Abstract
The basal ganglia are subcortical brain structures involved in motor control, cognition, and emotion regulation. We conducted univariate and multivariate genome-wide association analyses (GWAS) to explore the genetic architecture of basal ganglia volumes using brain scans obtained from 34,794 Europeans with replication in 4,808 white and generalization in 5,220 non-white Europeans. Our multivariate GWAS identified 72 genetic loci associated with basal ganglia volumes with a replication rate of 55.6% at P < 0.05 and 87.5% showed the same direction, revealing a distributed genetic architecture across basal ganglia structures. Of these, 50 loci were novel, including exonic regions of APOE, NBR1 and HLAA. We examined the genetic overlap between basal ganglia volumes and several neurological and psychiatric disorders. The strongest genetic overlap was between basal ganglia and Parkinson’s disease, as supported by robust LD-score regression-based genetic correlations. Mendelian randomization indicated genetic liability to larger striatal volume as potentially causal for Parkinson’s disease, in addition to a suggestive causal effect of greater genetic liability to Alzheimer’s disease on smaller accumbens. Functional analyses implicated neurogenesis, neuron differentiation and development in basal ganglia volumes. These results enhance our understanding of the genetic architecture and molecular associations of basal ganglia structure and their role in brain disorders.
Similar content being viewed by others
Introduction
The basal ganglia are a group of interconnected subcortical nuclei deep in the brain1. The major parts of the basal ganglia are located in the cerebrum and include the caudate nucleus, putamen, globus pallidus and the accumbens area2,3. Substantia nigra in the midbrain and the subthalamic nucleus in diencephalon may be regarded as associated structures. Although the basal ganglia comprise physically distinct entities, the individual nuclei exhibit a robust functional cohesion due to their intricate interconnections, and is often regarded as a functional unit4.
The basal ganglia integrate and modulate cortical information, and are involved in motor5, cognitive6 and limbic functions3,7. The functional unit is involved in motor functions through initiation, execution, and coordination of movements8, action selection6 and in learning and execution of procedural memory and habits, allowing actions to become automatic and efficient over time9,10. Beyond motor control, the basal ganglia play important roles in cognitive functions like decision-making6, the shifting of attention, updating information, and adapting behavior6,11,12,13, in addition to reward processing, which shapes behavior, decision-making14 and motivation15. The basal ganglia are involved in emotional processing and the regulation of affective states3,7, and integration of emotional information in decision-making processes3,16. However, there is strong evidence supporting that the different basal ganglia are integrated, as the function of the individual nuclei depends on input from neighboring nuclei, making basal ganglia an interconnected network4.
Likely due to their widespread connections to other parts of the brain, the basal ganglia have been implicated in several brain diseases, ranging from neurodegenerative to psychiatric and neurodevelopmental conditions. The most well-known disorder related to the basal ganglia is Parkinson’s disease (PD), where loss of dopaminergic projections from the substantia nigra to the basal ganglia leads to progressive motor symptoms. In Alzheimer’s disease (ALZ), evidence suggests that tau- and amyloid aggregation can affect the basal ganglia and contribute to cognitive and motor impairments17. Through pain processing and modulation, basal ganglia are also involved in the pathophysiology of the primary headache disorder migraine (MIG). Dysfunction within the basal ganglia circuits may contribute to stereotyped/repetitive movements, reduced attentional control12,13 and social and emotional processing7 often observed in individuals with autism spectrum disorder (ASD)18,19. Through its role in regulating motor hyperactivity20,21, reward processing22,23,24 and inhibitory control25,26, the basal ganglia is also central for the pathophysiology of attention-deficit/hyperactivity disorder (ADHD)25,26,27. Dysregulation of dopamine neurotransmission in the mesocorticolimbic pathway involving the basal ganglia has been implicated in ADHD28,29,30, but also related to hallucinations and delusions in schizophrenia (SCZ)31,32, while cortico-striatal circuitry abnormalities may contribute to the cognitive impairments observed in SCZ33,34. The basal ganglia also play a role in mood regulation and emotional processing35,36, and basal ganglia functions have been implicated in both bipolar disorder (BIP)36 and major depressive disorder (MDD)35,37. Studies of basal ganglia volumes have in many circumstances showed inconsistent results. For MIG38, PD39, MDD40, ASD13, and ADHD41,42 some studies report reduced basal ganglia volumes, while others report no significant differences43,44, or even striatal enlargement45,46,47,48,49.
While the functional aspects of the basal ganglia have been extensively studied, the genetic architecture underlying their structural characteristics remains largely unexplored. There are, however, genetic studies on volumetrics of subcortical structures which includes basal ganglia nuclei50,51, without focusing on the nuclei as a functional unit. Hibar and colleagues50 reported one locus significantly associated with caudate nucleus and four loci significantly associated with putamen, but none with accumbens area or globus pallidus. Satizabal and colleagues51, on the other hand, reported four loci associated with the accumbens area, 10 loci with the caudate nucleus, six loci with globus pallidus and nine loci with putamen. A comprehensive understanding of the genetic mechanisms shaping basal ganglia volumes is lacking. The current study represents the multivariate genome wide association study (GWAS) on the basal ganglia volumes, an analytical approach that can take advantage of basal ganglia as one functional unit. Thus, there are likely pleiotropic genetic variants shared across the different nuclei, which can be identified with the multivariate MOSTest method52. Still, we also include univariate GWASs for comparisons with earlier studies50,51. Understanding the genetic basis of basal ganglia volumes as a whole can provide insights into the pathogenesis and etiology of basal ganglia-related brain disorders, which can form the basis for the future development of targeted therapeutic interventions.
Results
Univariate GWASs reveal 47 novel genetic loci associated with basal ganglia
Univariate GWAS analyses on individual nuclei collectively known as the basal ganglia revealed 60 unique loci, 43 of which were novel. We identified 11 loci associated with accumbens area (Supplementary Data 1, Supplementary Fig. 1), where three out of the four loci reported by Satizabal et al.51 are replicated, leaving eight novel loci. We report 26 genetic loci significantly associated with the caudate nucleus (Supplementary Data 1, Supplementary Fig. 1), of which 18 loci are novel, due to replication of eight out of the ten loci reported by Satizabal et al.51, while the one locus reported by Hibar et al.50 is not replicated. For pallidum, we report 13 significantly associated genetic loci (Supplementary Data 1, Supplementary Fig. 1), nine of which are novel, while four are replications of the six loci reported by Satizabal et al.51. For putamen, we replicate all four loci reported associated with putamen by Hibar et al.50 and seven out of the nine loci reported by Satizabal et al.51, in addition to 15 novel loci, altogether 23 loci, significantly associated with putamen in our univariate GWAS (Supplementary Data 1, Supplementary Fig. 1). We have also performed a univariate GWAS on basal ganglia as a functional unit (one volume), resulting in 22 significant loci spanning 12 chromosomes (Supplementary Data 1, Supplementary fig. 1). Of these loci, 14 have not been previously reported from the univariate GWASs from the individual nuclei of which basal ganglia consists50,51. There is a large overlap between the loci identified in the univariate GWASs of the different basal ganglia nuclei (mean 22%, range 9–38%) and between the univariate GWAS of the basal ganglia as one volume and univariate GWASs of the different basal ganglia nuclei (mean 42%, range 27–65%), supporting the basal ganglia as a functional group of intertwingled subcortical nuclei.
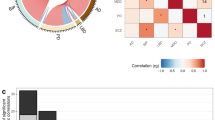
All basal ganglia nuclei showed significant single nucleotide polymorphism (SNP)-heritability (Fig. 1A), with highest estimates for the caudate (h2 = 0.34, se=0.025) and the whole basal ganglia (h2 = 0.31, se=0.026). Figure 1B shows a correlation matrix of basal ganglia volumes, with phenotypic correlation shown in upper-left section and genetic correlation using the univariate GWAS summary statistics shown in the lower-right section. As expected, genetic correlations mapped phenotypic correlations (Supplementary Data 2). The overall lowest genetic correlations were found between pallidum and accumbens (rg = 0.1153, see = 0.056), which are also the structures with the lowest estimated heritability.
