Abstract
SUMOylation regulates numerous cellular stress responses, yet targets in the apoptotic machinery remain elusive. We show that a single, DNA damage-induced monoSUMOylation event controls PIDDosome (PIDD1/RAIDD/caspase-2) formation and apoptotic death in response to unresolved DNA interstrand crosslinks (ICLs). SUMO-1 conjugation occurs on conserved K879 in the PIDD1 death domain (DD); is catalyzed by PIAS1 and countered by SENP3; and is triggered by ATR phosphorylation of neighboring T788 in the PIDD1 DD, which enables PIAS1 docking. Phospho/SUMO-PIDD1 proteins are captured by nucleolar RAIDD monomers via a SUMO-interacting motif (SIM) in the RAIDD DD, thus compartmentalizing nascent PIDDosomes for caspase-2 recruitment. Denying SUMOylation or the SUMO-SIM interaction spares the onset of PIDDosome assembly but blocks its completion, thus eliminating the apoptotic response to ICL repair failure. Conversely, removal of SENP3 forces apoptosis, even in cells with tolerable ICL levels. SUMO-mediated PIDDosome control is also seen in response to DNA breaks but not supernumerary centrosomes. These results illuminate PIDDosome formation in space and time and identify a direct role for SUMOylation in the assembly of a major pro-apoptotic device.
Similar content being viewed by others
Introduction
Caspase activation platforms (CAPs) are multi-oligomeric complexes essential for initiating cell death (apoptosis, necroptosis, pyroptosis) and inflammation through the activation of initiator caspases. Best understood are the apoptosome (cyt-c-APAF1-caspase-9), death-inducing signaling complexes (DR-FADD-caspase-8) and inflammasomes (NLR/AIM2-ASC-caspase-1)1,2,3. Less studied is the CAP for caspase-2 (C2), the PIDDosome, formed of p53-induced protein with DD (PIDD1, LRDD), which serves as core scaffold; the caspase adapter RIP-associated ICH-1/CED-3-homologous protein with DD (RAIDD, CRADD); and the C2 protease4. PIDDosome-activated C2 can trigger apoptotic cell death in response to a variety of DNA damage lesions5,6,7,8,9,10,11,12, or induce cell cycle arrest in response to polyploidy or supernumerary centrosomes13,14,15,16,17. Whether the levels or spatial localization PIDDosome assembly dictate the ultimate cellular outcome, or whether other mechanisms are at play, remains ill-defined18,19,20.
Central to PIDDosome formation is the homotypic (DD:DD) PIDD1-RAIDD interaction, implicated as both the initiating and rate-limiting step in platform assembly21,22. Full-length (FL) PIDD1 undergoes constitutive autoproteolysis at Phe445/Ser446 and Phe587/Ser588 to generate an N-terminal fragment (PIDD1-N) and two C-terminal fragments, PIDD1-C and PIDD1-CC (see Fig. 1a below)4,23,24. While both PIDD1-C and -CC harbor the DD, only the CC fragment can bind RAIDD and support PIDDosome assembly5,13,24,25,26. Initial observations suggested that the autoproteolytic cleavage generating PIDD1-CC occurs as a function of DNA damage levels, with increasing doses facilitating the event24. Thus, the PIDDosome would only assemble in response to severe DNA damage, whereas lesser DNA injury would permit cell survival through the formation of NF-κB-activating, PIDD1-C/RIP1 complexes. However, no changes in the net or relative abundances of either PIDD1 fragment are observed in verified settings of endogenous PIDDosome assembly5,11,12. Rather, DNA injury triggers PIDDosome formation through ATM/ATR-mediated phosphorylation of the PIDD1-CC DD on conserved threonine 788, with ATM and ATR carrying out the reaction in response to DSBs and ICLs, respectively5,11,27. In the context of ICLs, the pathway is initiated by the Fanconi anemia (FA) repair protein FANCI, which recruits PIDD1 to unresolved lesions to enable its recognition by ATR within a FANCI/PIDD1/ATR complex11. Regardless of stimulus, T788 phosphorylation is necessary and sufficient for the PIDD1-RAIDD interaction and ensuing PIDDosome formation5,28. While T788 phosphorylation has been proposed to “prime” the PIDD1-CC DD for RAIDD recruitment5, the precise mechanism by which phosphorylation enables platform formation has remained unknown.
a Domain structures of full-length (FL) PIDD1 and autocatalytic cleavage products. LRR, leucine-rich repeats; ZU5, domain present in Zona occludens, UNC5-like netrin receptors and Ankyrin; UPA, Uncharacterized protein domain in UNC5, PIDD and Ankyrin family; DD, death domain. b–g HeLa cells of indicated genotypes (c), transfected with indicated siRNAs (d) or stably expressing indicated shRNAs (e) were treated with indicated drugs, harvested at indicated time points and analyzed on 10% bis-tris NuPAGE gels with indicated antibodies. MMC, mitomycin C; CPT, camptothecin; TOPO, topotecan; 2-D08, SUMO E2 ligase inhibitor; thalidomide, CRBN inhibitor. h HCT116 cells stably expressing WT or ΔGG (non-conjugatable) FLAG-SUMO-1 were treated with MMC and/or Go6976, harvested at 24 h, immunoprecipitated with anti-FLAG antibody and analyzed by western blot. i, i’ HeLa cells grown on cover slips were treated with MMC and Chk1 inhibitor Go6976, fixed at indicated time points (hour), stained with indicated antibodies and imaged by confocal microscopy. At least 35 cells per time point were scored in each of n = 3 independent experiments. Representative images (i) quantified in (i’), with data expressed as means +/− SD; **, p < 0.005; ***p < 0.001; ns, non-significant; two-tailed Student’s t-test. j HeLa cells treated with indicated drugs were immunoprecipitated with anti-PIDD1pT788 antibody at indicated time points and analyzed by western blot. k Cartoon summarizing the modified PIDD1pT788 species. All drugs were given at 1 μM. Cells in (b–h, j) were TdR-synchronized as described in Supplementary Tables 1A (b, c, e–h) and 1B (d). Source data are provided as a Source data file.
Post-translational control of protein function via Small Ubiquitin-Related Modifier (SUMO) conjugation plays critical roles in a wide range of (mainly nuclear) processes29,30,31, especially stress responses such as the DNA damage response32,33,34. Conjugation of single SUMO units (typically SUMO-1) or chains thereof (typically made of SUMO-2/3) can alter protein-protein interactions or enable interactions with SUMO-Interacting Motif (SIM)-containing proteins, as well as impact the enzymatic activity, spatial distribution and turnover rate of substrates35. These effects are countered by a deSUMOylation machinery consisting of seven main SUMO proteases (SENPs 1–3 and 5–8, sentrin-specific proteases)36,37. While multiple SUMO targets have been described, including numerous DNA repair factors and cell cycle regulators, targets in the apoptotic machinery remain essentially unknown38. A direct role in the regulation of MCL1 stability was recently reported39, while potential SUMO target sites on caspase-740, caspase-841 and C2 itself42, all identified two decades ago, still await functional validation. Here we show that a single, phosphorylation-induced mono-SUMOylation event dictates the assembly of a vertebrate CAP, the PIDDosome, specifically in cells which fail to resolve DNA interstrand crosslinks (ICLs) and double-strand breaks (DSBs).
