase-3 in lysates from Dmr-PERK cells treated with the indicated dose of dimerizer (Dmr), DGAT1i (20 μM) or DGAT2i (20 μM), as indicated. Quantification of the number of LD (e) and cell number (f) simultaneously imaged by high-content live-cell microscopy in Dmr-PERK cells treated with vehicle, dimerizer (Dmr), DGAT1i (20 μM) or DGAT2i (20 μM), as indicated. Error bars show mean ± SD of three independent experiments. g Western blot for ATF4 in lysates from Dmr-PERK cells treated for 24 h with dimerizer (Dmr, 0.05 nM), DGAT1i (20 μM) or DGAT2i (20 μM), as indicated. h Average z-core of ISR GeneCLIC genes calculated from nCounter gene expression profiling of Dmr-PERK cells treated for 24 h with dimerizer (Dmr, 0.05 nM), DGAT1i (20 μM) or DGAT2i (20 μM), as indicated. Error bars show mean ± SD of three technical replicates. Statistical significance was evaluated by one-way ANOVA followed by Dunnett’s multiple comparison test. Source data are provided as a Source Data file.
Because DGAT enzymes have been reported to be modulated by posttranslational modifications, including phosphorylation55,56,57, we considered whether the downstream functions of DGAT1 and DGAT2 depended on the activity of the eIF2 kinase that initiated the ISR. We transiently knocked down expression of eIF2Bα in U2OS cells to trigger the ISR independently of eIF2 phosphorylation, in a manner analogous to the N208Y mutation (Supplementary Fig. 11a)43,58. As with Dmr-PERK activation, this resulted in a DGAT1-dependent increase in LD content (Supplementary Fig. 11b, c) and increased cell death when DGAT2 was inhibited (Supplementary Fig. 11d). We also observed an accumulation of LDs in N208Y MEFs compared to WT after 8 h of withdrawal from 2BAct, which was prevented by DGAT1i but not DGAT2i (Supplementary Fig. 11e, f). The distinctive roles of DGAT1 and DGAT2 in mediating LD formation and cell survival during ISR activation are therefore not a consequence of eIF2 kinase-dependent posttranslational modifications.
Finally, we examined whether DGAT1 and DGAT2 played similar roles in response to pleiotropic stress conditions. In both U2OS and HCT116 cells, LD buildup in response to oligomycin was only DGAT1-dependent (Supplementary Fig. 11g, h), whereas only inhibition of DGAT2 impacted proliferation of both cell types in the presence of a sub-lethal dose of thapsigargin (3.3 nM) (Supplementary Fig. 11i, j). Together, our results indicate that DGAT1 and DGAT2 have unique contributions downstream of ISR activation across multiple stress conditions and cell types.
Inhibition of DGAT2 activates the ISR
To better understand the relationship between DGAT activity, LD formation, and cell growth, we used high content live cell imaging to simultaneously track LD content and cell number over 24 h of dimerizer treatment (0.0048 nM, 0.024 nM, 0.12 nM) in the presence or absence of DGAT inhibitors. By imaging a large number of cells at higher time resolution, we found that the rapid accumulation of LDs over 8 h of Dmr-PERK activation was followed by a second, slower phase (Fig. 6e). This was accompanied by a decrease in cell number, with onset of cell death at the highest dimerizer doses (0.024, 0.12 nM) (Fig. 6f). Simultaneous inhibition of DGAT1 and DGAT2 ablated cellular LD content throughout the time course (Fig. 6e, f), whereas DGAT1i alone prevented the early peak of LD formation but did not completely inhibit the second phase (Fig. 6e, f, Supplementary Fig. 12a). This suggests that DGAT2 drives a later phase of LD biogenesis following ISR activation, which is consistent with the timing of its transcriptional induction (Supplementary Fig. 9a, b). DGAT2i treatment led to a striking amplification of the early peak of LD formation, followed by the expected loss of cell growth and viability at all dimerizer doses, which made it impossible to directly assess the effects of DGAT2 inhibition on the later phase of LD formation (Fig. 6e, f, Supplementary Fig. 12a). We found that the ISR-induced LDs formed in the presence of DGAT2i were on average smaller than with Dmr-PERK activation alone (Supplementary Fig. 12b, c), which reflects DGAT2’s role in promoting LD growth55,59. Together, these results suggest that TG synthesis and LD formation downstream of the ISR are dynamic processes that are controlled by time-dependent mechanisms to which DGAT1 and DGAT2 contribute differently and define distinct downstream cellular outputs.
We noted that the effect of DGAT2i on LD content and cell growth at low doses of dimerizer mimicked what was observed at higher dimerizer doses with no DGAT inhibition. We therefore examined the status of ISR activity when DGAT inhibitors were present. At a low dimerizer dose (0.05 nM), treatment with DGAT2i, but not DGAT1i, enhanced ATF4 protein levels (Fig. 6g) and elevated Dmr-PERK-induced expression of ISR GeneCLIC target genes, (Fig. 6h, Supplementary Fig. 12d, e), indicating that loss of DGAT2 activity amplifies ISR signaling. Strikingly, even in the absence of dimerizer, inhibition of DGAT2 was sufficient to drive ATF4 protein accumulation (Fig. 6g), promote LD formation (Fig. 6e), and induce an ISR gene expression profile comparable to that of Dmr-PERK (Fig. 6h, Supplementary Fig. 12e), suggesting that, even at baseline, DGAT2 is important for maintaining lipid homeostasis and that loss of its enzymatic activity triggers a cell stress response. Our findings are consistent with this endogenous response having additive effects on Dmr-PERK activation, leading to the observed increase in ISR signaling, enhanced DGAT1-dependent LD formation, and cell death. This suggests that the transcriptional induction of DGAT2 is a protective ISR mechanism that prevents additional cell stress and illustrates how coordination of transcriptional and metabolic ISR outputs is important in shaping downstream cellular outcomes.
Discussion
The use of a minimal pharmacogenomic system to activate the ISR allowed us to explore how the dose and timing of input into the pathway affects its various outputs. This approach uncovered a previously unappreciated metabolic remodeling that, in addition to the known effects on amino acid and glutathione synthesis, had profound effects on cellular bioenergetics and lipid content (Fig. 7). These metabolic responses were already evident at early time points and low levels of pathway input, in a regime when protein synthesis inhibition was minimal and SG formation was not yet detectable. Thus, we propose that this metabolic state is a primary output of the ISR that is critical for adaptation to physiological levels of stress.
The ISR increases glutathione and serine synthesis, while diverting glucose and glutamine carbon away from oxidation in mitochondria. Synthesis of aspartate, asparagine and acyl-CoA is maintained via reductive carboxylation of glutamine. These ATF4-dependent changes are accompanied by ATF4-independent inhibition of glycolysis, increased pentose phosphate pathway activity, and increased TG production and accumulation in lipid droplets. Changes in pathway utilization, metabolite abundance and enzyme expression are indicated by color (red = up, blue = down).