A Heritability of basal ganglia volumes (n = 34,794), where the numbers depict SNP-heritability estimates (h2). In the plot, the dots represent mean heritability for each subregion of the basal ganglia, while the error bars represent the standard error (SEM): Putamen in pink (h2 = 0.296, SEM = 0.0.026), Pallidum in light blue (h2 = 0.253, SEM = 0.021), Caudate in red (h2 = 0.342, SEM = 0.025), Accumbens area in green (h2 = 0.261, SEM = 0.02) and whole basal ganglia in gray (h2 = 0.304, SEM = 0.026). B LD-score regression-based genetic correlations (in lower-right section) and phenotypic correlation (in upper-left section) between each pair of regions, using the univariate GWAS summary statistics.
Multivariate GWAS reveals 72 genetic loci associated with basal ganglia
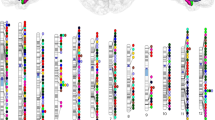
A multivariate GWAS deployed using the MOSTest framework53 identified 72 significant independent loci, including 50 novel loci (Supplementary Data 3). We here adopt a conservative definition of novelty, wherein we characterize it as loci identified through the multivariate MOSTest approach for the basal ganglia as a unified functional unit, ensuring non-overlapping with any loci previously identified in univariate GWASs for any of the individual basal ganglia nuclei.We thresholded based on genome-wide significance (P < 5 × 10−8) and identified a total of 12305 candidate SNPs, 294 independent significant SNPs and 89 lead SNPs across structures located in the 72 genomic loci, using the FUMA platform54 (Supplementary Data 4). Figure 2 illustrates the results of multivariate and univariate GWAS, where the upper part of the Miami plot shows the multivariate polygenic architecture across basal ganglia volumes. A distributed genetic architecture throughout the basal ganglia structure is supported by the higher multivariate statistics in comparison with the univariate statistics for various basal ganglia volumes in most of the identified loci (Fig. 1), which is further confirmed by genetic correlation analysis of the individual volumes (Supplementary Data 2).
A Schematic illustration of the basal ganglia regions, comprising the anatomically distinguishable subfields of putamen (pink), pallidum (light blue), caudate (red) and accumbens (green). Created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license B The upper part of the Miami plot illustrates the −log10(P) statistic from the multivariate GWAS across the entire basal ganglia (Multivariate Omnibus Statistical Test (MOSTest)), with 72 significant loci. For comparison, the lower part depicts for each of the 72 unique loci the corresponding −log10(P) statistics from univariate GWASs of single subregions (one color per subregion, black indicates non-significant SNPs, p-values are two-tailed), supporting a distributed genetic architecture across the basal ganglia structure. The color codes for the schematic illustration (A) and the Miami plot (B) are consistent and given in the bottom part of the figure.
Although the strongest associations among the 72 significant independent loci that were identified in the multivariate framework are also present in the results of univariate analysis, a considerable proportion of these loci demonstrated higher effects that were not significant at the genome-wide level. Leveraging these distributed effects across the various subregions, the multivariate approach resulted in enhanced discovery. Q–Q plots from MOSTest analysis (Supplementary Fig. 2) including one from permutation testing, showed successive validity of the multivariate test statistic with the LD-score regression intercept determined to be 1.024. The findings were supported by a multivariate replication study from white UK Biobank and a multivariate generalization (non-white) study that produced the same effect direction for 87.5% of the lead SNPs (Supplementary Fig. 3).
Functional annotation, gene mapping and genetic analyses
We functionally annotated SNPs associated with basal ganglia volumes that were in LD (r2 ≥ 0.6) with one of the independent significant SNPs with P < 5 × 10−8 in the discovery sample using FUMA v1.4.154. A majority of these SNPs were intronic (53.5%) or intergenic (29.1%) and 1.1% were exonic (Supplementary Fig. 4A and Supplementary Data 4). Supplementary Fig. 4B provides information for functional SNP categories for the basal ganglia volume. About 81.8% of the SNPs had a minimum chromatin state of 1–7, thus suggesting they were in open chromatin regions (Supplementary Fig. 4C)55,56. Three of the lead SNPs were exonic and combined annotation-dependent depletion (CADD) scores of those SNPs were 23.1 (rs13107325), 18.4 (rs2070835) and 12.64 (rs429358), thus indicating deleterious protein effects57 (Supplementary Data 3). rs13107325 and rs429358 are located in SLC39A8 and APOE, respectively, and have previously been associated with SCZ, PD and ALZ58,59,60,61.
Genome-wide gene-based association analyses (GWGAS; P < 2.622 × 10−6, i.e., 0.05/19073 genes) using MAGMA v1.0862 detected 149 unique genes across the basal ganglia (Supplementary Data 5). Supplementary Fig. 5 provides Manhattan and Q–Q plots for the GWGAS. Gene-set analyses using MAGMA identified significant Gene Ontology sets for neurogenesis, neuron differentiation and development (Supplementary Data 6).
Open target genetics
We additionally used Open Target Genetics to identify the target genes for each lead SNP of the 72 loci implied for the basal ganglia volumes (Fig. 3 and Supplementary Data 3).
We used the 73 uniquely mapped genes from Open Targets for gene set analysis, pathway analysis and differentially expressed genes (DEG) analysis. GO gene-set analysis for the target genes of the significant loci revealed 16 significantly associated biological processes, including ‘GO_NEURON_DIFFERENTIATION’, ‘GO_NEUROGENESIS’ and ‘GO_CELL_PART_MORPHOGENESIS’ (Supplementary Data 7). These genes were also significantly associated with four cellular component gene-sets, including ‘GO_NEURON_PROJECTION’ and ‘GO_SYNAPSE’ (Supplementary Data 7). There were 19 pathways significantly overrepresented among the target genes of the significant loci with ‘the DSCAM interactions’, ‘Axon guidance‘, ‘Nervous system development’ and ‘Netrin-1 signaling’ as the most significant (Supplementary Data 8).
We also tested the tissue specificity using the DEG sets defined for the target genes of the identified loci. The results show that these target genes are significantly expressed in the brain and specifically in basal ganglia (Supplementary Figs. 6,7).
In addition, we found enrichment for the mapped genes in the lymphatic system, nervous systems, and sensory systems (Supplementary Fig. 8A); the top enriched cell types were mostly related to the immune and nervous systems (natural killer cells, T cells, dendritic cells, bipolar neurons, inhibitory neurons, and endothelial cells) (Supplementary Fig. 8B). We also showed that the mapped genes expressed in the brain have cell-type-specific expression patterns in the main cell types of the human cerebral cortex (Supplementary Fig. 9).
We determined protein–protein and co-expression networks for the mapped genes. The genes have 14.17 and 16.41 physical interaction and gene co-expression, respectively (Supplementary Fig. 10). A gene drug interaction analysis shows that 13 out of 73 genes have interaction with 124 drugs (Supplementary Data 9), including antipsychotic (risperidone, chlorpromazine) and antiepileptic/mood stabilizing drugs (lamotrigine, carbamazepine). Within the UK Biobank, data on drug use are available for 60 out of the 124 drugs included in the gene-drug interaction analysis. The number of individuals in our cohort using any of these 60 drugs are illustrated for relevant characterization of our cohort (Supplementary Fig. 11).