Results
PIDD1 is mono-SUMOylated in response to ICL repair failure
We recently found that ICLs, such as induced by mitomycin C (MMC), trigger PIDDosome formation and apoptotic death when they fail to be repaired. This is seen when: (i) ICLs are introduced on FA repair-deficient backgrounds, such as in FANCP/SLX4 deficient cells; (ii) ICLs are forced into mitosis prior to repair completion by means of co-exposure to Chk1 inhibitors (Chk1i) such as Gö6976; or, (iii) excessive levels of ICLs are introduced in otherwise FA-proficient cells, thus mimicking repair failure (as in Supplementary Fig. 1a)11. While probing the timing of PIDDosome-mediated C2 activation in such contexts, we noted the concurrent appearance of slower migrating species of the ATR-phosphorylated PIDD1-CC fragment (CCpT788) on 10% bis-tris NuPAGE gels (Fig. 1b, c and Supplementary Fig. 1a; unless otherwise indicated, all experiments were performed in cells synchronized by double thymidine block (TdR)). Three species were detected which ran at approximately +6, +9 and +12 kDa relative to CCpT788, designated CCpT788-L ( ~44 kDa), CCpT788-XL ( ~47 kDa) and CCpT788-XXL ( ~50 kDa), respectively (Fig. 1b–d and S1a). All three species observed in HeLa cells were also detected in SV40-transformed mouse embryonic fibroblasts (MEF) and HCT116 cells of both WT and TP53 null genotypes (Supplementary Fig. 1b, c); in response to additional DNA crosslinkers (cisplatin and bendamustine, Supplementary Fig. 1d) as well as replication stressors known to activate ATR and the FA pathway (hydroxyurea and gemcitabine, Supplementary Fig. 1e); and were indeed encoded by PIDD1 (Fig. 1e). While L, XL and XXL were detected coincident with forced mitotic entry in Chk1i-treated cells (Supplementary Fig. 1f), progression into mitosis is not essential for the generation of either species in response to ICLs, as observed in FA repair-defective cells (Supplementary Fig. 1f’, note the lack of histone H3 phosphorylation).
The topoisomerase inhibitors camptothecin and topotecan also induced CCpT788 -L, -XL and -XXL when combined with Chk1i (Fig. 1f), indicating that DSBs can likewise signal these putative CCpT788 modifications. In contrast, levels of Plk4 overexpression sufficient for PIDDosome activation via centrosome amplification14 readily induced L and XL, but not XXL (Supplementary Fig. 1g). This indicated that of all three modified CCpT788 species, only XXL is DNA damage-specific. While L and XL, but not XXL, were detected in total lysates from irradiated cells (Supplementary Fig. 1d), XXL was readily detected in nuclear fractions from such cells (Supplementary Fig. 1h). Notably, C2 bifluorescence complementation (C2 BiFC; Supplementary Fig. 1i) revealed that IR was the sole DNA-damaging treatment tested which led to PIDDosome formation in the cytoplasm in addition to strictly in the nucleolus, the latter being the major site of PIDDosome assembly after DNA damage (Supplementary Fig. 1j-j’)6,43. These data suggested that the generation of the CCpT788 XXL species involves a nuclear modifier.
The likely reliance of XXL on a nuclear modifier, together with its ~12 kDa shift in size relative to native CCpT788, led us to test whether the species might reflect a single mono-SUMOylation event. In contrast to L and XL species, XXL was undetectable in cells exposed to the SUMO E2 ligase inhibitor 2-D08 (Fig. 1g)44. Indeed, XXL did reflect the conjugation of a SUMO-1 subunit onto CCpT788, as demonstrated in (i) FLAG pulldowns from HCT116 cells stably expressing FLAG-SUMO1 but not a non-conjugatable ΔGG mutant (Fig. 1h)34,45; and (ii) endogenous PIDD1-CCpT788 pulldowns probed with a monoclonal SUMO-1 antibody (Fig. 1j; note the inhibitory effect of 2-D08). Immunofluorescence studies demonstrated a staining overlap between CCpT788 and SUMO1 in the nuclei of ICL repair-failing cells (Fig. 1i-i’). Colocalization was first detectable at 12 h and peaked at 24 h, mirroring the dynamics of CCpT788-XXL in immunoblots (Fig. 1b, c). The signal overlap was notably restricted to discrete nuclear areas, which will become relevant later. CCpT788 was not detectably modified by SUMO2/3 (Fig. 1j). Collectively, these observations showed that phospho-PIDD1-CC is mono-SUMO1ylated in response to ICL repair failure. PIDD1pT788 is also likely the target of additional modifications reflected by the L and XL species (Fig. 1k). The latter modifications are not DNA damage-specific (see Supplementary Fig. 1f above) and will be studied elsewhere.
Mono-SUMOylation occurs on K879 in the PIDD1 DD
To identify the SUMO-1 target on PIDD1-CC, we disrupted candidate acceptor lysines in the fragment. We prioritized on those residues which are conserved across vertebrates: K606 in the N-terminal end; K639 and K702 in the Uncharacterized Protein domain in UNC5, PIDD and Ankyrin (UPA); and K879R at the distal end of the DD, a residue predicted to map in close proximity (11 Å) to the ATM/ATR target T788 in an AlphaFold2 PIDD1 model (Fig. 2a, b)46. Because PIDD1 SUMOylation and C2 activation occur with similar timing (Fig. 1b, c), we hypothesized that blocking conjugation would either negatively or positively impact PIDDosome assembly. We thus first interrogated the various mutants using the C2 BiFC reporter (Supplementary Fig. 1h).
a, b ClustalW alignment of human (Hs), mouse (Mm), rat (Rn), chicken (Gg), and zebrafish (Dr) PIDD1 C termini. UPA and DD boxed in light blue and dark blue, respectively. Conserved lysine residues and ATM/ATR target T788 also indicated both in alignment (a) and AlphaFold2 model of full-length PIDD1 (b). c, d C2.Pro-BiFC HeLa cells of indicated genotypes were transfected with indicated FLAG-PIDD1 variants, treated with or without MMC and Go6976 (5 μM each) and imaged at 24 h. Arrowheads mark Venus signal, reflecting dimerization of C2 Pro-VN and C2 Pro-VC (see Supplementary Fig. 1h). At least 40 cells per sample were scored in each of n = 3 independent experiments. Representative images (c) were quantified in (d). e HeLa cells stably expressing the indicated shRNAs were transfected with indicated FLAG-PIDD1 variants, treated with or without MMC (0.1 μM) and Go6976 (1 μM) and stained with the vital dye alamarBlue at 5 days. f HeLa cells of indicated genotypes were transfected with indicated FLAG-PIDD1 variants, treated with or without MMC and Chk1 inhibitor Go6976 (1 μM each), harvested at indicated time points and analyzed by western blot. g HeLa cells of indicated genotypes were transfected with indicated FLAG-PIDD1 variants, treated with or without MMC and Go6976 (1 μM each), harvested at indicated time points, immunoprecipitated with anti-PIDD1pT788 antibodies and analyzed by western blot. h Pidd1—/— MEF transfected with indicated FLAG-PIDD1 variants and treated with MMC and Go6976 (1 μM each) were harvested at 24 h, immunoprecipitated with anti-FLAG antibody and analyzed by western blot. Cells in (f–h) were TdR-synchronized as described in Supplementary Table 1B. Data in (d, e) expressed as means +/− SD of 3 independent experiments. **p < 0.005; ***p < 0.001; ns, non-significant; two-tailed Student’s t-test. Source data are provided as a Source data file.