Induction of amino acid synthesis and uptake by ATF4 has been well described downstream of pleiotropic ISR activating conditions such as ER stress8,60, mitochondrial dysfunction23,25,61, and amino acid depletion26. Here, we demonstrate that ISR activation is sufficient to drive this response, even in the absence of any cellular insult. The accumulation of amino acids may therefore be a hard-wired response preparing cells to resist adverse conditions where amino acid accumulation would be beneficial, such as ER stress and nutrient deprivation. It’s also notable that this metabolic response is initiated even at minimal levels of ISR activity and at very early time points. This may reflect a physiological condition where stress can still be resolved and these pools of amino acids would be available to support recovery of protein synthesis and to fuel cell growth62,63. The possibility that this ATF4-dependent process is supporting growth is consistent with reports of ATF4 activation downstream of mTORC1 activation by pro-growth signals9,64,65. Induction of ATF4 by low doses of insulin initiates a transcriptional response that mirrors the ATF4-dependent metabolic module defined here and is required to maintain increased protein synthesis9. This overlap also suggests that this transcriptional module represents a core ATF4 transcriptional program that may promote recovery when stress has subsided.
To our knowledge, this is the first report of ATF4-dependent slowing of oxidative metabolism. As we did not observe any gene expression changes that directly explain this effect, it appears to be a secondary consequence of the ATF4 transcriptional metabolic module. The accompanying upregulation of the reductive carboxylation of glutamine is a compensatory response typically seen in cells with impaired mitochondrial respiration42 and, in this case, is likely a direct response to the slowed TCA cycle. Mitochondrial dysfunction through inhibition of oxidative phosphorylation25,61, disruption of the TCA cycle23, or loss of membrane potential25 are major ISR triggers that induce the same ATF4-dependent changes in amino acid and glutathione metabolism as Dmr-PERK activation. Reductive carboxylation allows cells to route glutamine utilization away from oxidative pathways and maintain amino acid synthesis in the context of dysfunctional mitochondria. This would also alleviate metabolic demand and stress on already malfunctioning respiratory pathways. Thus, we propose that an additional central role for ATF4 during the ISR is to reroute biosynthetic pathways to enable amino acid synthesis during mitochondrial dysfunction.
This ATF4-dependent metabolic switch may also assist with matching cellular bioenergetics with reduced energy demand during stress. The maintenance of ATP levels despite slowing of the TCA cycle and glycolysis suggests that overall cellular anabolism is repressed by the ISR. This is also supported by the later-stage downregulation of genes that support growth processes (Class 4). It is possible that slowed growth also explains the decrease in cellular phospholipid content, as inhibition of anabolic processes would reduce the demand for membrane biogenesis66. Another possibility is that, in the context of inhibition of global protein synthesis, phospholipid levels may decrease to maintain the balance between membrane lipid and protein content67. Phospholipids and TG share a common precursor, DG, and both synthesis pathways draw from the same pool55,68. It’s possible that, in an effort to balance membrane phospholipid content, the ISR drives a conversion of phospholipids to TG. The predominance of PUFAs in the ISR-induced TG pool suggests that this process could have additional protective effects, such as resistance to oxidative stress. Sequestration of PUFAs in LDs can limit phospholipid peroxidation at cell membranes and protect from ferroptosis69.
The mechanisms underlying this ISR-induced shift towards TG accumulation have yet to be defined, as it was not a consequence of the ATF4-induced metabolic changes and was not dependent on fatty acid uptake or de novo synthesis. DGAT1-dependent conversion of phospholipids to TG has been reported downstream of mTORC1 inhibition, through a lysosome-dependent but autophagy-independent hydrolysis of phospholipid fatty acids52. However, this process does not seem to be occurring in this Dmr-PERK system, which did not inhibit mTORC1, nor demonstrate the reported accumulation in lyso-phospholipids. DGAT1-dependent TG accumulation is also seen in response to high levels of free fatty acids derived from mTORC1-induced autophagy during acute nutrient deprivation48. While we do not see any transcriptional evidence that autophagy is induced by Dmr-PERK activation, we cannot rule this out as a potential source of fatty acids contributing to TG synthesis by the ISR. Regardless of the mechanisms, they will undoubtedly be time-dependent and coordinated with baseline lipid homeostasis pathways, including TG synthesis by the DGATs. Similar to our findings with DGAT2, basal DGAT1 activity has been shown to protect the ER from lipotoxic stress in adipocytes by re-esterifying released fatty acids, a process whose upregulation becomes critical in conditions of enhanced lipolysis70. It’s possible that DGAT2 activity may be similarly necessary to prevent the accumulation of toxic, stress-inducing lipid species. Although the basal stress-protective function of DGAT2 is not yet known, nor is the nature of the stress sensors that are engaged in its absence, our findings highlight the extent to which the ISR is integrated with the state of the cellular metabolome and lipidome. Our findings add to a growing number of reports that LD formation, through various mechanisms, is a protective response to cell stress49 by preventing mitochondrial dysfunction from fatty acid overload during starvation48, sequestering misfolded proteins from the ER71, avoiding lipid peroxidation and oxidative stress72,73, maintaining membrane saturation74 or serving as a long-term supply of fatty acids for energy generation by β-oxidation48,52,75.
This pharmacogenomic approach establishes a footprint of ISR-sufficient transcriptional, metabolic and cellular events that promote early responses to cell stress. Although we used a construct derived from the kinase domain of PERK, the specificity of PERK, PKR, HRI, and GCN2 in targeting eIF2ɑ as their substrate, and the similarity of our transcriptomic signature with one obtained from a PKR-derived optogenetic tool76, together suggest that these downstream outputs are independent of the specific ISR kinase domain. Indeed, that many of the metabolic outputs downstream of Dmr-PERK activation were also observed in N208Y-EIF2α MEFs, where the is ISR activated without eIF2 phosphorylation, suggests that these responses are agnostic to the nature of the activating kinase.
Understanding the core response of the ISR will allow us to better understand the cellular state downstream of pleiotropic insults, where these core outputs are modulated by inputs via different signaling pathways. Although this footprint is derived from an immortalized human cell line, we found that the major ISR-induced changes were conserved in primary mouse fibroblasts, suggesting this may be a conserved response. Indeed, the sensitive ATF4-dependent transcriptional module is active in the astrocytes of Vanishing White Matter mice, where a mutation in eIF2B causes chronic engagement of the ISR and leukodystrophy, and is also represented in the ISR-associated GeneCLIC signature30,43. Data from the minimal cell line system generates a hypothesis for what transcriptional and metabolic changes to expect in vivo during ISR activation, including the newly uncovered role of the ISR in LD biogenesis, an important finding to help dissect the mechanisms of LD formation and function in stressed tissues27,77,78. Intriguingly, many of the neuropathologies associated with ISR activation are also associated with the accumulation of LDs79,80 and it will be critical to parse the contribution of the ISR to their formation and whether this is a protective or maladaptive response to these diseases. Thus, our results using a minimal pharmacogenomic system will help us resolve the mechanistic basis for the ISR’s contribution to cell- and tissue-specific outcomes to injury, aging and disease.
Methods
Cloning
The Dmr-PERK expression vector was generated using the Lenti-X iDimerizer Inducible Homodimerizer System (Takara). The mouse PERK kinase domain (amino acids 537-1114) was obtained as a custom gBlock from Integrated DNA Technologies. The pLVX-Hom-1 vector (Takara) contains a CMV promoter driving expression of a dimerization domain, and a multiple cloning site followed by an IRES sequence driving expression of a Puromycin resistance cassette. pLVX-Hom-1 was digested with BamHI and ligated with the mouse PERK kinase domain amplified with corresponding BamHI overhangs. Ligation and transformation was performed using the In-Fusion Cloning System with Stellar competent cells (Takara). Clones expressing the correct vector sequence were confirmed by Sanger sequencing.