Genetic overlap between the basal ganglia and common brain disorders
To further examine the polygenic architecture of basal ganglia volumes and the potential genetic overlap between basal ganglia and common brain disorders, we used GWAS summary statistics for ADHD, ASD, BIP, MDD, SCZ, ALZ, MIG, and PD. Genetic correlations of the disorders with individual basal ganglia subregions revealed only a significant association with PD after Bonferroni correction (Supplementary Fig. 12 and Supplementary Data 10). Conditional Q–Q plots conditioning the multivariate statistic of basal ganglia on the disorders and vice versa clearly demonstrated a pattern of pleiotropic enrichment in both directions (Supplementary Fig. 13) as a leftward shift in the group of SNPs with higher significance. This shift towards significance in the conditional Q-Q plot implies that the genetic variants associated with one phenotype (either basal ganglia or brain disorders) are also enriched for association with the other phenotype, supporting the notion of shared genetic factors or pleiotropy, as outlined63,64,65. Conjunctional FDR analysis allowed us to test for shared loci between the basal ganglia and each of the disorders. We identified 3 loci significantly overlapping with ADHD, 2 loci with ASD, 20 with BIP, 83 with SCZ, 15 with MDD, 33 with MIG, 21 with ALZ and 28 with PD (Fig. 4 and Supplementary Data 11-18).
Conjunctional FDR Manhattan plots, showing the –log10 transformed conjunctional FDR values for each SNP on the y-axis and chromosomal positions along the x-axis. The dotted horizontal line represents the threshold for significant shared associations (conjFDR < 0.05). Independent lead SNPs are encircled in black. ASD autism spectrum disorder, ADHD attention deficit hyperactivity disorder, SCZ schizophrenia, BIP bipolar disorder, MIG migraine, MDD, major depression, PD Parkinson’s disease, ALZ Alzheimer’s disease.
Gene mapping and overlapping shared significant loci and genes
A full list of loci overlapping between basal ganglia and the eight disorders is provided in Supplementary Datas 11–18. Figure 5 illustrates the number of significantly shared loci and genes across each combination of disorders. First, we identified significant shared loci according to the FUMA protocol54 for each conjFDR analyses. Across all phenotypes, we grouped physically overlapping loci, resulting in a total number of 159 distinct loci including 103 loci that were associated with psychiatric disorders and 73 loci associated with neurological disorders. In total, 17 loci were shared between neurological and psychiatric disorders.
A The figure shows the total number of genes overlapping for each combination of disorders. For example, 8 of the genes overlapping between basal ganglia and SCZ were also found to overlap between basal ganglia and MDD. B All mapped genes to overlapped loci between basal ganglia and disorders and genes that were implied for more than 4 disorders (shown in red). Genes TMEM161B and HP were mapped for four disorders. ASD autism spectrum disorder, ADHD attention deficit hyperactivity disorder, SCZ schizophrenia, BIP bipolar disorder, MIG migraine, MDD, major depression, PD Parkinson’s disease, ALZ Alzheimer’s disease.
Next, we mapped each of these shared loci to significantly associated protein-coding genes using Open target mapping (Supplementary Data 11–18). Overall, the extent of gene pleiotropy was similar to that observed for loci. A total of 165 distinct genes were identified across all phenotypes. Of these, 106 and 75 genes were associated with psychiatric and neurological disorders, respectively, of which 16 were overlapping (Table 1).
We also examined genes that were implicated in multiple disorders. By far the strongest overlap was found between SCZ and MDD, where 8 of the genes overlapping between basal ganglia and SCZ were also found to overlap between basal ganglia and MDD (Fig. 5A). We also found large overlap between other combinations of disorders, such as SCZ and PD (7 genes), and SCZ and BIP (6 genes) (Fig. 5A).
Some of the genes were implicated in more than two disorders. Figure 5B shows all genes overlapping between the basal ganglia volumes and the 8 disorders. The most frequently mapped genes were the HP and TMEM161B genes, which overlapped between basal ganglia and at least four disorders. (Fig. 5B).
Causal interference between the basal ganglia nuclei and common brain disorders
To investigate the potential causal links between basal ganglia nuclei and eight common brain diseases, Mendelian randomization was performed using the same GWAS summary statistics as for conjFDR: ADHD, ASD, BIP, MDD, SCZ, ALZ, MIG, and PD. Individual Mendelian randomization analyses of the disorders with each basal ganglia nuclei (Supplementary Data 19 and 20) identified that the greater genetic liability to ALZ may cause lower accumbens volume (weighted-median: beta = −5.36, padj = 7.31 × 10−3; weighted-mode: beta = −6.14, padj = 3.40 × 10−3; MR-PRESSO: beta = −4.81, padj = 4.02 × 10−2). Meanwhile, there was weak evidence that greater volume of the caudate (IVW: beta = 0.0011, padj = 6.74 × 10−3) and putamen (Egger: beta = 0.0012, padj = 4.67 × 10−2) may increase risk of PD. As both ALZ and PD exhibit a long preclinical disease trajectory, sensitivity analyses were performed using two alternative age cut-offs: 75 and 65 years old, encompassing n = 32,455 and n = 17,591 individuals, respectively. The results remained robust when using the 75-year age cut-off. However, for the 65-year age cut-off, only the association between greater volume of caudate (IVW: beta=0.0011, padj = 1.43 × 10−2; weighted-median: beta = 0.0014, padj = 1.06 × 10−2) and the risk of PD remained significant after adjustment for multiple testing (Supplementary Data 21).
Discussion
In summary, our multivariate GWAS of basal ganglia volumes uncovered 72 genomic loci, 50 of which were not previously identified. Overall, the results indicate that the genetic effects on basal ganglia structures are distributed across basal ganglia structures in line with their function as one unit. The mapped genes have pivotal roles in neurotransmission, neuronal differentiation and synaptogenesis, underscoring their significance in shaping neural circuitry and facilitating brain development. The overlapping genetic architecture and shared genes between basal ganglia and common neurological and psychiatric disorders suggest potential disease-independent drug targets.
Our findings align with and expand upon earlier reports from univariate analyses of studies including basal ganglia volumes together with other subcortical structures. Previous univariate GWASs including basal ganglia nuclei reported five50 and 2951 significant loci, respectively, associated with basal ganglia volumes. We replicated 21 loci in our multivariate basal ganglia GWAS and 23 significant loci in our univariate GWASs of accumbens area, caudate, pallidum and putamen. With a total sample size of 30,717, spanning 48 cohorts, Hibar et al.50 reported reported one locus associated with caudate, four loci with putamen, and none with accumbens or globus pallidus. Satizabal et al.51, analyzed a sample of 32,562–37,571, depending on the structure (53 cohorts), and reported four loci associated with accumbens, 10 loci with caudate, six loci with globus pallidus and nine loci with putamen. In the present study, we identified 11 loci associated with accumbens, 26 loci with caudate, 13 loci with globus pallidus and 23 loci with putamen in univariate GWASs on a sample size of 34,794 from the UK Biobank. Of the 95 loci from the five different univariate GWASs, 60 represent unique loci due to a large overlap of loci between the nuclei within the basal ganglia, supporting the interconnectivity between these nuclei. Thus, by leveraging the overlap and integration of function, we were able to increase gene discovery even further with a multivariate GWAS applying the MOSTest. Both our univariate and multivariate GWAS results, represent a large increase in gene discovery from previous work. Our improved results are, beyond the multivariate GWAS approach, likely due to less methodological biases since our study sample is from one cohort, all genotyped with same procedure66, harmonized imaging data from only four MRI scanners (identical 3 Tesla scanner models, type of coils and protocols), and the same postprocessing protocol across all individuals67.
Our two most significant loci associated with the basal ganglia have been consistently reported in the previous univariate GWASs on subcortical volumes. One of these loci is located at the 11q14-3 region near the FAT3 gene, which plays a crucial role in neuronal morphogenesis and cell migration as a conserved cellular adhesion molecule68. The second locus is situated at the 14q22 region near the KTN1 gene, which encodes a kinesin-binding protein involved in the transport of cellular components along microtubules69. Earlier univariate GWASs have previously reported associations between FAT3 and both caudate nucleus50,51 and putamen51 volumes, and between KTN1 and putamen50,51, in addition to accumbens, caudate nucleus and the globus pallidus51. The wide range of associations with basal ganglia volumes contributes to the unquestionable significance of these two loci in our multivariate GWAS analysis. Additional findings that provide support for previous univariate GWAS results include 208 candidate SNPs annotated to the 8p11.21 region associated with the present basal ganglia GWAS (see Supplementary Data 4), and previously seen associated with putamen in univariate GWAS51. Among these, 73 SNPs are mapped to SLC20A2, previously linked to the familiar basal ganglia calcification70,71,72, although the lead SNP is mapped to SMIM19 gene, located in close proximity to SLC20A2.