As expected, deletion of PIDD1 completely blocked C2 BiFC signals in repair-failing cells, which was rescued by a WT FLAG-PIDD1 construct (Fig. 2c, d)11. Strikingly, whereas a quadruple K606R;K639R;K702R;K879R (4xK/R) variant failed to restore C2 BiFC signals, a triple K606R;K639R;K702R mutant (3xK/R) retained full PIDDosome-inducing activity (Fig. 2c, d). This pointed to K879 as the putative acceptor. Indeed, the single K879R variant failed to rescue C2 BiFC in PIDD1—/— cells while PIDD1K639R was fully effective (Fig. 2c, d). These observations in MMC-treated cells co-treated with Chk1i were confirmed on a FANCP/SLX4-depleted background (Supplementary Fig. 2a, b). Cell viability assays validated the inability of PIDD1K879R to restore PIDDosome function in PIDD1—/— cells while PIDD1K639R and even PIDD13xKR tested in parallel were as effective as WT PIDD1 (Fig. 2e). Running lysates from the above experiments on 10% bis-tris NuPAGE gels showed that K879R specifically prevented mono-SUMOylation whereas the CCpT788 -L and -XL species were unaffected (Fig. 2f). In contrast, all three species were detected in cells expressing K639R (Fig. 2f) or 3xK/R (Supplementary Fig. 2c). Furthermore, FLAG-PIDD1K879R was not reactive to SUMO-1 antibodies, in contrast with WT, K639R and 3xK/R variants (Fig. 2g and S2c). Importantly, whereas PIDD1-CCK639R recruited RAIDD as efficiently as WT PIDD1-CC in stimulated Pidd1—/— MEF47, PIDD1-CCK879R failed to do so (Fig. 2h). These observations, which were confirmed on HCT116 WT and TP53—/— backgrounds (Supplementary Fig. 2d), both provided a mechanism for the C2 BiFC phenotype observed above and provided first evidence that PIDD1 mono-SUMOylation is essential for PIDDosome formation in response to ICL repair failure. Collectively, these data identified K879 as the SUMOylation target on PIDD1.
PIDD1 SUMOylation is catalyzed by PIAS1 and reversed by SENP3
We next sought to identify the E3 ligase responsible for PIDD1 SUMOylation, as well as a possibly associated SUMO protease. Nine SUMO E3 ligases have been formally identified: PIAS1-4, NSE/MMS21, RanBP2 and ZNF451/1-348,49. We excluded ZNF451 E3s because they conjugate SUMO-2/3 chains, not single SUMO-1 subunits50. A C2 BiFC screen of the remaining six SUMO E3s, first performed in cells treated with MMC+Chk1i, identified PIAS1 as required for PIDDosome assembly (Fig. 3a–c). This was confirmed on a FANCP/SLX4 deficient background (Supplementary Fig. 3a). PIAS1, but not PIAS3, was detected in complex with CCpT788 from 6 h onwards, including during the time window of SUMOylation (15–24 h post-stimulus) (Fig. 3d). Most importantly, depletion of PIAS1 specifically eliminated CCpT788 SUMOylation without affecting the CCpT788 -L and -XL species (Fig. 3e), and SUMOylation was restored by WT but not catalytically inactive Myc-PIAS1 (PIAS1-CI, mutated at C346S;C351S;H353A;C356S) (Fig. 3e, f)51. Conversely, overexpression of PIAS1, but not PIAS1-CI or WT PIAS3, enhanced PIDD1 SUMOylation, including the generation of a di-SUMOylated form (Fig. 3g and S3b). Collectively, these results identified PIAS1 as the PIDD1 K879 E3 SUMO ligase.
a–c C2.Pro-BiFC HeLa cells transfected with indicated siRNAs were treated with or without MMC and Go6976 (5 μM each) and analyzed by confocal microscopy at 24 h. At least 40 cells per sample were scored in each of n = 3 independent experiments. Representative images (a) were quantified for percentage of C2 BiFC positive cells (b) and overall signal intensity (c). d HeLa cells treated with MMC and Go6976 and harvested at indicated time points were immunoprecipitated with anti-PIDD1pT788 antibody and analyzed by western blot. e HeLa cells transfected with indicated siRNAs were treated with MMC and Go6976, harvested at indicated time points and analyzed by western blot. f HeLa cells transfected with indicated siRNAs, co-transfected with indicated Myc-PIAS1 constructs and treated and analyzed as in (e). C1, catalytically inactive. (g) HeLa cells transfected with indicated Myc-PIAS constructs and treated with MMC and Go6976 were analyzed as in (e). CCpT788di-SUMO1, di-SUMO-1ylated CCpT788. h FANCP—/— HeLa cells treated with MMC were harvested at indicated time points and PIDD1pT788 immunoprecipitates were analyzed by western blot. i–k Cells treated and analyzed as in (a) were quantified for percentage of C2 BiFC positive cells (j) and average number of Venus puncta per cell (k). l Parent and SENP3—/— HeLa cells treated with MMC and Go6976 at indicated doses were analyzed as in (i–k). At least 40 cells per sample were scored in each of n = 2 independent experiments (representative images shown in Supplementary Fig. 3c) (m) Cells as in (l) were stained with the vital dye alamarBlue at 5 days. n HeLa cells transfected with indicated siRNAs and treated with indicated drugs were immunoprecipitated with anti-PIDD1pT788 antibody and analyzed by western blot. Cells in (d–h, n) were TdR-synchronized as described in Supplementary Tables 1A (d, h), 1B (e–g), 1C (n). Unless otherwise indicated, drugs were given at 1μM. Data in (b, c, j, k, m) are expressed as means +/− SD, with *p < 0.05, **p < 0.005, ***p < 0.001, ns, non-significant, two-tailed Student’s t-test. Source data are provided as a Source data file.
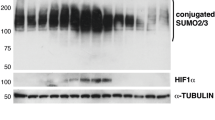
Next, we asked whether a SUMO protease might act to counter PIAS1 function. Seven SUMO proteases have been identified52, of which one, SENP3, has previously been detected in FLAG-PIDD1 pulldowns analyzed by mass spectrometry53. SENP3 also localizes specifically to the nucleolus54,55, which is the major site of PIDDosome formation after DNA damage6,11. We found that SENP3 associated with CCpT788 starting at approximately 18 hr post-stimulus (Fig. 3h), that is, ~3 h after the onset of SUMOylation (Fig. 1b, c). The CCpT788/SENP3 interaction persisted until at least 24 h, when PIDDosomes actively assemble in the cell (see C2 BiFC signals in Figs. 2c, 3a). SENP7 was also detected in the pulldowns at similar time points while the only SENP other than SENP3 that localizes to the nucleolus, SENP555, was not detected, similar to SENPs −1, −2, −6 and −8 (Fig. 3h). Depletion of SENP3, but not SENP7, enhanced C2 BiFC signals in repair-failing cells (Fig. 3i), both in percentage of positive cells (Fig. 3j) and number of Venus signals per cell (Fig. 3k), suggesting the formation of supernumerary PIDDosomes. These results were confirmed in a CRISPR/Cas9-generated SENP3—/— HeLa cell line, in which the increase in BiFC signals (Fig. 3l and S3c) correlated with reduced cell viability at 5 days post-stimulus (Fig. 3m). Most importantly, silencing SENP3 led to increased levels of mono-SUMOylated CCpT788 proteins (Fig. 3n), validating SENP3 as the PIDD1 K879 SUMO protease and indicating that SENP3 actively deSUMOylates CCpT788 as PIDDosomes assemble in the cell. Collectively, these results identified PIAS1 and SENP3 as the major constituents of the SUMOylation machinery responsible for PIDDosome control in response to DNA damage.