Cell culture
U2OS cells were grown in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Sigma) and 1X antibiotic-antimycotic solution (Gibco). Cells were housed in an incubator at 37 °C, 5% CO2, 20% O2 and passaged every 2–3 days with trypsin. In experiments comparing WT and ATF4 KO cells, cells of both genotypes were grown in media supplemented with 1X NEAA (Sigma) and 55 nM beta mercaptoethanol (Gibco) for at least 24 h prior to experimentation. For experiments in delipidated media, cells were washed 3× and treated in DMEM supplemented with 10% lipid-depleted FBS (Biowest LLC) and 1X antibiotic-antimycotic solution. Where indicated, cells were treated with AP20187 dimerizer (Sigma Aldrich), 100 nM oligomycin (Sigma Aldrich), 100 nM thapsigargin (Sigma Aldrich), 500 nM ISRIB (Sigma Aldrich), 500 nM 2BAct (synthesized in house), 100 nM Torin (Tocris).
Wild-type and N208Y MEFs were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Sigma) and 1X antibiotic-antimycotic solution (Gibco) and 500 nM 2BAct. Cells were housed in an incubator at 37 °C, 5% CO2, 3% O2 and passaged every 2–3 days with trypsin. For withdrawal studies, MEFs were acclimatized to 20% O2 for 24 h then washed 3× with PBS and replenished with fresh media with or without 2BAct. For metabolomics, media was supplemented with 1X NEAA (Sigma) and 55 nM beta mercaptoethanol (Gibco) for at least 24 h to mimic U2OS media conditions in these experiments. All downstream assays were performed in normoxic conditions.
Confluence measurements were performed by plating 0.012 × 106 cells per well, in duplicates, in 96 well plates and treatment started the next day. As indicated, cells were incubated with 10 μM Avasimibe (Selleckchem), 20 μM T863 (Sigma Aldrich), 20 μM PF-06424439 (Sigma Aldrich) or 2 μg/mL TOFA (Sigma Aldrich) immediately before transferring cells to the Incucyte SX-5 Live-Cell Analysis (Sartorius). Cells were imaged every 2 h for 96 h and confluence was quantified with the Incucyte software using label-free segmentation methods.
Cell line generation
The U2OS Dmr-PERK cell line was generated by lentiviral transduction. Dmr-PERK lentivirus was generated by delivering the pLVX-Hom-1-Dmr-PERK-IRES-Puro vector to HEK293T using the Takara Lenti-X Packaging Single Shot (VSV-G) system. Supernatant was collected after 48 h and concentrated using Lenti-X Concentrator (Takara). U2OS cells were transduced with varying concentrations of viral supernatant and stable integrants were selected with media containing 3 µg/mL puromycin. Stable clones were confirmed by Western blot analysis of PERK at the expected protein size and by induction of ISR outputs after treatment with dimerizer, including p-eIF2 AlphaLISA, and ATF4, GADD34, and CHOP protein expression (Fig. 1 and Supplementary Fig. 1).
U2OS Dmr-PERK ATF4 KO cells were generated using Alt-R CRISPR-Cas9 reagents from Integrated DNA Technologies according to the manufacturer’s protocol. Predesigned crRNAs targeting ATF4 were obtained from IDT (Hs.Cas9.ATF4.1.AA) and annealed with ATTO 550-labeled tracrRNA. RNPs were generated by combining with Cas9 protein (IDT) and reverse transfected into U2OS Dmr-PERK cells with Lipofectamine RNAiMAX reagent. 24 h after transfection, cells were trypsinized and sorted for ATTO 550-positive cells into single wells of a 96-well plate. ATF4KO cells were grown in supplemented DMEM that included 1X NEAA (Sigma) and 55 uM beta-mercaptoethanol (Gibco). Individual clones were verified to be ATF4-null by sequencing and confirmed by Western blot for ATF4 after ISR induction.
siRNA knockdown
U2OS Dmr-PERK cells were transfected with Silencer Select siRNA (ThermoFisher) using the reverse transfection method. As indicated, 10 pmol of siRNA for DGAT1 (s16569), DGAT2 (112269) or eIF2B1 (s4558 or s4559) was diluted in 100 μL Opti-MEM (Gibco) and added to the wells of a 24 well plate followed by 1 μL of Lipofectamine RNAiMAX (ThermoFisher) and incubated for 20 min at room temperature. 0.5 mL of suspension of 100,000 cells/mL in complete DMEM was added to each well, incubated for 48 h, then split into vessels for various downstream assays.
AlphaLISA assay
Dmr-PERK U2OS cells were plated on 96-well plates and left to recover overnight. Cells were treated in triplicate with indicated compounds for 2 h and lysed in 50 μL 1X AlphaLisa lysis buffer. Lysates were assayed in technical duplicates to measure the level of eIF2α phosphorylation using the Total eIF2α AlphaLISA SureFire Ultra Detection Kit (Perkin Elmer) following the manufacturer’s instructions. For p70 S6K phosphorylation, lysates were assayed with the Alpha SureFire Ultra Multiplex phospho/total p70 S6K Assay (Perkin Elmer). Plates were read using a CLARIOstar Plus plate reader (BMG Labtech) with standard AlphaLISA settings.
Immunofluorescence
Cells were plated in CellCarrier 96 well plates (PerkinElmer) to achieve 80% confluence at the time of fixation. After treatment, cells were washed once with PBS and fixed with 4% paraformaldehyde for 15 min. Cells were washed 3 times with PBS then permeabilized and blocked in blocking buffer (PBS, 5% goat serum and 0.5% Triton X-100) for 1 h. The following primary antibodies were diluted in labeling buffer (PBS, 0.1% Tween 20 and 5% goat serum) (1:1000) and incubated overnight at 4 °C with gentle rocking: anti-G3BP1 (BD), anti-CHOP (1:1000, Cell Signaling L63F7). Following three 5-min PBST washes, secondary antibodies were applied (1:2500) in blocking buffer and incubated for 1 h. After three PBST washes for 5 min each, cells were stained with DAPI (1:10,000) in PBS for 5 min. When co-staining with BODIPY, Triton X-100 and Tween 20 were replaced with 0.05% saponin and 0.01% saponin, respectively. All steps were performed at room temperature unless otherwise noted. For lipid droplet imaging, fixed cells were labeled with 50 ng/mL BODIPY 493/503 and DAPI (1:5000) in PBS for 15 min followed by three PBS washes. All labeling steps were performed at room temperature unless otherwise noted and protected from light throughout.
Cells were imaged in fresh PBS. Imaging was performed on the Opera Phenix High Content Screening system (Perkin Elmer). Plates were imaged with the 20× or 40× water objective with 9–25 fields of view imaged per well. Nucleus, cell, and spot segmentation was performed using Harmony software (Perkin Elmer). For quantification of LD content, eight random fields were selected and a total of >100 cells were scored per conditions for each of >3 independent experiments Percentage of cells with SGs, mean number of spots per cell was calculated in R. Data plotting, EC50 determination, and statistics were performed on GraphPad Prism software.