The two most significant SNPs, and the above-mentioned SNPs mapped to SLC20A2 and SMIM19, are all located in intergenic regions of the genome. However, four of the 72 basal ganglia associated loci have lead SNPs in the exonic regions (see Supplementary Fig. 4 and Supplementary Data 3): SLC39A8 on chr 4, TRIM10 and HLA-A on chr 6, VAT1 and NBR1 on chr 17 and APOE on chr 19. SLC39A8 has also previously been linked to basal ganglia51, is a transmembrane protein known to cotransport divalent cations with bicarbonate73, of which transport of manganese is suggested to be of greatest importance74, especially for dopaminergic projections in the basal ganglia75. The remaining three, however, are novel for this basal ganglia GWAS. rs3094134 on chr 6 located in the exon on TRIM10 and mapped to HLA-A, both important for the immune functioning in the brain76,77,78. The ALZ-linked APOE gene on chr 19, known to promote amyloid degradation79, was associated with basal ganglia volumes, which is a novel finding. Several significant SNPs are annotated to the same region on chr 17 where rs8482 is in the exon of NBR1, encoding a autophagic adapter protein involved in Lewy body formation in PD80 and targeting ubiquitinated protein for degradation in general81. The lead SNP in this locus, rs2070835, is located in the exonic region of VAT1 (vesicle amine transport protein 1), which encodes a protein that plays a crucial role in the process of vesicular neurotransmitter transport82. Vat1 is primarily involved in the packaging and storage of monoamine neurotransmitters, such as dopamine, norepinephrine, and serotonin, all neurotransmitters distributed in the basal ganglia. VAT1 and NBR1 are located in close proximity to each other on 17q21, on each side of the tumor suppressor gene BRCA1. Mapped with Open Target NBR1 is ranked first and VAT1 second.
The current boost in discovery with the multivariate MOSTest method supports shared genetic underpinnings53,83 underlying the interconnected nature of the basal ganglia nuclei. These structures don’t operate in isolation, and the finely tuned internal connections within the basal ganglia that are paramount for its function are probably also related to genetic variation, as suggested by the current findings of polygenic pleiotropy across the basal ganglia volumes. By treating multiple interconnected nuclei within the basal ganglia as a collective unit, we capture a wider range of genetic variation, allowing for the detection of loci missed in traditional univariate analyses. This approach not only enhances sensitivity to identify loci with subtle effects but also aligns with the biological rationale of the basal ganglia functioning as a coordinated network, influencing various aspects of motor control, cognition, and emotion. Thus, it is likely that genetic overlap of basal ganglia with various neurological, psychiatric, and developmental conditions could indicate potential dysregulation. There is clear pleiotropic enrichment between basal ganglia volume and neurological (PD, AD, migraine), neurodevelopmental (ADHD, ASD) and psychiatric (BIP, MDD, SCZ) disorders, shown by Q-Q plots (Supplementary Fig. 13), with many shared loci (Fig. 4, Supplementary Datas 11–18). Notably, the sole significant correlation revealed through LD-score regression-based genetic correlations was a positive association between genetic risk of PD and genetic architecture of basal ganglia (Supplementary Fig. 12 and Supplementary Data 10). Through Mendelian randomization, two independent methods show that genetically determined larger caudate and putamen volume is associated with a slight increase in the risk of PD, supporting the LD-score regression results. As a response to learning, locally increased brain volumes have been reported, possibly representing synaptogenesis and increased dendritic arborization84,85. Gene ontology analysis (Supplementary Data 8) support these mechanisms in the genetic architecture of basal ganglia volume, with gene ontology on cell part morphogenesis, neuron part and synapse, but details on cell structure changes representing the positive correlation between the genetic pattern of basal ganglia and PD need separate studies. Additionally, Mendelian randomization showed potential causal effect of ALZ on smaller nucleus accumbens volume and higher striatal volume on PD. The results indicating that genetic predisposition to ALZ influence nucleus accumbens volume is supported by multiple Mendelian randomization methods, increasing the robustness of the findings. In addition to the hallmark feature of medial temporal lobe atrophy, accumbens atrophy has also been found in ALZ patients, correlating with lower cognitive scores86. As a limitation, individuals with ALZ and PD are included in the imaging sample, but the confounding effect is negligible as only five and 17 of the 34,794 individuals included in the current study are diagnosed with ALZ and PD, respectively (Supplementary Fig. 14). While implementing sensitivity analysis (Supplementary Data 21) with stringent age filters, we observed an anticipated decline in statistical power; nevertheless, the persistent association between higher striatal volumes and PD genetic markers underscores the potential linkage, as detailed in the supplementary material. Despite the diminished cohort size, the consistency of our PD-related results across age brackets bolsters their validity. Conversely, the non-significant correlation between accumbens volume and AD risk in participants under 65 suggests that our findings may be influenced by preclinical neuropathology affecting this result, challenging the notion of genetic AD risk as a stand-alone causal factor for reduced accumbens volume.
Our results suggest significant genetic pleiotropy between the basal ganglia and especially PD, SCZ and migraine, but also between the basal ganglia and ALZ, BIP and MDD, and less with ASD and ADHD. Out of more than 150 genes, only 16 genes were mapped to basal ganglia and both neurological and psychiatric disorders, indicating distinct genetic architecture underlying each of the disorder’s relationship with the basal ganglia. These 16 genes that show overlapping involvement in both neurological and psychiatric disorders, demonstrate important shared biology between brain disorders (Table 1). Notably, TMEM161B (transmembrane protein 161B) and HP (haptoglobin) is overlapping between basal ganglia and MDD, PD and SCZ, in addition to ADHD and BIP, respectively. Haptoglobin is involved in binding and transporting hemoglobin, and it plays a role in modulating the immune response and oxidative stress87. TMEM161B has been mapped to MDD in mouse models and humans88,89 and has also been coupled to basal ganglia activation during reward processing90. Although functionally linked to basal ganglia, TMEM161B have previously only been linked to neocortex structurally, through gyrification in neocortical development91.
In summary, our findings indicate a shared polygenic architecture across the basal ganglia, spanning the putamen, caudate, pallidum, and accumbens, with high correlation between the basal ganglia nuclei and a robust 72 loci when analyzed together, of which 50 are novel. The genetic overlap observed with different brain disorders highlights the interconnectedness of the basal ganglia in neurological (PD, ALZ, migraine) and psychiatric (SCZ, BIP, MDD) conditions, but less for neurodevelopmental conditions (ASD and ADHD). By recognizing the distributed nature of genetic effects across the basal ganglia, functional brain unit, we can gain valuable insights into the mechanisms underlying brain disorders and pave the way for future advancements in this field. Moreover, our study reveals that some of these genetic findings align with known treatment targets, supporting the value of our approach. This suggests shared underlying mechanisms and potential targets for therapeutic interventions. Further research is necessary to fully understand the complex relationship between the basal ganglia and brain disorders and apply this to develop more effective treatments.
Methods
Sample and pre-processing of imaging and genetic data
We employed T1 weighted MRI data to investigate basal ganglia volumes. Our primary analysis was based on a dataset consisting of 34,794 genotyped individuals of white British descent from the UK Biobank (age range: 45-82 years, mean: 64.3 years, s.d.: 7.5 years, 52.3% females)66. Within our cohort, only a few have registered major neurological and psychiatric conditions (Supplementary Fig. 14). To ensure the robustness of our findings, we also conducted a replication analysis using an independent dataset from UK Biobank consisting of 4808 individuals of British descent (age range: 45-81, mean: 69.1, s.d.: 7.8 years, 50.1 % females) and a generalization analysis using an independent dataset from UK Biobank consisting of 5220 individuals with non-white ethnicity (age range: 45–81, mean: 62.9, s.d.: 7.4 years, 54.1% females).