DNA damage-induced PIDD1 phosphorylation is necessary and sufficient for SUMOylation
Having identified the SUMO E3 ligase and protease regulating PIDD1 SUMOylation at K879, we next investigated the mechanism by which DNA damage signals to this SUMO machinery. Thus far, we have detected PIDD1 SUMOylation exclusively on ATM/ATR-phosphorylated PIDD1-CC, suggesting that conjugation requires T788 phosphorylation. To test this, we transfected PIDD1—/— cells with non-phosphorylatable (T788A) and phosphomimetic (T788D) FLAG-PIDD1 variants5 and analyzed lysates at 24 h post-treatment (when the bulk of CCpT788 is in its SUMOylated form, see Fig. 1b, c). In damaged PIDD1—/— cells, FLAG-PIDD1WT and FLAG-PIDD1T788D, but not FLAG-PIDD1T788A, restored a CCpT788 species of ~ 54 kDa which was eliminated by SUMOi 2-D08 (Fig. 4a). These observations indicated that T788 phosphorylation is necessary for PIDD1 SUMOylation. PIDD1T788D, but not PIDD1WT or PIDD1T788A, produced this same band in unstimulated cells (Fig. 4b). This indicated that T788 phosphorylation is not only necessary but also sufficient for SUMOylation. Strikingly, the K879R mutation abrogated the ability of PIDD1-CCT788D to constitutively bind RAIDD (Fig. 4c) and enable PIDDosome formation (Fig. 4d). Thus, DNA damage-induced PIDD1-CC phosphorylation promotes PIDDosome formation strictly through its induction of PIDD1 SUMOylation.
a, b PIDD1—/— HeLa cells transfected with indicated FLAG-PIDD1 variants, treated with MMC and Go6976 with or without SUMO E2 ligase inhibitor 2-D08 were harvested at 24 h and analyzed by western blot. c Pidd1—/— MEF transfected with indicated FLAG-PIDD1 variants and treated with or without MMC and Go6976 were harvested at 24 h, immunoprecipitated with anti-FLAG antibody and analyzed by western blot. d PIDD1—/— C2.Pro-BiFC HeLa cells were transfected with indicated FLAG-PIDD1 variants, treated with or without MMC and Go6976 (5 μM each) and imaged at 24 h. At least 40 cells per sample were scored in each of n = 3 independent experiments. Representative images were quantified (right), with data expressed as means +/− SD. **p < 0.005; ***p < 0.001; ns, non-significant, two-tailed Student’s t-test. e, f PIDD1—/— HeLa cells transfected with indicated FLAG-PIDD1 variants and treated with MMC and Go6976 (e) with or without ATR inhibitor BAY-1895344 (BAY, 0.75 mM) (f) were harvested at 24 h, immunoprecipitated with anti-PIDD1pT788 antibody and analyzed by western blot. Cells in (a–c, e, f ) were TdR-synchronized as described in Supplementary Tables 1B and 1C. Unless otherwise indicated, drugs were given at 1 μM. Source data are provided as a Source data file.
We reasoned that phosphorylation might promote SUMOylation by (i) enabling PIDD1 recognition by PIAS1; (ii) preventing PIDD1 recognition by SENP3; and/or (iii) dislodging SENP3 from its substrate. Abrogating PIDD1 phosphorylation with T788A completely blocked PIAS1 recruitment to PIDD1 (Fig. 4e). Likewise, blocking T788 phosphorylation with ATR inhibitor BAY-189534456,57 prevented PIDD1 recognition by PIAS1 (Fig. 4f, lanes 4–6, and Supplementary Fig. 4a). This effect specifically resulted from loss of T788 phosphorylation because BAY-1895344 failed to block PIAS1 binding in PIDD1—/— cells reconstituted with PIDD1T788D (Fig. 4f, lanes 10–12). Mutationally or pharmacologically blocking T788 phosphorylation did not enable but rather prevented SENP3 binding, similar to PIAS1 (Fig. 4e, f). Lastly, forcing phosphorylation through T788D failed to dislodge SENP3 from PIDD1 (Fig. 4f). We therefore conclude that ATR-mediated phosphorylation of the PIDD1 DD triggers its SUMOylation by enabling PIAS1 docking.
PIDD1 SUMOylation sustains its interaction with RAIDD and enables C2 recruitment
Our earlier results indicated that PIDD1 SUMOylation is essential for RAIDD recruitment and PIDDosome assembly (Figs. 2h, 4d). As K879 is located in the DD, we first examined whether SUMOylation enables the PIDD1-RAIDD interaction itself. We scanned vertebrate RAIDD sequences for potential SUMO-interacting motifs (SIMs). GPS-SUMO58 predicted three conserved SIMs, of which two, 57LLDI60 (in the caspase recruitment domain (CARD)) and 135VLSL138 (in the DD) were likewise predicted by JASSA59 in all species examined (Fig. 5a). While the putative SIMs have relatively low probability scores (Fig. 5a), a SIM of identical sequence to that identified in the RAIDD DD (VLSL) was previously validated in the viral helicase BBLF359 and is reasonably surface-exposed in an AlphaFold2 RAIDD model (Fig. 5b)46. Because mutation of a single leucine to alanine can be sufficient to disrupt SIM function60,61, we targeted individual conserved L residues in each predicted SIM. VSV-RAIDD L57A and L138A variants were expressed at levels similar compared to parental WT construct and were used for analyses (Supplementary Fig. 5a).
a, b ClustalW alignment of human (Hs), mouse (Mm), xenopus (Xl), and zebrafish (Dr) RAIDD homologs with GPS- SUMO and JASSA -predicted SUMO-interacting domains (SIM) boxed in red and green, respectively (probability scores also indicated) (a), with conserved putative SIMs boxed in black on AlphaFold2 RAIDD model (b). Caspase recruitment domain (CARD) and DD underlined in orange and blue, respectively. (c-d) RAIDD—/— C2.Pro-BiFC HeLa cells were transfected with indicated VSV-RAIDD variants, treated with or without MMC and Go6976 (5 μM each) and imaged at 24 h. At least 40 cells per sample were scored in each of n = 3 independent experiments. Representative images (c) were quantified (d), with data expressed as means +/− SD. ***p < 0.001, two-tailed Student’s t-test. e RAIDD—/— HeLa cells transfected with indicated VSV-RAIDD variants, and treated with or without MMC and Go6976 (1 μM each) were harvested at 24 h, immunoprecipitated with anti-VSV antibodies and analyzed by western blot. f SV40-transformed MEF of indicated genotypes treated with MMC and Go6976 (1 μM each) were harvested at indicated time points and analyzed by western blot. g PIDD1—/— HeLa cells transfected with indicated siRNA and cotransfected with indicated FLAG-PIDD1 variants were harvested at 24 h, immunoprecipitated with anti-FLAG antibody and analyzed by western blot. h, i PIDD1—/— HeLa cells transfected with indicated FLAG-PIDD1 variants, were treated MMC and Go6976 (1 μM each), harvested at indicated time points, immunoprecipitated with anti-FLAG antibody and analyzed by western blot. Cells in (e–i) were TdR-synchronized as described in Supplementary Tables 1A (f) and 1B (e, g–i). Source data are provided as a Source data file.
Whereas VSV-RAIDDL57A retained intact PIDDosome-inducing activity in response to unrepaired ICLs, VSV-RAIDDL138A failed to restore C2 BiFC signals when likewise introduced in RAIDD—/— cells (Fig. 5c, d). These results were confirmed by C2 cleavage assay whereby VSV-RAIDDL57A, but not VSV-RAIDDL138A, restored normal levels of the fully mature C2 p19 subunit in RAIDD—/— cells (Supplementary Fig. 5b). Co-immunoprecipitation showed that VSV-RAIDDL138A failed to interact with PIDD1, in contrast to WT and L57A variants (Fig. 5e). RAIDDL138A was otherwise not affected in its ability to bind C2, indicating that the mutation selectively blocks the PIDD1-RAIDD interaction (Fig. 5e). Consistent with the C2 BiFC assay, RAIDDL138A also failed to restore PIDDosome formation at the endogenous level (see triple immunofluorescence experiments below). Confirming that 135VLSL138 supports a SIM59, the L138A mutation, but not L57A, reduced the affinity of RAIDD for free SUMO-1 in cells devoid of PIDD1 (Supplementary Fig. 5c). Collectively, these results indicated that a SUMO-SIM interaction involving PIDD1 K879SUMO1 and the 135VLSL138 motif in RAIDD is critical for the DD:DD interaction between PIDD1 and RAIDD in response to ICL repair failure.