High-content microscopy
U2OS Dmr-PERK cells were seeded in a 384-well PhenoPlate (PerkinElmer) at a density of 4000 cells/well in DMEM supplemented with 10% FBS, 1X antibiotic-antimycotic solution and 1X GlutaMAX (Gibco). The following morning cells were washed using a BioTek EL406 Washer and labeled with 114 nM BODIPY 493/503 and 1X SPY650-DNA (Cytoskeleton, Inc) in FluoroBrite DMEM (Gibco) supplemented as above. 3 h later cells were dosed, as indicated, with AP20187 dimerizer (Sigma Aldrich), 20 μM T863 (Sigma Aldrich) or 20 μM PF-06424439 (Sigma Aldrich) using an Echo 555 or 655 liquid handler (Thermo Scientific) and all volumes normalized with vehicle (DMSO). Six replicates per condition were arranged randomly across 2 plates performed on different days.
Following compound dosing, cells were immediately transferred to a pre-warmed Opera Phenix High Content Screening System running Harmony version 5.1 for imaging. Cells were imaged every 20 min for 12 h and then every 2 h up to 24 h using a 40× water objective with four fields of view imaged per well. The imaging chamber was kept at 37 °C and 5% CO2 for the duration of imaging. Nuclei, cytoplasm and spot segmentation were performed in the Harmony software on maximum intensity projections of three slices for each field. Data plotting and statistical analysis were performed in Python.
LD radius quantification was performed in Python 3.10. LDs were first identified by creating maximum intensity projections of three slices for each imaging field. We then ran a multi-scale Laplacian of a Gaussian (LoG) filter over five sigma values ranging from 1 to 2.67 pixels in size to enhance spherical structures, keeping only those pixels with a post-filter value above 0. The first LoG filtered image corresponding to a sigma of 1 underwent a triangle threshold to create a definitive LD semantic segmentation mask. A maximum filter was then applied across the LoG stack, and local peak maxima were identified as those pixels in the maximum filtered image whose intensity value matched those same pixels in the LoG stack, corresponding to the midpoint of individual lipid droplets. A border mask was then created by performing a binary dilation of the LD semantic segmentation mask, followed by a binary NOT operation on the original mask, leaving behind only the outline of the LD mask. These border mask pixel coordinates were then queried using a k-d tree to find each local max peaks’ nearest border pixel neighbor, giving the effective radius for each LD in a computationally efficient and accurate way, as previously described81. Any lipid droplet whose local max peak coordinates’ pixel was below a threshold value of 4000 was removed from the analysis. The code is open-source and publicly available82.
Puromycin incorporation assay
To measure puromycin incorporation, the Click-iT Plus OPP Alexa Fluor 488 Protein Synthesis assay kit was used and manufacturer’s instructions were followed, with a 30-min puromycin labeling. For puromycin incorporation measurements by Western blot, cells were labeled with 10 µg/mL puromycin for 30 min prior to harvesting and protein analysis was performed as described below.
Protein expression analysis
Lysates were collected by washing cells once with PBS and harvesting in RIPA buffer (ThermoScientific) with phosphatase and protease inhibitors (Pierce). 4X Laemmli buffer was added to samples and boiled at 95 °C for 5 min. For signaling time courses, cells were directly lysed in 2X Laemmli sample buffer (BioRad) and boiled for 10 min.
Samples were run on a precast PAGE gel (Mini-PROTEAN TGX, BioRad) using the Mini-PROTEAN Tetra Cell and then transferred to a nitrocellulose membrane at 100 volts for 1 h using a Mini Trans-Blot Cell (BioRad). Membranes were incubated in blocking buffer (5% milk or 5% BSA in TBST) for 1 h at room temperature. The following primary antibodies was diluted in blocking buffer and added to the membrane, incubating overnight at 4 °C with gentle rocking: anti-p70S6K (Cell Signaling 9202) (1:2000), anti-phospho-p70S6K (T389) (Cell Signaling 9205) (1:1000), anti-cleaved caspase-3 (Cell Signaling 9662s) (1:2000), anti-cleaved caspase-8 (Cell signaling 9746t) (1:2000), anti-eIF2B1 (Proteintech 18010-1-AP), anti-actin (Cell signaling 3700) (1:2000). Membranes were washed with TBST three times for 5 min each, and then secondary antibody diluted in blocking buffer was added for 1 h at room temperature. Membrane was washed three times with TBST and then visualized with the Odyssey Infrared Imaging System (LI-COR Biosciences) or detection reagents were applied, and imaging was performed on a ChemiDoc imager (BioRad).
For automated Western blot analysis (Wes), samples were diluted to 0.4 mg/mL and 3 μL of sample was run on a ProteinSimple Wes capillary system using a 12–230 kDa separation module with the Chemiluminescent Detection module and using following primary antibodies: anti-ATF4 (1:100, Cell Signaling 11815), anti-GADD34 (1:100, Proteintech 104491-1-AP), anti-phospho-eIF2a (S51) (1:200, Cell Signaling 3398), anti-eIF2a (1:200, Cell Signaling 5324).
nCounter gene expression analysis
Multiplex transcript expression levels were measured by nCounter (Nanostring Technologies, Seattle, WA) with a custom panel containing the 95 ISR GeneCLIC genes. 100–500 ng of purified total RNA was used for nCounter gene expression analysis as instructed by the manufacturer. Briefly, reporter and capture probes to the genes of interest were hybridized to total RNA at 65 °C for 16 h. Hybridized probes were then captured to the nCounter cartridge prior to imaging and quantification.
Raw counts were background subtracted against negative control probes then normalized against housekeeping genes (B2m, Gapdh, Hprt, Rpl19) and positive control samples. The counts were log2-transformed and a Z-score for each gene was calculated by subtracting the overall mean of the control group from the sample and dividing that result by the SD of all of the measured intensities. ISR pathway activation was measured by taking the averages of the Z-score within each sample. GraphPad Prism (La Jolla, CA) was used to perform statistical analyses utilizing one way ANOVA with Dunnett’s Test performed post-hoc to correct for multiple comparisons. Log2 fold changes of ISR GeneCLIC genes were clustered by correlation-based distance using the heatmap.2 function from the gplots package.
Bioenergetics
1 × 104 WT or ATF4 KO U2OS cells were seeded per well in a Seahorse 96-well assay plate in DMEM with 10% FBS, 1% NEAA and 55 µM β-mercaptoethanol. Medium was exchanged the following day with either 0.05 nM AP20187 dimerizer or ethanol vehicle. 16 h later, media was exchanged to Seahorse XF DMEM with 2% dialyzed FBS, 2 mM glutamine, 25 mM glucose, 1X NEAA, and 55 µM β-mercaptoethanol ±0.05 nM dimerizer. Oligomycin, FCCP, and Rotenone/Antimycin A were diluted in Seahorse XF DMEM and added to the calibrant plate according to manufacturer protocol. Mito Stress tests were run on a Seahorse Bioanalyzer with Wave software (Agilent). Data was exported to GraphPad prism for visualization.
RNA extraction and library preparation
Cells were seeded at 2.5 × 105 cells/mL in 24-well plates one day prior to treatment. Treatments added to wells at indicated time points prior to sample collection. Each condition and time point were measured in triplicate. RNA was extracted using the MagMAX mirVana Total RNA Isolation Kit in 96-well plate format and processed on the KingFisher Flex Magnetic Particle Processor using the A27828_FLEX_Tissue_Cells protocol. RNA quality and concentration was determined with the Fragment Analyzer Standard Sense RNA kit. RNA-seq libraries were prepared using the NEBNextUltra II Directional RNA Library Prep Kit for Illumina with 1000 ng of input material. Samples were pooled for sequencing on the NovaSeq instrument.