We used Freesurfer v5.3 to extract the volumes of the accumbens, caudate, pallidum, and putamen92. For genetic analyses, we used the UK Biobank v3 imputed genetic data and followed the standard quality control procedures to remove SNPs with an imputation quality score of 0.5, a minor allele frequency <0.005, missingness in more than 10% of individuals, and failing the Hardy-Weinberg equilibrium tests at a P value < 1 × 10−9.
Univariate and multivariate genome-wide association analysis
We calculated the average volume between the left and right hemispheres for each of the accumbens, caudate, pallidum, and putamen as well as the total basal ganglia volume. To address potential confounding factors, we controlled for age, age squared, sex, scanning site, Euler score (which serves as a proxy for image quality), total intracranial volume (ICV), and the 20 genetic principal components for each region comprising the basal ganglia. Furthermore, we conducted additional analyses, controlling for the total volume defined as the sum of the four basal ganglia volumes. We controlled for the same covariates for both white and non-white ethnicity samples. For each basal ganglia volume, we used standard univariate GWAS procedure and annotated the significant loci using FUMA. For each basal ganglia volume, we applied standard univariate GWAS procedure using a linear model in PLINK v1.9, with a MAF filter of 0.005 and annotated the significant loci using FUMA. The min-P approach takes the smallest p-value of each SNP across multiple univariate GWAS, and corrects this for the effective number of traits studied93. SNP-based heritability estimates for the basal ganglia volumes, as well as the genetic correlations between these volumes, were estimated using linkage disequilibrium (LD) score regression94,95.
We further conducted a multivariate GWAS on pre-residualized basal ganglia volumes for our main analysis using Multivariate Omnibus Statistical Test (MOSTest)53, which is a powerful statistical tool used for joint genetic analysis of multiple traits in large-scale data. It aims to identify genetic variants associated with complex phenotypes influenced by multiple genetic factors with small effects. By analyzing the traits jointly, MOSTest takes a multivariate approach, considering multiple traits simultaneously and leveraging the genetic overlap across different regions and measures of the brain. This approach enhances the statistical power to detect associations by avoiding the stringent multiple comparison correction required in mass-univariate approaches. Van der Meer et al.53 provide detailed description of the method, while github.com/precimed/mostest provides information on the software implementation. We also conducted a univariate GWAS of the four basal ganglia volumes and whole basal ganglia (extracted from the univariate stream of MOSTest53) for comparison to conventional univariate techniques (GWAS).
Multivariate replication and generalization analysis
We used a multivariate replication approach developed to test if the multivariate pattern of genetic associations identified in discovery analysis is consistent between discovery, replication and generalization samples. This procedure generates a composite score from mass-univariate z-statistics for each locus identified in the multivariate analysis of the discovery sample, and tests for associations of the composite score with genotype in the replication and generalization samples (for mathematical formulation see Loughnan et al.96). We report the percent of loci replicating at P < 0.05 and the percent of loci showing the same effect direction.
Functional annotation, gene-based association, gene-set, tissue, and pathway analysis
We submitted the summary statistic to the FUMA platform v1.4.154 in order to identify independent genetic loci. We discovered independent significant SNPs at the statistical significance level P < 5 × 10−8 using the 1000GPhase3 EUR as the reference panel. A portion of the independent significant SNPs in linkage equilibrium with one another at r2 < 0.1 were termed lead SNPs, and all SNPs at r2 < 0.6 with each other were considered independent significant SNPs. If two or more lead SNPs located within one LD block (in 250 kb), we merged them into one genomic risk locus. Combined Annotation Dependent Depletion (CADD) scores, which forecast the deleteriousness of SNPs on protein structure/function57, RegulomeDB scores, which forecast regulatory functions55, and chromatin states, which illustrate the transcriptional/regulatory effects of chromatin states at the SNP locus, are all used by FUMA to annotate associated SNPs.
We conducted genome-wide gene-based association and gene-set analyses using MAGMA v.1.0862 in FUMA. MAGMA performs the gene-based association analysis and assigns p-values to individual genes based on the association between genetic variants within or near the gene and the trait of interest. This analysis is conducted using a multiple regression model, which improves statistical performance compared to single-marker analysis62. Furthermore, the gene-set analysis using MAGMA enables the assessment of groups of genes that share common biological functions or pathways, which provides insights into the functional and biological mechanisms underlying complex traits. All variants in the major histocompatibility complex (MHC) region were excluded before running the MAGMA analyses.
Gene mapping is a crucial process for understanding the relationship between genetic variants and traits or diseases. To map the significant SNPs to genes, we used variant to gene (v2g) from Open Targets Genetics97, which provides a comprehensive and systematic approach to prioritize causal variants and identify likely causal genes associated with various phenotypes and diseases. By integrating positional data on the distance between the variant and each gene’s canonical transcription start site, eQTL, pQTL, splicingQTL, epigenomic data, and functional prediction, Open Targets maps lead SNPs to genes.
We also conducted Gene Ontology gene-set analysis based on FUMA’s gene ontology classification system54,98, and pathway analysis using Consensus PathDB99.
Cell specificity, protein–protein interaction and gene-drug interaction analyses
To investigate the context-specific expression of mapped genes associated with basal ganglia we used WebCSEA (Web-based Cell-type-Specific Enrichment Analysis of Genes). WebCSEA incorporates a curated collection of 111 scRNA-seq panels from various human tissues and 1,355 tissue-cell types across 11 organ systems100. We also determined the cell-specific expression within the human cerebral cortex including neurons, fetal and mature astrocytes, oligodendrocytes, microglia/macrophages, and endothelial cells for each mapped gene separately101.
In addition, we employed GENEmania101 for functional genomics analysis, to investigate protein-protein interactions (PPIs) between protein-encoded basal ganglia-linked genes and exported the network to Cytoscape 3.10. We identified potential protein clusters and gain insights into the functional relationships between the proteins of interest. Finally, we performed gene-drug interaction analysis using the Drug Gene Interaction Database to gain insights into the potential associations between mapped genes to basal ganglia and drugs, including known drug targets and potential off-target effects102 to uncover drug targets that specifically influence the basal ganglia.
Genetic overlap between the basal ganglia and common brain disorders
We studied the genetic overlap between basal ganglia and ASD, ADHD, SCZ, BIP, MDD, MIG, PD, and ALZ. We used GWAS summary statistics from the Psychiatric Genomics Consortium for ADHD103, ASD104, BIP105, MDD106, SCZ107 and ALZ108, from International headache genetics Consortium for MIG109, and from the International Parkinson Disease Genomics Consortium110,111 for PD data. All the studies included in the original GWAS were approved by local ethics authorities.
First, we used linkage disequilibrium (LD) score regression94,95 to estimate genetic correlations between basal ganglia volumes and each of the brain disorders. Then we created conditional quantile-quantile (Q-Q) plots to evaluate the presence of cross-phenotype polygenic enrichment65,112, conditioning basal ganglia on each brain disorder and vice versa. The conditional FDR method65,112,113 builds on an empirical Bayesian statistical framework and leverages polygenic overlap between two traits to improve the discovery of genetic loci associated with the traits of interest. The condFDR method estimate the posterior probability that a SNP has no association with the primary trait, given that the P values for that SNP in both the primary and conditional traits are as small as or smaller than the observed P value. Thus, the method improves the detection of genetic variants associated with the primary trait via reranking the test statistics of the primary phenotype based on the strength of the association with the secondary phenotype. Next, we used the conjunctional FDR (conjFDR) approach at FDR 0.0565,112,113 to identify common genetic loci between basal ganglia and the included brain disorders. This approach is an extension of the conditional FDR and is defined as the maximum of the 2 conditional FDR statistics for a specific SNP and estimates the posterior probability that a SNP is null for either trait or both, given that the P values for both phenotypes are as small as or smaller than the P values for each trait individually. It takes the highest of the two condFDR statistics for a particular SNP, i.e. for trait A conditional on trait B and trait B conditional on trait A. If the p values for both phenotypes are equal to or less than the p values for each trait separately, this reflects an estimate of the posterior probability that a SNP is null for one or both characteristics. More details are found in the original112,113 and subsequent publication65. The conjFDR analysis does incorporate effect directions, in contrast to genetic correlation analysis, and may thus be used to analyze summary data from multivariate GWAS that lack effect directions. Because complex correlations in regions with intricate LD can bias FDR estimation, two genomic regions—the extended major histocompatibility complex genes region (hg19 location Chr 6: 25119106-33854733) and chromosome 8p23.1 (hg19 location Chr 8: 7242715-12483982) for all phenotypes and MAPT region for PD and APOE region for ALZ and ASD, respectively—were excluded from the FDR-fitting procedures. We used the FUMA protocol to annotate independent genomic loci jointly associated with basal ganglia and each brain disorder. We submitted the results of the conjFDR analysis to FUMA v1.5.154. The lead SNPs and candidate SNPs within each genomic locus were used to identify the genomic loci as significant SNPs with an LD r2 >= 0.6 and conjFDR 0.1 and at least one associated independent significant SNP. ConjFDR <0.05 and r2 < 0.6 were used to determine independent significant SNPs, while lead SNPs were defined if they were in approximate linkage equilibrium with each other (r2 < 0.1). Finally, the significant SNPs were mapped to genes using Open Targets97 as described above.