Unexpectedly, we found that RAIDD was required for PIDD1 SUMOylation itself (Fig. 5f). The third PIDDosome subunit, C2, was not, and if anything acted to suppress SUMOylation (Fig. 5f). Mechanistically, we found that RAIDD is required for PIAS1 binding to phosphorylated PIDD1 (Fig. 5g). These observations led to an apparent paradox: How can RAIDD be necessary for PIDD1 SUMOylation when SUMOylation is necessary for the PIDD1-RAIDD interaction in the first place?
We reasoned that PIDD1 SUMOylation might not be required for the initiation of the PIDD1-RAIDD interaction per se, but rather, its maintenance. To test this, we analyzed PIDD1 immunoprecipitates over time. Disrupting SUMOylation with PIDD1K879R did not affect the initial recruitment of RAIDD to PIDD1 (at 12 h post-stimulus) but failed to sustain PIDD1-RAIDD complexes (Fig. 5h). These observations were confirmed in experiments with the PIDD1 T788D phosphomimetic, in which the variant constitutively recruited RAIDD, as expected5, but failed to maintain the interaction when SUMOylation was denied (Fig. 5i, compare T788D, lanes 9–12, and T788D;K879R, lanes 13–16). In contrast, disrupting SUMOylation had no effect on the maintenance of PIDD1/NPM1 complexes— NPM1 (nucleophosmin) is a nucleoplasmic and nucleolar protein chaperone required for PIDDosome assembly after DNA damage6. Unlike SUMOylation, PIDD1 phosphorylation was necessary for the initiation of the PIDD1-RAIDD interaction (Fig. 5i, lanes 5–8), consistent with previous studies5,8. Lastly, failure to maintain the PIDD1-RAIDD interaction, whether with FLAG-PIDD1K879R or FLAG-PIDD1T788D;K879R, prevented the completion of PIDDosome assembly: C2 was not mobilized to the complex, regardless of time point (Fig. 5h, i). Collectively, these experiments revealed a three-step model for PIDDosome assembly whereby: 1) damage-induced PIDD1 phosphorylation on T788 in the DD initiates the PIDD1-RAIDD interaction; 2) this “priming” step enables PIAS1 binding and SUMOylation of PIDD1 K879; and 3) the SUMO-SIM stabilized PIDD1/RAIDD complex recruits C2 to the platform.
The SUMO-SIM interaction between PIDD1 and RAIDD enables the nucleolar internalization of PIDD1 and PIDDosome formation therein
Lastly, we investigated the mechanism by which the SUMO-SIM interaction between PIDD1 and RAIDD acts to sustain the interaction and, critically, how this maintenance enables C2 recruitment and completion of PIDDosome formation. A key distinctive feature of SUMO-1 is the localization of itself and/or many its conjugates to the nucleolus62,63,64, as confirmed herein (Supplementary Fig. 6a, b). Whether SUMO-1 conjugation acts as nucleolar localization signal (NoLS) or occurs preferentially within the organelle is not known. The nucleolus also defines the major site of PIDDosome assembly in damaged cells (Supplementary Fig. 1j-j’)6,11,27,65, and pro-C2 is the only caspase zymogen present in the nucleus6,66,67,68. Additionally, a specific monoclonal antibody revealed that RAIDD is a nucleolar resident protein whose spatial distribution does not change in response to ICL repair failure (Supplementary Fig. 7b), regardless of time point examined (Fig. 6a, b). Within the nucleolus, however, persistent ICLs led to RAIDD relocating from the dense fibrillar center (DFC, as marked by fibrillarin) to the granular component (GC) at the inner nucleolar periphery (Supplementary Fig. 7b). In contrast to nucleolus-residing RAIDD, PIDD1-CCpT788 molecules originate from ATM/ATR phosphorylation at damage sites distributed throughout the nucleoplasm and, in turn, are incorporated in the nucleolus (Fig. 6a, b)11,12. Therefore, we asked whether the SUMO-SIM interaction between PIDD1 and RAIDD might be responsible for localizing CCpT788 to the nucleolus via RAIDD-mediated capture.
a–i HeLa cells of indicated genotypes and transfected with indicated FLAG-PIDD1 or VSV-RAIDD variants (c–i) were treated with MMC and Go6976 (1 μM each), fixed at indicated time points (hours; (a, b)) and stained with indicated antibodies and DAPI to mark nuclei (contours marked by dashed white line in (a)). At least 40 cells per sample were scored in each of n = 2 independent experiments. Representative images (a, c) were analyzed for percentage of cells with nucleolar PIDD1pT788 signals (d, f), Pearson’s coefficient (b, e, g) and distribution of PIDD1pT788 signal intensity (h, i). Source data are provided as a Source data file.
Strikingly, PIDD1—/— cells reconstituted with FLAG-PIDD1K879R failed to localize nucleoplasmic CCpT788 molecules to the nucleolar GC (marked by nucleolin, NCL), as observed in independent settings of ICL repair failure (Fig. 6c–e, h and Supplementary Fig. 7a). Likewise, RAIDD—/— cells reconstituted with SIM-defective VSV-RAIDDL138A failed to localize CCpT788 to the organelle (Fig. 6c, f, g, i, and Supplementary Fig. 7a). Importantly, VSV-RAIDDL138A properly localized to the GC in these cells (Supplementary Fig. 7c), and the overall intensity and distribution of SUMO-1 immunoreactivity were unaffected regardless of stimulus, repair status or genotype of interest (Supplementary Fig. 6a, b). Thus, the SUMO-SIM interaction between PIDD1 and RAIDD is specifically required for the nucleolar incorporation of CCpT788.
Triple stainings with a specific antibody to cleaved C2 (Supplementary Fig. 8a–c) definitively showed that the SUMO-SIM interaction between PIDD1 and RAIDD is essential for completion of PIDDosome formation in the nucleolus (Fig. 7a–e). To our knowledge, the CCpT788/RAIDD/cleaved-C2 triple-positive foci detected in the GCs of stimulated WT cells and RAIDDWT-rescued RAIDD—/— cells, but not that reconstituted with RAIDDL138A, are the first direct observations of fully assembled PIDDosomes in cells. Endogenous PIDDosomes were also detected in stimulated WT and TP53—/— HCT116 cells (Supplementary Fig. 9). These results conclusively identify the nucleolus, and specifically the peripheral GC therein, as the major cellular location of PIDDosome formation in response to DNA repair failure, in line with previous C2 BiFC imaging studies of cells exposed to a variety of DNA damage stimuli6.
a–e Parent and RAIDD—/— HeLa cells transfected with indicated VSV-RAIDD variants were treated with or without MMC and Go6976 (1 μM each), fixed at 24 h and stained with indicated antibodies and DAPI. At least 20 cells per sample were scored in each of n = 3 independent experiments. Representative images (a) were analyzed for triple colocalization count (b), triple colocalization area (c), percent area of triple signal overlaps (d) and overall cleaved C2 signal intensity (e). Data expressed as means +/− SD. *p < 0.05; **p < 0.005; ***p < 0.001; ns, non-significant, two-tailed Student’s t-test. f Three-step model for PIDDosome formation (see text). Source data are provided as a Source data file.
Altogether, we conclude that PIDD1 SUMOylation simultaneously serves two essential roles in PIDDosome formation: (1) At the biochemical level, the modification acts to sustain otherwise unstable nascent PIDD1/RAIDD complexes, a process we refer to as “locking” step; and (2) at the cellular level, it acts as a bona fide NoLS which enables the incorporation of PIDD1 by RAIDD into the nucleolus, thus compartmentalizing PIDD1/RAIDD scaffolds for C2 recruitment and completion of PIDDosome formation (Fig. 7f). Altogether, these results identify a direct role for SUMOylation in the assembly of a vertebrate CAP.