Transcriptomics processing and quantification
RNA-seq analysis was executed and visualized using an in-house, web-based platform, consisting of the following steps. Sequencing quality control was performed using FastQC (v0.11.5). Transcript expression was then quantified using Salmon83 (v0.9.1) in pseudo-alignment mode, without adapter trimming, producing transcript-per-million estimates, using the Ensembl human GRCh38 transcriptome. For the dose response study, samples were corrected by removing the second principal component that captured variation due to batch. For visualization and time course analyses, low count genes (<25) were excluded. Differential expression analysis was performed using the DESeq2 package in R producing q values and effect sizes for the comparison between basal states of WT and ATF4 KO Dmr-PERK cells.
Transcriptomics time course analysis
To identify genes that were significantly changing over time in response to ISR induction, we used the method described in ref. 84 First, each gene measurement was normalized to its mean expression level at t = 0. Then, genes were selected if they showed a log2 fold change greater than 1.5 in any condition. Of these, the expression profile for each gene was fit to a quadratic model:
Time-dependent genes required a p value of the F statistic of the model to be <0.001. To perform pairwise comparisons of gene expression time courses between genotypes (WT vs. ATF4 KO), a multiple regression analysis was performed. We fit an additional model that took into account the genotype with the variable \({D}_{G}\):
where \({D}_{G}=0\) for WT and \({D}_{G}\,\)= 1 for ATF4 KO. We computed the p value of the F statistic between the original and genotype-aware models. Genes were classified as significantly different if the Benjamini and Yekutieli FDR-corrected p value < 0.05. The same procedure was performed to identify genes differentially expressed in two treatment conditions, with a p value cutoff of 1e-5 (dimerizer vs. thapsigargin conditions in Fig. 1f and the dimerizer vs. arsenite conditions in Fig. 1g).
To generate heatmaps, protein-coding genes were included (non-protein coding genes are included in raw data files and Supplementary Data) and hierarchical clustering and visualization was performed using the heatmap.2 function in R. Overrepresentation analysis of Gene Ontology-Biological Process terms for the genes in each cluster was performed using the R function enrichGO from the ClusterProfiler library85.
LC-MS/MS for polar metabolites
Cells were washed with room temperature PBS followed by quenching in 80% methanol containing internal standards (D4-L-tyrosine, 15N4-L-arginine and D5-benzoic acid; Sigma-Aldrich) maintained at −20 °C. Cells were scraped, transferred to microcentrifuge tubes, then centrifuged at 16,000 × g at 4 °C for 15 min. Supernatants were dried down under nitrogen at 4 °C. Dried extracts were resuspended at 5× concentration in 40% methanol/40% acetonitrile/20% water containing additional standards (D5-L-phenylalanine, D4-L-lysine and D5-succinate; Sigma-Aldrich). Metabolomics samples were analyzed in both positive and negative ESI-LC-MS methods on Vanquish UPLCs coupled to Q-Exactive Plus mass spectrometers. Metabolites were separated using a SeQuant® ZIC-pHILIC column (5 μm, 200 Å, 150 × 2.1 mm) where the mobile phase A was 20 mM ammonium carbonate in water (pH 9.2) and mobile phase B was 95:5 (v/v) acetonitrile/mobile A at a flow rate of 150 μL/min and the gradient was t = −6, 84.2% B; t = 0, 84.2% B; t = 2.5, 76.8% B; t = 5, 68.4% B, t = 7.5, 60% B; t = 10, 52.6% B; t = 15, 36.8% B; t = 20; 21% B; t = 22, 15.8% B; t = 22.5, 84.2% B; t = 24; 84.2% B. Data were acquired using data-dependent acquisition (DDA) mode with the following parameters: resolution = 70,000, AGC target = 3.00 × 10 maximum IT (ms) = 100, scan range = 70–1050. The MS2 parameters were as follows: resolution = 17,500, AGC target = 1.00 × 105, maximum IT (ms) = 50, loop count = 6, isolation window (m/z) = 1, (N)CE = 20, 40, 80; underfill ratio = 1.00%, Apex trigger(s) = 3–10, dynamic exclusion(s) = 25. For negative mode, (N)CE = 20, 50, 100.
For 13C-glucose and 13C-glutamine tracing studies, media was exchanged to glucose-free DMEM (Gibco) with addition of 25 mM U-13C6-glucose (Cambridge Isotopes) or glutamine-free DMEM (Gibco) with addition of 2 mM U-13C5-glutamine (Cambridge Isotopes), respectively, at different timepoints prior to harvest to determine steady state labeling of intracellular metabolites. Dialyzed fetal bovine serum (Corning) was utilized to reduce unlabeled glucose and glutamine derived from serum.
Relative fatty acid synthesis rates
For 2H2O tracing into lipid palmitate (C16:0), 8% deuterium oxide (Sigma) or 8% water was added to standard culture medium and media was exchanged 24 h prior to harvest. 13C-glucose and 13C-glutamine tracing were performed by replacing standard medium with medium containing 100% U-13C-glucose or U-13C-glutamine in separate cultures, 24 h before harvest. Lipids were extracted with isopropanol, and saponified by incubating in 0.6 M 75% KOH for 45 mins at 60 °C, neutralized with 25% acetic acid, dried under nitrogen and resuspended in 200 µL of ethanol.
FA analysis was performed in negative ion mode using LC-MS method of a Vanquish UPLC-Q-Exactive Plus. Compounds were separated using a Phenomenex Kinetex C8 (2.6 µm, 100 ×2.1 mm) with a column guard UHPLC C8 2.1 mm. Method was adapted from ref. 86. Label incorporation of label was determined using MAVEN287, which identified isotopic envelopes using expected MS1 m/z shifts, and quantified isotopic peaks either by standard peak quantitation approaches or by full integration of observed intensity within the [M + 0] RT peak bounds applied to each isotopic m/z. Natural 13C isotope abundance was corrected using MAVEN2’s natural isotopic abundance correction functionality and the IsoCorrectoR R package88.
MTBE-LLE extraction for lipidomics
5e5 500,000 cells per well was washed with warm PBS and quenched with 0.8 mL of 50% MeOH/H2O containing internal standards (100 µL of LipidSplash standards (Avanti Polar Lipids)) for 15 min at −20 °C. Samples were scraped and collected to a 2 mL glass vial and extracted for lipidomics analysis using methyl tert-butyl ether liquid-liquid extraction (MTBE-LLE) adopted from Matyash et al.89 In short, 800 μL of MTBE was added to the mixture, vortexed for 30 s and incubated on ice for another 15 min and centrifuged at 3000 g for 10 min at 4 °C. Lipids partitioned in the top layer were collected into a separate vial, and the extraction process was repeated with an addition of 600 µL of MTBE. After incubating and centrifuging, the second organic layer was collected and combined in the first lipid vial, dried under nitrogen at 4 °C, resuspended in 200 µL of ButOH/MeOH/H2O (2:1:1, v/v/v) for analysis.