Mendelian randomization
To study potential relationships between basal ganglia nuclei and eight common brain disorders, a set of Mendelian randomization analyses were performed. The Mendelian randomization approaches included weighted mode and weighted median114, Egger regression (Egger)115 and the inverse-variance weighted model (IVW)116, in addition to the outlier-corrected MR-PRESSO approach117. For these analyses, only genome-wide significant SNPs (p < 5 × 10−8) for the basal ganglia volumes or disorders were used as instruments. Furthermore, we performed a sensitivity analysis to assess the robustness of our Mendelian randomization findings. This analysis involved young individuals with two alternative age cut-offs: 75 and 65 years old, including n = 32,455 and n = 17,591 individuals, respectively. We used the TwoSampleMR package in R (0.4.26; github.com/MRCIEU/TwoSampleMR)118.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
In this study we used brain imaging and genetics data from the UK Biobank [https://www.ukbiobank.ac.uk/], and GWAS summary statistics obtained from the Psychiatric Genomics Consortium [https://www.med.unc.edu/pgc/shared-methods/], 23andMe,inc. [https://www.23andme.com/], International headache genetics Consortium (IHGC) [http://www.headachegenetics.org/content/datasets-and-cohorts], the International Genomics of Alzheimer’s Project [https://ctg.cncr.nl/software/summary_statistics], and the International Parkinson Disease Genomics Consortium [https://pdgenetics.org/resources]. The latter included 23andMe data, which was made available through 23andMe under an agreement with 23andMe that protects the privacy of the 23andMe participants [https://research.23andme.com/collaborate/#dataset-access/]. The summary statistics for basal ganglia derived in this study is available in GWAS Catalog [https://www.ebi.ac.uk/gwas/studies/GCST90432154]. FUMA results are available online [https://fuma.ctglab.nl/browse/371].
Code availability
All code and software needed to generate the results is available as part of public resources, specifically MOSTest (https://github.com/precimed/mostest), FUMA, conjunctional FDR and LD score regression (https://github.com/bulik/ldsc).
References
Alexander, G. E. & Crutcher, M. D. Functional architecture of basal ganglia circuits: neural substrates of parallel processing. Trends Neurosci. 13, 266–271 (1990).
Parent, A. Extrinsic connections of the basal ganglia. Trends Neurosci. 13, 254–258 (1990).
Haber, S. N. & Knutson, B. The reward circuit: linking primate anatomy and human imaging. Neuropsychopharmacology 35, 4–26 (2010).
Nelson, A. B. & Kreitzer, A. C. Reassessing models of basal ganglia function and dysfunction. Annu. Rev. Neurosci. 37, 117–135 (2014).
DeLong, M. R. & Wichmann, T. Circuits and circuit disorders of the basal ganglia. Arch. Neurol. 64, 20–24 (2007).
Grahn, J. A., Parkinson, J. A. & Owen, A. M. The cognitive functions of the caudate nucleus. Prog. Neurobiol. 86, 141–155 (2008).
Delmonte, S., Gallagher, L., O’hanlon, E., McGrath, J. & Balsters, J. H. Functional and structural connectivity of frontostriatal circuitry in Autism Spectrum Disorder. Front. Hum. Neurosci. 7, 430 (2013).
Redgrave, P. et al. Goal-directed and habitual control in the basal ganglia: implications for Parkinson’s disease. Nat. Rev. Neurosci. 11, 760–772 (2010).
Yin, H. H. & Knowlton, B. J. The role of the basal ganglia in habit formation. Nat. Rev. Neurosci. 7, 464–476 (2006).
Graybiel, A. M. Habits, rituals, and the evaluative brain. Annu. Rev. Neurosci. 31, 359–387 (2008).
Frank, M. J. Dynamic dopamine modulation in the basal ganglia: a neurocomputational account of cognitive deficits in medicated and nonmedicated Parkinsonism. J. Cogn. Neurosci. 17, 51–72 (2005).
D’Cruz, A.-M. et al. Reduced behavioral flexibility in autism spectrum disorders. Neuropsychology 27, 152 (2013).
McAlonan, G. M. et al. Brain anatomy and sensorimotor gating in Asperger’s syndrome. Brain: J. Neurol. 125, 1594–1606 (2002).
O’doherty, J. P. Reward representations and reward-related learning in the human brain: insights from neuroimaging. Curr. Opin. Neurobiol. 14, 769–776 (2004).
Schultz, W. Behavioral theories and the neurophysiology of reward. Annu. Rev. Psychol. 57, 87–115 (2006).
Haber, S. N. Corticostriatal circuitry. Dialogues Clin. Neurosci. 18 (2022).
Cogswell, P. M. et al. Associations of quantitative susceptibility mapping with Alzheimer’s disease clinical and imaging markers. Neuroimage 224, 117433 (2021).
Mostofsky, S. H. & Ewen, J. B. Altered connectivity and action model formation in autism is autism. Neuroscientist 17, 437–448 (2011).
Langen, M. et al. Changes in the development of striatum are involved in repetitive behavior in autism. Biol. psychiatry 76, 405–411 (2014).
Mostofsky, S. H. & Simmonds, D. J. Response inhibition and response selection: two sides of the same coin. J. Cogn. Neurosci. 20, 751–761 (2008).
Castellanos, F. X., Sonuga-Barke, E. J., Milham, M. P. & Tannock, R. Characterizing cognition in ADHD: beyond executive dysfunction. Trends Cogn. Sci. 10, 117–123 (2006).
Luman, M., Tripp, G. & Scheres, A. Identifying the neurobiology of altered reinforcement sensitivity in ADHD: a review and research agenda. Neurosci. Biobehav. Rev. 34, 744–754 (2010).
Volkow, N. D. et al. Motivation deficit in ADHD is associated with dysfunction of the dopamine reward pathway. Mol. Psychiatry 16, 1147–1154 (2011).
Plichta, M. M. et al. Neural hyporesponsiveness and hyperresponsiveness during immediate and delayed reward processing in adult attention-deficit/hyperactivity disorder. Biol. Psychiatry 65, 7–14 (2009).
Cubillo, A., Halari, R., Smith, A., Taylor, E. & Rubia, K. A review of fronto-striatal and fronto-cortical brain abnormalities in children and adults with Attention Deficit Hyperactivity Disorder (ADHD) and new evidence for dysfunction in adults with ADHD during motivation and attention. Cortex 48, 194–215 (2012).
Nigg, J. T. Neuropsychologic theory and findings in attention-deficit/hyperactivity disorder: the state of the field and salient challenges for the coming decade. Biol. Psychiatry 57, 1424–1435 (2005).