Discussion
The oligomeric and multi-oligomeric (5xPIDD1/7xRAIDD/7xC2) structures of the PIDDosome were revealed by Wu and colleagues in 200722, yet the mechanisms governing platform formation had remained elusive. Our results support a three-step assembly process which is tightly regulated in both space and time by stress-induced, sequential post-translational modifications of the PIDDosome core scaffold, the PIDD1-CC DD (Fig. 7f). Each modification is reversible, via an as-yet unidentified phosphatase and, as identified herein, the SUMO protease SENP3. The data suggest a highly dynamic assembly system which allows for fine regulation of signal intensity and for reversal of commitment to cell death.
Shortly after the PIDDosome structure was solved, a biochemical study identified the PIDD1-RAIDD interaction, not the RAIDD-C2 interaction, as the rate limiting step in PIDDosome formation in vitro21. RAIDD was found to exist in a closed conformation, whereby exposure of its C2-recruiting CARD domain required prior interaction of the RAIDD DD with that of PIDD1. In our cell-based assays, we found that nascent PIDD1/RAIDD dimers fail to recruit C2 unless PIDD1 undergoes SUMOylation, indicating that the endogenous PIDD1/RAIDD interaction is in of itself insufficient for C2 recruitment. Whether PIDD1 SUMOylation and ensuing SUMO-SIM interaction with RAIDD acts to expose the RAIDD CARD will require detailed structural analyses. Alternatively, the impact of the SUMO-SIM interaction may only be observed the cellular setting, i.e., should it solely be required to properly localize PIDD1/RAIDD complexes to the nucleolus. Unexpectedly, we found that SIMDD-deficient RAIDDL138A, which failed to bind PIDD1 in response to ICL repair failure, associated with C2 as efficiently as WT RAIDD in this context. This observation is at odds with the closed state model for RAIDD monomers discussed above and suggests that RAIDD and C2 may be recruited to PIDD1 as preassembled heterodimers.
Interestingly, the C2 CARD harbors a SUMO-1 target at K63 (previously annotated as K60) whose physiologic significance has remained unknown42. It is possible that a SUMO-SIM dimerization between the C2 and RAIDD CARD domains, similar to that we identified between the PIDD1 and RAIDD DDs, is additionally required for PIDDosome formation. Such poly(SUMO)/poly(SIM) interactions drive the assembly of stimuli-responsive condensates such as PML bodies69,70, ATR signaling hubs71,72 and SLX4 repair foci73. We note, however, that K63 is moderately conserved among vertebrate C2 paralogs, and mutation of the only conserved putative SIM located in the RAIDD CARD (57LLDI60) did not prevent C2 recruitment (Fig. 5e). It is possible that the L57A mutation we generated is insufficient for SIMCARD disruption, unlike the L138A mutation which effectively altered the RAIDD SIMDD.
Whether SUMO-SIM interactions support the assembly of CAPs other than the PIDDosome remains to be seen. While none of the apoptosome subunits (cyt c, APAF1, caspase-9) have been identified as SUMO targets, acceptor lysines have been identified in two of three DISC subunits, FADD and caspase-841,74. The putative caspase-8 acceptor, K156, is not conserved including in mice, but the three acceptors identified in FADD are conserved in mammals and birds and are all located in the DD74. Whether FADD SUMOylation impacts DISC assembly remains to be investigated, however, SUMOylated FADD formed a complex with dynamin-related protein 1 (Drp1) and caspase-10 in vitro, a function potentially relevant to mitochondrial fragmentation during regulated necrosis74. There is also emerging evidence that SUMOylation regulates inflammasome formation, with both the NLRP3 scaffold and ASC adapter implicated as targets75,76,77,78. Future identification of SUMO-SIM interactions between inflammasome subunits, similar to that we detected between PIDD1 and RAIDD, would solidify SUMO as a general driver of CAP assembly in vertebrate cells.
Until now, two mechanisms of phosphorylation-induced SUMOylation had been described. First, through phosphorylation-dependent SUMO motifs (PDSM) on the target substrate, in which phosphorylation of a serine at position +5 from the acceptor lysine augments conjugation79,80,81. Second, via phosphorylation of the SUMO ligase, resulting in its targeting to a specific cellular location housing the substrate82. Here we identify a third mechanism for phosphorylation-dependent SUMOylation, whereby phosphorylation of the substrate at a distant site from the SUMO acceptor enables substrate recognition by the SUMO E3 ligase. It should be noted that the SUMOylation-inducing phosphorylation event is predicted to occur at a distance of 11 Å from the acceptor lysine in an AlphaFold2 model (Fig. 2b), which may reconstitute a PDSM-like motif in 3D. However, RAIDD is essential for phosphorylation-induced PIAS1 docking, suggesting that RAIDD recruitment acts as a critical intermediate in PIDD1 recognition by PIAS1. Whatever the precise mechanism, ATR/ATM-phosphorylation control of PIDD1 SUMOylation is critical as it enables cells to restrict PIDDosome formation to the occurrence of unrepaired DNA lesions, thus ensuring an opportunity to repair prior to committing to death. Likewise, SENP3 may integrate DNA damage/repair inputs to limit or block PIDDosome assembly should the lesions eventually resolve through the dedicated repair machinery or backup pathways, thus reversing the death decision. Consistent with this, SENP3—/— cells committed to cell death when treated with otherwise tolerated levels of ICLs (Fig. 3l, m).
A central question in the PIDDosome field is how the same CAP can dictate distinct outcomes depending on context. In response to DNA damage stress, such as studied herein, the PIDDosome signals apoptosis5,11,12, whereas in response to overduplicated centrosomes, it induces cell cycle arrest13,14,15. These distinct outputs may contribute to the platform’s ambiguous cancer genetics17,83,84,85. When considered alongside recent studies, our data suggest that PIDDosome output is dictated by subcellular location of platform assembly rather than CAP levels or signaling intensity. Two groups showed that PIDD1 is recruited to centrioles via ANKRD26, such that centrosome amplification results in a local increase in PIDD1 levels13,14. Ensuing PIDDosome activation, through mechanisms not yet entirely clear, releases active C2 molecules in the cytoplasm, resulting in MDM2 cleavage and p53-dependent cell cycle arrest. In response to DNA damage, by contrast, PIDD1 proteins are compartmentalized in the nucleolus via ATM/ATR phosphorylation-induced SUMOylation, which ultimately leads to the release of active C2 in the nucleolar GC and nucleoplasm, but not the cytoplasm (Fig. 7 and Supplementary Figs. 8, 9). C2’s substrate(s) in the nucleus, whose cleavage triggers death, are a key open question. We were surprised to find that ectopic Plk4 levels sufficient for centrosome overduplication also led to PIDD1 phosphorylation on T788, along with two of three CCpT788 modifications, CCpT788-L and CCpT788-XL (Supplementary Fig. 1g). Despite PIDD1 phosphorylation being sufficient for SUMOylation, SUMOylated PIDD1 was notably undetectable in these cells. Whether this was achieved through downregulation of PIAS1, upregulation of SENP3 or other mechanisms, and how these cells phosphorylated PIDD1 in the first place, remain to be investigated. Clearly, the SUMO switch to PIDDosome-mediated death can be evaded, which may prove relevant to tumor development and treatment resistance.