LC-MS for lipidomics
Cells were cultured in six-well plates, and ~500,000 cells per well were washed with warm PBS and quenched with 0.8 mL of 50/50 MeOH/H2O containing 100 µL of LipidSplash internal standards (Avanti Polar Lipids), followed by incubation at −20 °C for 15 min. Samples were scraped and collected to a 2 mL glass vial and extracted for lipidomics analysis using an MTBE-LLE extraction adopted from Matyash et al.89. In brief, 800 μL of MTBE was added to the mixture, vortexed for 30 s and incubated on ice for another 15 min and centrifuged at 3000 g for 10 min at 4 °C. Lipids partitioned in the top layer were collected into a separate vial, and the extraction process was repeated with an addition of 600 µL of MTBE. After incubating and centrifuging, the second organic layer was collected and combined in the first lipid vial, dried under nitrogen at 4 °C, resuspended in 200 µL of ButOH/MeOH/H2O (2:1:1, v/v/v) for lipid analysis.
Lipidomics samples were analyzed in both positive and negative ion modes using the same LC-MS method consisting of a Vanquish UPLC coupled to a Q-Exactive Plus mass spectrometer. Lipids were separated using a Thermo Scientific Accucore C30 column (2.6 μm, 150 Å, 2.1 × 250 mm) at a flow rate of 200 μL/min. Mobile phase A was 20 mM ammonium formate in 60:40 acetonitrile:water +0.25 μM medronic acid, mobile phase B was 20 mM ammonium formate in 90:10 isopropanol:acetonitrile + 0.25 μM medronic acid. The gradient was t = −7, 30% B, t = 0, 30% B, t = 7, 43% B, t = 12, 65% B, t = 30, 70% B, t = 31, 88% B, t = 51, 95% B, t = 53, 100% B, t = 55, 100% B, t = 55.1, 30% B, t = 60, 30% B. The mass spectrometer settings were as follows: DDA was performed with the following parameters: resolution = 140,000, AGC target = 3.00 × 106, maximum IT (ms) = 100, scan range = 200–2000. The MS2 parameters were as follows: resolution = 17,500, AGC target = 3.00 × 106, maximum IT (ms) = 150, loop count = 8, isolation window (m/z) = 1, (N)CE = 20, 30, 40; underfill ratio = 1.00%, Apex trigger(s) = 5–30, dynamic exclusion(s) = 15 s.
Raw files were converted to mzML files using msconvert from ProteoWizard90, using vendor centroiding on all scans, and analyzed using MAVEN2 software87. Identification was performed by spectral matching using in-house spectral libraries87.
Statistical analysis for metabolomics and lipidomics
Analysis of the lipidomics and metabolomics data was performed using Calico Lipidomics and Metabolomics Analysis (clamanR) package (https://github.com/calico/claman) in R. Raw files were converted to the mzML format using ProteoWizard Ver 390. Compound identifications were detected and grouped using the OpenCLaM R package (https://github.com/calico/open_clam) and manually inspected using the MAVEN2 peak analysis program (https://github.com/eugenemel/maven)87 with the criteria of a precursor ion tolerance of 10 ppm and a product ion tolerance of 20 ppm, comparing fragmentation and retention time to an in-house library generated from authentic standards for metabolomics, and in-house generated in-silico libraries for lipidomics (https://github.com/calico/CalicoLipidLibrary). The sum of the the raw scan intensity of the smoothed maximum, as well as the scan immediately preceding and following the smoothed maximum scan, was taken as the peak quantity, hereafter referred to as “peak area top”. Peak area top of each feature underwent log2 transformation, peaks below the limit of detection were assigned a value of 12. Each compound was normalized to the average signal of that compound in the reference condition at t0. Heatmaps were generated using the pheatmap package in R. Relative abundance is shown as log2 fold change (lipids) or raw z scores (metabolites) as calculated using the row scaling function in pheatmap.
A linear model was fit to the data and a pairwise comparison between treatments at individual time points was performed for each identified feature using this model: \({{y}}_{{ik}}=\,\alpha+{\beta }_{1}{g}_{i}+{\beta }_{2,k}{t}_{k}+{\beta }_{3,k}{t}_{k}{g}_{i}\), where \({{y}}_{{ij}}\) are the normalized log2 to the reference condition for the annotated lipids and metabolites. Observation indenced by i = 1 are from WT U2OS, and i = 2 from ATF4-KO, so that \({\beta }_{1}\) are expected to be the difference between genotypes. k = 0, 1, 2, 4, 8 or 24 indexes the time-course study of cells in dimerizer. For lipids, the model excludes out the genotype differences since the experiment was done only in WT-U2OS. P values from these regressions were False Discovery Rate controlled on a term-by-term basis using the qvalue R package adapted from the Storey lab91. Significant changes were reported for q values < 0.01.
Absolute quantification of lipid species and lipid classes was done by normalizing peak area top of each species to the lipid standards included in LipidSplash spiked into samples prior to LLE. For lipid enrichment analysis, we ranked all identified species using the estimated values calculated from the regression model. Identified lipid species were grouped by lipid class and acyl chain composition. Relative enrichment of differentially abundant lipid species was measured using the R package FGSEA92,93. We report the −log10 Benjamini-Hochberg corrected p value and use the normalized enrichment score to assign the direction of the change.
Relative metabolic flux to glutathione biosynthesis was calculated from 13C-tracing data by determining the first order rate constants for disappearance of unlabeled gamma-Glu-Cys and glutathione, as previously described94, but using the relative, mass spectrometry-derived, peak sizes in place of the absolute pool sizes.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data generated in the current study are available in the supplementary information or in the accompanying Source Data file. Raw RNA-seq data and annotated gene counts are available in Gene expression omnibus under accession codes GSE273599, GSE273600, and GSE273601. Raw untargeted LC-MS metabolomics and lipidomics data are available in Metabolomics Workbench under study ID ST003405 and ST003398, respectively. Source data are provided with this paper.
References
Pakos-Zebrucka, K. et al. The integrated stress response. EMBO Rep. 17, 1374–1395 (2016).
Harding, H. P. et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108 (2000).
Donnelly, N., Gorman, A. M., Gupta, S. & Samali, A. The eIF2α kinases: their structures and functions. Cell. Mol. Life Sci. 70, 3493–3511 (2013).
Merrick, W. C. & Pavitt, G. D. Protein synthesis initiation in eukaryotic cells. Cold Spring Harb. Perspect. Biol. 10, a033092 (2018).
Hinnebusch, A. G. & Lorsch, J. R. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb. Perspect. Biol. 4, a011544 (2012).
Adomavicius, T. et al. The structural basis of translational control by eIF2 phosphorylation. Nat. Commun. 10, 2136–2136 (2019).
Krishnamoorthy, T., Pavitt, G. D., Zhang, F., Dever, T. E. & Hinnebusch, A. G. Tight binding of the phosphorylated alpha subunit of initiation factor 2 (eIF2alpha) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Mol. Cell Biol. 21, 5018–5030 (2001).
Harding, H. P. et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633 (2003).
Torrence, M. E. et al. The mTORC1-mediated activation of ATF4 promotes protein and glutathione synthesis downstream of growth signals. Elife 10, e63326 (2021).
Lu, P. D., Harding, H. P. & Ron, D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 167, 27–33 (2004).
Vattem, K. M. & Wek, R. C. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl Acad. Sci. USA 101, 11269–11274 (2004).
Alam, O. A single-cell-type transcriptomics map of human tissues. Nat. Genet. 53, 1275–1275 (2021).