Sonuga-Barke, E. J. & Fairchild, G. Neuroeconomics of attention-deficit/hyperactivity disorder: differential influences of medial, dorsal, and ventral prefrontal brain networks on suboptimal decision making? Biol. Psychiatry 72, 126–133 (2012).
Solanto, M. V. Dopamine dysfunction in AD/HD: integrating clinical and basic neuroscience research. Behav. Brain Res. 130, 65–71 (2002).
Swanson, J. M. et al. Etiologic subtypes of attention-deficit/hyperactivity disorder: brain imaging, molecular genetic and environmental factors and the dopamine hypothesis. Neuropsychol. Rev. 17, 39–59 (2007).
Castellanos, F. X. & Tannock, R. Neuroscience of attention-deficit/hyperactivity disorder: the search for endophenotypes. Nat. Rev. Neurosci. 3, 617–628 (2002).
Howes, O. D. & Kapur, S. The dopamine hypothesis of schizophrenia: version III—the final common pathway. Schizophr. Bull. 35, 549–562 (2009).
Abi-Dargham, A. & Moore, H. Prefrontal DA transmission at D1 receptors and the pathology of schizophrenia. Neuroscientist 9, 404–416 (2003).
Fornito, A., Yoon, J., Zalesky, A., Bullmore, E. T. & Carter, C. S. General and specific functional connectivity disturbances in first-episode schizophrenia during cognitive control performance. Biol. Psychiatry 70, 64–72 (2011).
Andreasen, N. C. et al. Progressive brain change in schizophrenia: a prospective longitudinal study of first-episode schizophrenia. Biol. Psychiatry 70, 672–679 (2011).
Russo, S. J. & Nestler, E. J. The brain reward circuitry in mood disorders. Nat. Rev. Neurosci. 14, 609–625 (2013).
Phillips, M. L., Ladouceur, C. D. & Drevets, W. C. A neural model of voluntary and automatic emotion regulation: implications for understanding the pathophysiology and neurodevelopment of bipolar disorder. Mol. Psychiatry 13, 833–857 (2008).
Pizzagalli, D. A. Depression, stress, and anhedonia: toward a synthesis and integrated model. Annu. Rev. Clin. Psychol. 10, 393–423 (2014).
Magon, S. et al. Morphological abnormalities of thalamic subnuclei in migraine: a multicenter MRI study at 3 tesla. J. Neurosci. 35, 13800–13806 (2015).
Lewis, M. M. et al. The pattern of gray matter atrophy in Parkinson’s disease differs in cortical and subcortical regions. J. Neurol. 263, 68–75 (2016).
Beyer, J. L. et al. Caudate volume measurement in older adults with bipolar disorder. Int. J. Geriatr. Psychiatry 19, 109–114 (2004).
Ellison-Wright, I., Ellison-Wright, Z. & Bullmore, E. Structural brain change in attention deficit hyperactivity disorder identified by meta-analysis. BMC Psychiatry 8, 1–8 (2008).
Nakao, T., Radua, J., Rubia, K. & Mataix-Cols, D. Gray matter volume abnormalities in ADHD: voxel-based meta-analysis exploring the effects of age and stimulant medication. Am. J. Psychiatry 168, 1154–1163 (2011).
Kim, J. et al. Regional grey matter changes in patients with migraine: a voxel-based morphometry study. Cephalalgia 28, 598–604 (2008).
Rojas, D. C. et al. Hippocampus and amygdala volumes in parents of children with autistic disorder. Am. J. Psychiatry 161, 2038–2044 (2004).
Strakowski, S. M. et al. Ventricular and periventricular structural volumes in first-versus multiple-episode bipolar disorder. Am. J. Psychiatry 159, 1841–1847 (2002).
DelBello, M. P., Zimmerman, M. E., Mills, N. P., Getz, G. E. & Strakowski, S. M. Magnetic resonance imaging analysis of amygdala and other subcortical brain regions in adolescents with bipolar disorder. Bipolar Disord. 6, 43–52 (2004).
Wilke, M., Kowatch, R. A., DelBello, M. P., Mills, N. P. & Holland, S. K. Voxel-based morphometry in adolescents with bipolar disorder: first results. Psychiatry Res.: Neuroimaging 131, 57–69 (2004).
Estes, A. et al. Basal ganglia morphometry and repetitive behavior in young children with autism spectrum disorder. Autism Res. 4, 212–220 (2011).
Sears, L. L. et al. An MRI study of the basal ganglia in autism. Prog. Neuropsychopharmacol. Biol. Psychiatry 23, 613–624 (1999).
Hibar, D. P. et al. Common genetic variants influence human subcortical brain structures. Nature 520, 224–229 (2015).
Satizabal, C. L. et al. Genetic architecture of subcortical brain structures in 38,851 individuals. Nat. Genet. 51, 1624–1636 (2019).
van der Meer, D. et al. Making the MOSTest of imaging genetics. Biol. Psychiatry 87, S304–S305 (2020).
van der Meer, D. et al. Understanding the genetic determinants of the brain with MOSTest. Nat. Commun. 11, 1–9 (2020).
Watanabe, K., Taskesen, E., Van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1–11 (2017).
Boyle, A. P. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22, 1790–1797 (2012).
Kundaje, A. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Kircher, M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014).
Smeland, O. B. et al. Genome-wide association analysis of Parkinson’s disease and schizophrenia reveals shared genetic architecture and identifies novel risk loci. Biol. Psychiatry 89, 227–235 (2021).
Pickrell, J. K. et al. Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet. 48, 709–717 (2016).
Leonenko, G. et al. Identifying individuals with high risk of Alzheimer’s disease using polygenic risk scores. Nat. Commun. 12, 4506 (2021).
Kulminski, A. M. et al. Genetic and regulatory architecture of Alzheimer’s disease in the APOE region. Alzheimer’s Dement. (Amst., Neth.) 12, e12008 (2020).
de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11, e1004219 (2015).
Smeland, O. B., Frei, O., Dale, A. M. & Andreassen, O. A. The polygenic architecture of schizophrenia—rethinking pathogenesis and nosology. Nat. Rev. Neurol. 16, 366–379 (2020).
Andreassen, O. A., Hindley, G. F. L., Frei, O. & Smeland, O. B. New insights from the last decade of research in psychiatric genetics: discoveries, challenges and clinical implications. World Psychiatry 22, 4–24 (2023).
Smeland, O. B. et al. Discovery of shared genomic loci using the conditional false discovery rate approach. Hum. Genet. 139, 85–94 (2020).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Littlejohns, T. J. et al. The UK Biobank imaging enhancement of 100,000 participants: rationale, data collection, management and future directions. Nat. Commun. 11, 2624 (2020).
Deans, M. R. et al. Control of neuronal morphology by the atypical cadherin Fat3. Neuron 71, 820–832 (2011).
Santama, N., Er, C. P., Ong, L.-L. & Yu, H. Distribution and functions of kinectin isoforms. J. Cell Sci. 117, 4537–4549 (2004).
Mu, W., Tochen, L., Bertsch, C., Singer, H. S. & Barañano, K. W. Intracranial calcifications and dystonia associated with a novel deletion of chromosome 8p11.2 encompassing SLC20A2 and THAP1. BMJ Case Rep. 12, e228782 (2019).
Hsu, S. C. et al. Mutations in SLC20A2 are a major cause of familial idiopathic basal ganglia calcification. Neurogenetics 14, 11–22 (2013).
Taglia, I., Bonifati, V., Mignarri, A., Dotti, M. T. & Federico, A. Primary familial brain calcification: update on molecular genetics. Neurol. Sci. 36, 787–794 (2015).
He, L. et al. ZIP8, member of the solute-carrier-39 (SLC39) metal-transporter family: characterization of transporter properties. Mol. Pharmacol. 70, 171–180 (2006).
Choi, E.-K., Nguyen, T.-T., Gupta, N., Iwase, S. & Seo, Y. A. Functional analysis of SLC39A8 mutations and their implications for manganese deficiency and mitochondrial disorders. Sci. Rep. 8, 3163 (2018).