Our identification of SENP3 as the PIDD1 deSUMOylase was in part unexpected because the protease has a known preference for poly(SUMO-2/3) chains in vitro86. However, SENPs exhibiting specificity for SUMO chains in biochemical assays, such as SENP5 and SENP3 itself, have been reported to cleave monoSUMO-1 conjugates in several cellular settings78,87. SENP3 serving as the PIDD1 deSUMOylase is also consistent with its previous detection in FLAG-PIDD1 pulldowns analyzed by mass spectrometry53 as well as with the singular presence of the protease in nucleoli54,55. Inactivation of SENP3 markedly sensitized cells to MMC-induced PIDDosome formation, suggesting a new strategy for activating the PIDDosome in treatment-resistant tumors5,10. The development of SENP inhibitors and especially SENP3-specific inhibitors is still in its infancy88,89. The emergence of the SUMO machinery as a key regulator of apoptotic and inflammatory CAPs is suggestive of broad therapeutic applications for such drugs in the clinic.
Methods
Cell culture and reagents
HeLa (cervical, p53-defective via HPV-E6), TP53—/— HCT116 (colon) cancer cell lines90 were cultured in DMEM medium (Life Technologies) supplemented with 10% Fetal Bovine Serum (FBS) (Sigma- Aldrich) and 1% penicillin/streptomycin (P/S) (Life Technologies). HeLa shPIDD1, shRAIDD and shCASP2 cells (Supplementary Table 7) were cultured as previously described5. Caspase-2—/—, Raidd—/—, Pidd1—/—, and corresponding WT, SV40-transformed MEFs, kindly provided by Andreas Villunger, Douglas Green were cultured as previously described91. HeLa.C2-Pro-BiFC cells of WT, PIDD1—/— and RAIDD—/— genotypes6 were provided by Lisa Bouchier-Hayes and were cultured in DMEM medium (Life Technologies) supplemented with 10% Fetal Bovine Serum (FBS) (Sigma- Aldrich) and 1% penicillin/streptomycin (P/S) (Life Technologies). RPE‐1 PLK4Dox cells of WT, ANKRD26—/— and PIDD1—/— genotypes14 were generously provided by Andrew J. Holland and were cultured in DMEM-F12 medium (Life Technologies) supplemented with 10% Fetal Bovine Serum (FBS) (Sigma- Aldrich) and 1% penicillin/streptomycin (P/S) (Life Technologies). Gö6976, MMC, hydroxyurea, cisplatin, camptothecin, topotecan, gemcitabine and doxycycline were purchased from Sigma-Aldrich. 2-D08 and thalidomide were purchased from Selleckchem. BAY1895344 was purchased from MedChem Express. Bendamustine was a gift from Dr. Joshua Brody.
RNAi
siRNA transfections were performed using X-tremeGENE siRNA transfection reagent (Roche) and 20 nM siRNA according to the manufacturer’s instructions. Cells were treated with Gö6976 + / − MMC at 48 hrs post-transfection (except for TdR experiments, see Supplementary Table 1B). Previously validated siRNAs were siLACZ92, siRAIDD (RAIDD-2)4,5,93, and siFANCP (BTBD12)11 (Qiagen). siRNAs to DNAPKcs, PIAS1, PIAS2, PIAS3, PIAS4, NSMCE2, RANBP2, SENP1, SENP2, SENP3, SENP5, SENP6, SENP7, SENP8 and ZMIZ1 were purchased from Qiagen. HeLa cells stably expressing shGFP and shPIDD1 have been previously described5. See Supplementary Tables 5, 7 for siRNA and shRNA sequences.
Expression vectors, DNA transfections and site-directed mutagenesis
Plasmid DNA was transfected into HeLa parental, shPIDD1 and shRAIDD cells using X-tremeGENE HP (Roche) according to the manufacturer’s instructions. Transfected cells were treated with or without Gö6976 at 24 hrs post-transfection and MMC was added one hour later. C-terminally Flag-tagged PIDD1-FL, cloned in pcDNA5/FRT4,24,53 and N-terminally VSV-tagged RAIDD (VSV-RAIDD) cloned in pCR34 were a kind gift from Dr Emanuelle Logette. Myc-tagged PIAS1 WT, PIAS1-C1 and PIAS3 cDNA were gifted by Dr. Weibin Wang51. FLAG-tagged SUMO1 and SUMO1ΔGG cDNAs45 were a gift from Stephen P. Goff. The T788A, T788D, K639R, K879R, K3R (K639/702/879 R) K4R(K606/639/702/879R) mutations were introduced into Flag-PIDD1-FL and L138A and L57A mutations were introduced in VSV-RAIDD-FL according to the manufacturer’s instructions using a Q5® Site-Directed Mutagenesis Kit (NEB, E0554S). See Supplementary Table 4 for primer sequences.
Generation of stable FLAG-SUMO-1 WT and ΔGG HCT116 cells
Phoenix cells were seeded at a density of 3 × 105 cells in a 10-cm plate. The following day, they were transfected with 8 μg of either FLAG- tagged SUMO1 or SUMO1ΔGG cDNA, along with 4 μg of pCMVR8.2 DNA and 4 μg of pVSV-G DNA using polyethylenimine (PEI). Cell supernatants were collected after 48 h for virus preparations. Virus was then added along with polybrene to HCT116 cells. Media was changed after 48 h and the cells were selected with puromycin (1 μg/ml).
CRISPR–Cas9 gene editing
Plasmid lentiCRISPR v2 was digested with BsmBI-v2 (New England Biolabs, R0580) according to the manufacturer’s recommendations. Briefly, 1 μg of plasmid was digested for 1 h at 55 °C, and then digested plasmid was gel-purified using a QIAquick Gel Extraction kit and eluted in water. Single-guide RNA (sgRNA) oligonucleotides for cloning were annealed by mixing them in equal 10 μM concentrations with the addition of 1 × T4 DNA ligase buffer, and the mixture was incubated at 37 °C for 30 min and at 95 °C for 5 min and then ramped down to 25 °C at 5 °C min−1. Hybridized oligonucleotides were diluted 1:200 with H2O. BsmBI-v2 digested lentiCRISPR v2 plasmid (50 ng) was ligated with 1 μM final concentration oligo duplex using T4 ligase (NEB, M0202) according to the manufacturer’s recommendations and incubated overnight at 4 °C. XL-1 blue competent Escherichia coli were transformed with 1 μl of ligation reaction according to the manufacturer’s protocol (Agilent Technologies, catalog no. 200249). Single clones were sequence-verified using Sanger sequencing. Lentivirus particles containing sgRNA construct of FANCP and SENP3 were generated by transfecting Phoenix packaging cells with lentiCRISPR v2 containing corresponding sgRNAs (Eurofins Genomics) and a combination of the lentiviral helper plasmids pCMV-dR8.91 and pMD.G at a ratio of 2:1:1, respectively. jetPEI (Polyplus, 101-10 N) was used as the transfection ant. After 24 h, medium containing viral particles was collected and concentrated using Lenti-X Concentrator according to the manufacturer’s protocol (Clontech, 631231). Briefly, one volume of Lenti-X Concentrator was mixed with three volumes of 0.45-μm filtered viral particle–containing medium. The solution was then incubated overnight at 4 °C. The samples were centrifuged at 1500 g for 45 min at 4 °C, the supernatant was collected and was added in HeLa cell culture medium. For infection, 2 × 105 HeLa cells were plated into six-well plates. The next day, 5 μM polybrene (Millipore, tr-1003-g) and 200 μl of concentrated viral particles were added per well. The medium was replaced the next day with medium containing puromycin (1 μg ml−1) for selection. See Supplementary Table 6 for sgRNA sequences.