Zinszner, H. et al. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 12, 982–995 (1998).
Oyadomari, S. & Mori, M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ. 11, 381–389 (2003).
Novoa, I. et al. Stress-induced gene expression requires programmed recovery from translational repression. EMBO J. 22, 1180–1187 (2003).
Novoa, I., Zeng, H., Harding, H. P. & Ron, D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J. Cell Biol. 153, 1011–1022 (2001).
Kedersha, N., Ivanov, P. & Anderson, P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci. 38, 494–506 (2013).
Glauninger, H., Wong Hickernell, C. J., Bard, J. A. M. & Drummond, D. A. Stressful steps: progress and challenges in understanding stress-induced mRNA condensation and accumulation in stress granules. Mol. Cell 82, 2544–2556 (2022).
Buchan, J. R. & Parker, R. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36, 932–941 (2009).
Pavitt, G. D. & Ron, D. New insights into translational regulation in the endoplasmic reticulum unfolded protein response. Cold Spring Harb. Perspect. Biol. 4, a012278 (2012).
Walter, P. & Ron, D. The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011).
Blais, J. D. et al. Activating transcription factor 4 is translationally regulated by hypoxic stress. Mol. Cell Biol. 24, 7469–7482 (2004).
Ryan, D. G. et al. Disruption of the TCA cycle reveals an ATF4-dependent integration of redox and amino acid metabolism. eLife 10, e72593 (2021).
Bao, X. R. et al. Mitochondrial dysfunction remodels one-carbon metabolism in human cells. eLife 5, e10575 (2016).
Quirós, P. M. et al. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 216, 2027–2045 (2017).
Ye, J. et al. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 29, 2082–2096 (2010).
Oyadomari, S., Harding, H. P., Zhang, Y., Oyadomari, M. & Ron, D. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 7, 520–532 (2008).
Lu, P. D. et al. Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 23, 169–179 (2004).
Miyake, M. et al. Ligand-induced rapid skeletal muscle atrophy in HSA-Fv2E-PERK transgenic mice. PLoS One 12, e0179955 (2017).
Wong, Y. L. et al. eIF2B activator prevents neurological defects caused by a chronic integrated stress response. eLife 8, e42940 (2019).
Li, Y., Jourdain, A. A., Calvo, S. E., Liu, J. S. & Mootha, V. K. CLIC, a tool for expanding biological pathways based on co-expression across thousands of datasets. PLoS Comput. Biol. 13, e1005653 (2017).
Adamson, B. et al. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167, 1867–1882.e21 (2016).
Scheuner, D. et al. Double-stranded RNA-dependent protein kinase phosphorylation of the α-subunit of eukaryotic translation initiation factor 2 mediates apoptosis. J. Biol. Chem. 281, 21458–21468 (2006).
Sidrauski, C., McGeachy, A. M., Ingolia, N. T. & Walter, P. The small molecule ISRIB reverses the effects of eIF2α phosphorylation on translation and stress granule assembly. eLife 4, e05033 (2015).
Zyryanova, A. F. et al. ISRIB blunts the integrated stress response by allosterically antagonising the inhibitory effect of phosphorylated eIF2 on eIF2B. Mol. Cell 81, 88–103.e6 (2021).
Tsai, J. C. et al. Structure of the nucleotide exchange factor eIF2B reveals mechanism of memory-enhancing molecule. Science 359, eaaq0939 (2018).
Lee, Y.-Y., Cevallos, R. C. & Jan, E. An upstream open reading frame regulates translation of GADD34 during cellular stresses that induce eIF2alpha phosphorylation. J. Biol. Chem. 284, 6661–6673 (2009).
Fusakio, M. E. et al. Transcription factor ATF4 directs basal and stress-induced gene expression in the unfolded protein response and cholesterol metabolism in the liver. Mol. Biol. Cell 27, 1536–1551 (2016).
Lomelino, C. L., Andring, J. T., McKenna, R. & Kilberg, M. S. Asparagine synthetase: function, structure, and role in disease. J. Biol. Chem. 292, 19952–19958 (2017).
Fan, J. et al. Glutamine-driven oxidative phosphorylation is a major ATP source in transformed mammalian cells in both normoxia and hypoxia. Mol. Syst. Biol. 9, 712 (2013).
Metallo, C. M. et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481, 380–384 (2011).
Mullen, A. R. et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 481, 385–388 (2011).
Lee, J. J. et al. eIF2B activator rescues neonatal lethality of an eIF2Bα sugar phosphate binding mutation associated with vanishing white matter disease. https://doi.org/10.1101/2023.05.06.539602 (2023).
Schwörer, S. et al. Proline biosynthesis is a vent for TGFβ‐induced mitochondrial redox stress. EMBO J. 39, e103334 (2020).
Olzmann, J. A. & Carvalho, P. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 20, 137–155 (2019).
Walther, T. C. & Farese, R. V. Jr Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem 81, 687–714 (2012).
Amen, T. & Kaganovich, D. Stress granules inhibit fatty acid oxidation by modulating mitochondrial permeability. Cell Rep. 35, 109237 (2021).
Nguyen, T. B. et al. DGAT1-dependent lipid droplet biogenesis protects mitochondrial function during starvation-induced autophagy. Dev. Cell 42, 9–21.e5 (2017).
Jarc, E. & Petan, T. A twist of FATe: lipid droplets and inflammatory lipid mediators. Biochimie 169, 69–87 (2020).
Sidrauski, C. et al. Pharmacological dimerization and activation of the exchange factor eIF2B antagonizes the integrated stress response. eLife 4, e07314 (2015).
Condon, K. J. et al. Genome-wide CRISPR screens reveal multitiered mechanisms through which mTORC1 senses mitochondrial dysfunction. Proc. Natl. Acad. Sci. USA 118, e2022120118 (2021).
Hosios, A. M. et al. mTORC1 regulates a lysosome-dependent adaptive shift in intracellular lipid species. Nat. Metab. 4, 1792–1811 (2022).
Brugarolas, J. et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 18, 2893–2904 (2004).
Ye, J. et al. GCN2 sustains mTORC1 suppression upon amino acid deprivation by inducing Sestrin2. Genes Dev. 29, 2331–2336 (2015).
Yen, C.-L. E. et al. Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J. Lipid Res. 49, 2283–2301 (2008).
Yu, J., Li, Y., Zou, F., Xu, S. & Liu, P. Phosphorylation and function of DGAT1 in skeletal muscle cells. Biophys. Rep. 1, 41–50 (2015).
Caldo, K. M. P. et al. Diacylglycerol acyltransferase 1 is activated by phosphatidate and inhibited by SnRK1‐catalyzed phosphorylation. Plant J. 96, 287–299 (2018).
Schoof, M. et al. eIF2B conformation and assembly state regulate the integrated stress response. eLife 10, e65703 (2021).
Stone, S. J. et al. The endoplasmic reticulum enzyme DGAT2 is found in mitochondria-associated membranes and has a mitochondrial targeting signal that promotes its association with mitochondria. J. Biol. Chem. 284, 5352–5361 (2009).
Han, J. et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 15, 481–490 (2013).
Mick, E. et al. Distinct mitochondrial defects trigger the integrated stress response depending on the metabolic state of the cell. Elife 9, e49178 (2020).