Horning, K. J., Caito, S. W., Tipps, K. G., Bowman, A. B. & Aschner, M. Manganese is essential for neuronal health. Annu. Rev. Nutr. 35, 71–108 (2015).
Kong, L. et al. The ubiquitin E3 ligase TRIM10 promotes STING aggregation and activation in the Golgi apparatus. Cell Rep. 42 (2023).
Tamouza, R., Krishnamoorthy, R. & Leboyer, M. Understanding the genetic contribution of the human leukocyte antigen system to common major psychiatric disorders in a world pandemic context. Brain, Behav., Immun. 91, 731–739 (2021).
Endres, D. et al. Immunological causes of obsessive-compulsive disorder: is it time for the concept of an “autoimmune OCD” subtype? Transl. Psychiatry 12, 5 (2022).
Jiang, Q. et al. ApoE promotes the proteolytic degradation of Aβ. Neuron 58, 681–693 (2008).
Odagiri, S. et al. Autophagic adapter protein NBR1 is localized in Lewy bodies and glial cytoplasmic inclusions and is involved in aggregate formation in α-synucleinopathy. Acta Neuropathol. 124, 173–186 (2012).
Lange, S. et al. The kinase domain of titin controls muscle gene expression and protein turnover. Science 308, 1599–1603 (2005).
Smith, T. M. et al. Complete genomic sequence and analysis of 117 kb of human DNA containing the gene BRCA1. Genome Res. 6, 1029–1049 (1996).
van der Meer, D. et al. The genetic architecture of human cortical folding. Sci. Adv. 7, eabj9446 (2021).
Trachtenberg, J. T. et al. Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature 420, 788–794 (2002).
Kozorovitskiy, Y., Saunders, A., Johnson, C. A., Lowell, B. B. & Sabatini, B. L. Recurrent network activity drives striatal synaptogenesis. Nature 485, 646–650 (2012).
Nie, X. et al. Subregional structural alterations in hippocampus and nucleus accumbens correlate with the clinical impairment in patients with Alzheimer’s disease clinical spectrum: parallel combining volume and vertex-based approach. Front. Neurol. 8, 399 (2017).
Carrascoza, F. & Silaghi-Dumitrescu, R. The dynamics of hemoglobin-haptoglobin complexes. Relevance for oxidative stress. J. Mol. Struct. 1250, 131703 (2022).
Hyde, C. L. et al. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat. Genet. 48, 1031–1036 (2016).
Yue, S. et al. Gene-gene interaction and new onset of major depressive disorder: Findings from a Chinese freshmen nested case-control study. J. Affect. Disord. 300, 505–510 (2022).
Muench, C. et al. The major depressive disorder GWAS-supported variant rs10514299 in TMEM161B-MEF2C predicts putamen activation during reward processing in alcohol dependence. Transl. Psychiatry 8, 131 (2018).
Wang, L. et al. TMEM161B modulates radial glial scaffolding in neocortical development. Proc. Natl Acad. Sci. USA 120, e2209983120 (2023).
Fischl, B. et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33, 341–355 (2002).
Grasby, K. L. et al. The genetic architecture of the human cerebral cortex. Science 367 (2020).
Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236 (2015).
Loughnan, R. J. et al. Generalization of cortical MOSTest genome-wide associations within and across samples. Neuroimage 263, 119632 (2022).
Ochoa, D. et al. The next-generation Open Targets Platform: reimagined, redesigned, rebuilt. Nucleic Acids Res. 51, D1353–d1359 (2023).
Consortium, G. O. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 49, D325–d334 (2021).
Herwig, R., Hardt, C., Lienhard, M. & Kamburov, A. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat. Protoc. 11, 1889–1907 (2016).
Dai, Y. et al. WebCSEA: web-based cell-type-specific enrichment analysis of genes. Nucleic Acids Res. 50, W782–w790 (2022).
Franz, M. et al. GeneMANIA update 2018. Nucleic Acids Res. 46, W60–w64 (2018).
Freshour, S. L. et al. Integration of the Drug-Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 49, D1144–d1151 (2021).
Demontis, D. et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 51, 63–75 (2019).
Grove, J. et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 51, 431–444 (2019).
Mullins, N. et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat. Genet. 53, 817–829 (2021).
Wray, N. R. et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 50, 668–681 (2018).
Pardiñas, A. F. et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet. 50, 381–389 (2018).
Wightman, D. P. et al. A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer’s disease. Nat. Genet. 53, 1276–1282 (2021).
Gormley, P. et al. Meta-analysis of 375,000 individuals identifies 38 susceptibility loci for migraine. Nat. Genet. 48, 856–866 (2016).
Nalls, M. A. et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 46, 989–993 (2014).
Nalls, M. A. et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 18, 1091–1102 (2019).
Andreassen, O. A. et al. Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. Am. J. Hum. Genet. 92, 197–209 (2013).
Andreassen, O. A. et al. Improved detection of common variants associated with schizophrenia and bipolar disorder using pleiotropy-informed conditional false discovery rate. PLoS Genet. 9, e1003455 (2013).
Bowden, J., Davey Smith, G., Haycock, P. C. & Burgess, S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40, 304–314 (2016).
Bowden, J., Davey Smith, G. & Burgess, S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44, 512–525 (2015).
Relton, C. L. & Davey Smith, G. Two-step epigenetic Mendelian randomization: a strategy for establishing the causal role of epigenetic processes in pathways to disease. Int. J. Epidemiol. 41, 161–176 (2012).
Verbanck, M., Chen, C.-Y., Neale, B. & Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698 (2018).
Hemani, G. et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 7, e34408 (2018).
Acknowledgements
The authors were funded by Norwegian Health Association (S.B.: 22731, K.N.: 25598), the South-Eastern Norway Regional Health Authority (O.A.A.: 2013-123, 2017-112, 2019-108, L.T.W.: 2014-097, 2015-073, 2016-083), the Research Council of Norway (T.K.: 276082, 323961. O.A.A.: 213837, 223273, 248778, 273291, 262656, 229129, 283798, 311993, 324499. L.T.W.: 204966, 249795, 273345). Stiftelsen Kristian Gerhard Jebsen (O.A.A., L.T.W.), the European Research Council under the European Union’s Horizon 2020 Research and Innovation program (L.T.W.: 802998, O.A.A.: 847776), NIH (O.A.A.: 1R01MH129742) and the Department of Neurology at Oslo University Hospital (K.N.). The funding bodies had no role in the analysis or interpretation of the data; the preparation, review or approval of the manuscript; nor in the decision to submit the manuscript for publication. This work was performed on the Tjenester for Sensitive Data (TSD) facilities, owned by the University of Oslo, operated and developed by the TSD service group at the University of Oslo, IT-Department (USIT) and on resources provided by UNINETT Sigma2—the National Infrastructure for High Performance Computing and Data Storage in Norway. The research has been conducted using the UK Biobank Resource (access code 27412) and using summary statistics for various brain disorders that partly included 23andMe data. We would like to thank the research participants and employees of UK Biobank, the 23andMe, the Psychiatric Genomics Consortium, International Headache Genetics Consortium, the International Genomics of Alzheimer’s Project and International Parkinson Disease Genomics Consortium for contributing summary statistics for this work.
Author information
Authors and Affiliations
Contributions
S.B., K.N., and O.A.A. conceived the study. S.B., J.R., and Z.R. analyzed the data. S.B, and K.N. interpreted the results and spearheaded the writing. S.B. drafted the online methods. S.B., K.N., J.R, A.S., Z.R., O.S., P.P.J., N.P., P.P., K.O.C., T.E., M.T., S.D., A.D., L.T.W., T.K., and O.A.A. gave conceptual input on the methods and/or results and contributed to and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
Dr. Andreassen has received a speaker’s honorarium from Lundbeck, Janssen and a consultant for Cortechs.ai. Other authors report no conflicts.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bahrami, S., Nordengen, K., Rokicki, J. et al. The genetic landscape of basal ganglia and implications for common brain disorders. Nat Commun 15, 8476 (2024). https://doi.org/10.1038/s41467-024-52583-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-52583-0