Guan, B.-J. et al. A unique ISR program determines cellular responses to chronic stress. Mol. Cell 68, 885–900.e6 (2017).
Rutkowski, D. T. et al. Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLoS Biol. 4, e374 (2006).
Ben-Sahra, I., Hoxhaj, G., Ricoult, S. J. H., Asara, J. M. & Manning, B. D. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science 351, 728–733 (2016).
Park, Y., Reyna-Neyra, A., Philippe, L. & Thoreen, C. C. mTORC1 balances cellular amino acid supply with demand for protein synthesis through post-transcriptional control of ATF4. Cell Rep. 19, 1083–1090 (2017).
Nunnari, J. & Walter, P. Regulation of organelle biogenesis. Cell 84, 389–394 (1996).
Cox, J. S., Chapman, R. E. & Walter, P. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol. Biol. Cell 8, 1805–1814 (1997).
Bagnato, C. & Igal, R. A. Overexpression of diacylglycerol acyltransferase-1 reduces phospholipid synthesis, proliferation, and invasiveness in Simian virus 40-transformed human lung fibroblasts. J. Biol. Chem. 278, 52203–52211 (2003).
Jiang, X., Stockwell, B. R. & Conrad, M. Ferroptosis: mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 22, 266–282 (2021).
Chitraju, C. et al. Triglyceride synthesis by DGAT1 protects adipocytes from lipid-induced ER stress during lipolysis. Cell Metab. 26, 407–418.e3 (2017).
Vevea, J. D. et al. Role for lipid droplet biogenesis and microlipophagy in adaptation to lipid imbalance in yeast. Dev. Cell 35, 584–599 (2015).
Bailey, A. P. et al. Antioxidant role for lipid droplets in a stem cell niche of drosophila. Cell 163, 340–353 (2015).
Bensaad, K. et al. Fatty acid uptake and lipid storage induced by HIF-1α contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep. 9, 349–365 (2014).
Ackerman, D. et al. Triglycerides promote lipid homeostasis during hypoxic stress by balancing fatty acid saturation. Cell Rep. 24, 2596–2605.e5 (2018).
Rambold, A. S., Cohen, S. & Lippincott-Schwartz, J. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev. Cell 33, 489–490 (2015).
Batjargal, T. et al. Optogenetic control of the integrated stress response reveals proportional encoding and the stress memory landscape. https://doi.org/10.1101/2022.05.24.493309 (2022).
Teske, B. F. et al. The eIF2 kinase PERK and the integrated stress response facilitate activation of ATF6 during endoplasmic reticulum stress. Mol. Biol. Cell 22, 4390–4405 (2011).
Kim, J. M., Kim, H. G. & Son, C. G. Tissue-specific profiling of oxidative stress-associated transcriptome in a healthy mouse model. Int. J. Mol. Sci. 19, 3174 (2018).
Ralhan, I., Chang, C.-L., Lippincott-Schwartz, J. & Ioannou, M. S. Lipid droplets in the nervous system. J. Cell Biol. 220, e202102136 (2021).
Costa-Mattioli, M. & Walter, P. The integrated stress response: from mechanism to disease. Science 368, eaat5314 (2020).
Lefebvre, A. E. Y. T. et al. Nellie: automated organelle segmentation, tracking, and hierarchical feature extraction in 2D/3D live-cell microscopy. Preprint at http://arxiv.org/abs/2403.13214 (2024).
Austin Epiphane Yann Tung-Shan Lefebvre. aelefebv/LD_quantification: Paper accepted. Zenodo https://doi.org/10.5281/ZENODO.12768009 (2024).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
McIsaac, R. S., Petti, A. A., Bussemaker, H. J. & Botstein, D. Perturbation-based analysis and modeling of combinatorial regulation in the yeast sulfur assimilation pathway. Mol. Biol. Cell 23, 2993–3007 (2012).
Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Koch, E., Wiebel, M., Hopmann, C., Kampschulte, N. & Schebb, N. H. Rapid quantification of fatty acids in plant oils and biological samples by LC-MS. Anal. Bioanal. Chem. 413, 5439–5451 (2021).
Seitzer, P., Bennett, B. & Melamud, E. MAVEN2: an updated open-source mass spectrometry exploration platform. Metabolites 12, 684 (2022).
Heinrich, P. et al. Correcting for natural isotope abundance and tracer impurity in MS-, MS/MS- and high-resolution-multiple-tracer-data from stable isotope labeling experiments with IsoCorrectoR. Sci. Rep. 8, 17910–17910 (2018).
Matyash, V., Liebisch, G., Kurzchalia, T. V., Shevchenko, A. & Schwudke, D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 49, 1137–1146 (2008).
Chambers, M. C. et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 (2012).
Storey, J. D. False discovery rate. International Encyclopedia of Statistical Science (ed. Lovric, M.) 504–508. https://doi.org/10.1007/978-3-642-04898-2_248 (Springer Berlin Heidelberg, 2011).
Kessler, N. et al. MeltDB 2.0–advances of the metabolomics software system. Bioinformatics 29, 2452–2459 (2013).
Xia, J., Mandal, R., Sinelnikov, I. V., Broadhurst, D. & Wishart, D. S. MetaboAnalyst 2.0—a comprehensive server for metabolomic data analysis. Nucleic Acids Res. 40, W127–W133 (2012).
Yuan, J., Fowler, W. U., Kimball, E., Lu, W. & Rabinowitz, J. D. Kinetic flux profiling of nitrogen assimilation in Escherichia coli. Nat. Chem. Biol. 2, 529–530 (2006).
Acknowledgements
We thank our Calico colleagues Twaritha Vijay, Hannah DeBaets, Irene Lam, Amy Jo Johnson, Ikenna Anigbogu, Andrew Keyser, David Hendrickson, and Margaret Roy for their support for RNA sequencing. We thank Scott McIsaac for assistance with analysis of time course gene expression data. This work was supported by Calico Life Sciences LLC.
Author information
Authors and Affiliations
Contributions
K.L., L.L., B.K., B.B. and C.S. conceptualized the work. K.L., L.L., B.K., N.V., E.H.S., N.L., A.E.Y.T.L., P.S., S.K. and J.H. performed experiments and data analysis. K.L., L.L., B.K., N.V., B.B. and C.S. wrote the manuscript. All authors contributed to review and editing.
Corresponding authors
Ethics declarations
Competing interests
K.L., L.L., BK., N.V., E.H.S., N.L., A.E.Y.T.L., P.S., B.B., and C.S. are employees of Calico Life Sciences LLC and declare no other competing interests. C.S. is an inventor on U.S. Patent 9708247 describing ISRIB and its analogs. Rights to the invention have been licensed to Calico Life Sciences LLC from the University of California, San Francisco.
Peer review
Peer review information
Nature Communications thanks Martin Giera and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Labbé, K., LeBon, L., King, B. et al. Specific activation of the integrated stress response uncovers regulation of central carbon metabolism and lipid droplet biogenesis. Nat Commun 15, 8301 (2024). https://doi.org/10.1038/s41467-024-52538-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-52538-5
This article is cited by
-
Mammalian mitohormesis: from mitochondrial stressors to organismal benefits
The EMBO Journal (2025)
-
Arginine depletion potentiates standard-of-care chemo-immunotherapy in preclinical models of high-risk neuroblastoma
Journal of Experimental & Clinical Cancer Research (2025)