Western blotting and Antibodies
All western blot experiments were performed after synchronizing the cells with double thymidine block (TdR) (See Supplementary Table 1) Cells were seeded at a density of 1 × 106 in 10 cm plates. For experiments with Gö6976 and MMC, cells were treated with Gö6976 (1 μM). After 1 h, cells were treated with MMC (1 μM). For experiments with DNA damaging agents, cells were treated with Gö6976 (1 μM). After 1 h, camptothecin (1 μM), topotecan (1 μM), cisplatin (5 μM), bendamustine (50 μM), gemcitabine (10 μM) or hydroxyurea (0.4 mM) was added. RPE1 cell lines were treated with doxycycline (1ug/ml). Cells were then harvested at indicated time points and lysed using 1% NP-40 buffer (Boston BioProducts) with protease and phosphatase inhibitors. Lysates (180 – 200ug) were incubated at 70 °C for 10 min after adding NuPAGE LDS Sample Buffer (4X) (Life Technologies) and 5% 2-Mercaptoethanol (Sigma Aldrich). For detection of the CCpT788(L, XL, and XXL) bands, samples were run on 10% Bis-Tris gels using 1X MOPS buffer (Life Technologies) for 75 min at 170 V. After electrophoresis, samples were transferred on a nitro-cellulose membrane (Bio-rad) at 94 V for 100 min. Membranes were then blocked with 5% Bovine serum albumin (BSA, Sigma Aldrich) in TBS with 0.1% Tween and probed with primary antibodies at 4 °C overnight. Membranes were then rinsed with TBS-Tween (5 × 5 min) and probed with specific HRP-linked secondary antibody in 5% milk or BSA (in TBS-Tween) for 1 h at room temperature. Membranes were washed as described earlier and placed in SuperSignal West Pico Chemiluminescent Substrate or SuperSignal West Dura Extended Duration Substrate (Pierce Biotechnology). The membrane was then developed with photographic film. A full list of antibodies and protocols can be found in Supplementary Table 2.
Co-Immunoprecipitation
All co-immunoprecipitation experiments were performed in synchronized cells (See Supplementary Table 1). Cells were seeded at a density of 3 × 106 in 15 cm plates. Lysates for immunoprecipitation (IP) were prepared in 1 or 0.1% NP-40 buffer (1 or 0.1% NP-40, 50 mM Tris-HCl [pH 8.0], 150–250 mM NaCl, 5 mM EDTA, protease inhibitors cocktail [Complete Mini, Roche] and phosphatase inhibitor cocktail [PhosSTOP, Roche]). For endogenous IPs, whole-cell lysates (1–5 mg) were mixed with Protein-G magnetic beads (Invitrogen, 20 µl of a 50% slurry) and α-pPIDD1 (pT788) (5 µg, 1 ml final volume) for 10 min to 2 h at room temp on a rotating wheel. Beads were then washed three times with PBS-Tween20 (0.02%), resolved by SDS-PAGE, and probed with primary antibodies detected with the corresponding secondary antibodies or mouse TrueBlot HRP-conjugated secondary antibodies [eBioscience]). For α-Flag or α-VSV IPs, whole-cell lysates (0.15–2 mg) were mixed with 20 µL beads (50% slurry) and mouse α-Flag (M2) antibody (3 µg) or α–VSV (3ug) in 1% NP-40 buffer (500 µL final volume) for 10 min on a rotating wheel. Beads were then washed three times with PBS-T, resolved by SDS-PAGE and analyzed by western blot.
Cell fractionation
HeLa cells, seeded at a density of 2 × 106 cells/plate were treated with or without IR (10 Gy) and harvested at 24 hrs post-IR. Hela RAIDD -/- cells, seeded at a density of 3 × 106 cells/plate were transfected with indicated cDNAs for 24 h, treated with Gö6976 (1 μM) and MMC (1 μM) and harvested at 24 h post-MMC. Cells were then fractionated using the Subcellular Protein Fractionation kit (ThermoFisher Scientific) following the instruction manual and analyzed by Western blotting or immunoprecipitation.
Caspase-2 Bimolecular Fluorescence Complementation (C2 BiFC) imaging
Bimolecular fluorescence complementation (BiFC) assay was used as a read-out for PIDDosome assembly, as previously described43. Pro-BiFC cells of parental, PIDD—/— and RAIDD—/— genotypes (1 × 105 cells)6 were seeded directly on coverslips, transfected with or without cDNA (1 ug/ml) for 24 h or with or without siRNAs (20 nmol) for 48 h and then treated with qVD-OPH (20 μM) and Gö6976 (5 μM) and MMC (5 μM) and fixed 24 h post-treatment. For experiments with DNA damaging agents, cells were treated with the compounds at the same concentrations as western blot experiments and fixed 24 h post-treatment, Cells expressing the BiFC components were identified by fluorescence of the linked mCherry protein in stable cell lines. Venus channel image data was analyzed to determine the cells positive for C2 BiFC. More than 100 cells were counted over three independent experiments.
Confocal microscopy
Parental, PIDD1—/— and RAIDD—/— HeLa.C2 Pro-BiFC cells (see above), and parental HeLa, along with PIDD1—/— and RAIDD—/— cells (5 × 104) were seeded directly onto coverslips, transfected with indicated cDNAs for 24 h or indicated siRNAs for 48 h, treated with Gö6976 (1 μM) and MMC (1 μM) and harvested 24 h post-treatment (unless otherwise indicated) fixed in 1% paraformaldehyde, permeabilized in 0.5% Triton-X100, and stained as described. Confocal microscopy was performed using a Leica TCS SP5 II Confocal over an inverted microscope. Images were acquired using LAS software. See Supplementary Table 3 for a list of antibodies used and specific staining methods.
Image analysis
For all BiFC experiments, more than 100 cells were counted for Venus signal, over 3 independent experiments. For Fig. 3a, total intensity of the C2 BiFC signal for each condition was calculated using FIJI software. For Fig. 3K, average number of BiFC dots/ cell were also counted over 3 independent experiments. Two-channel Colocalization analyses were performed with Just Another Co-localization Plugin (JaCoP) (https://imagej.net/plugins/jacop), downloaded as a plug-in for FIJI. Pearson’s coefficient was calculated as a colocalization indicator. Triple channel colocalization analyses were performed using FIJI software. Images were opened, channels were arranged into blue (DAPI), magenta (RAIDD), green (pT788) and red (cleaved C2). All the channels were split, and then converted into 8-bit (except DAPI). Colocalization steps were carried out starting with the magenta channel first. Threshold was adjusted on both magenta and red channels first to have binary images built for both the channels. From the Image Process in the toolbar, value of 254 was subtracted using MATH option from both the binary images to make the signal value 1 and background value 0. Next, Image Calculator was selected from the Image Process toolbar and both the channels were added. Threshold was adjusted to 2–255 and applied on the newly generated image. This resulted in a composite image showing only the colocalized pixels between magenta (RAIDD) and red (cleaved Caspase2). Next, threshold was adjusted on the green channel (pT788) and using MATH option from the Image Process, value of 254 was subtracted from the channel. This image was then added to the composite magenta-red channel using the Image Calculator Add function. Threshold for this 3-channel image was SET (not APPLY) to 0–0. The colocalized particles among all the three channels were analyzed using Analyze Particles function on FIJI.
Cell viability assays
AlamarBlue-based cell viability assays were performed as described11,12, with several modifications. HeLa shGFP and shPIDD1 cells were seeded into 96-well plates at a density of 400 cells/well. After 16 h, cells were transfected with the indicated cDNAs (1 μg/ml) for 24 h, and then treated with DMSO (1 μM), Gö6976 (1 μM), and an hour later MMC (0.1 μM) was added. HeLa WT and SENP3−/− cells were seeded on a 96-well plate at a density of 400 cells/well. After 16 h, the cells were treated with indicated doses of Gö6976 and MMC. 3 days post-treatment, cells were incubated with alamarBlue (Thermo Fisher) at a final concentration of 10%. After 24 h, absorbance was measured at a wavelength of 570 nm with a 600 nm reference wavelength. Relative fluorescence (RFU) was calculated using cell free wells as a control reference and percent survival was calculated compared to non-treated shGFP and HeLa WT controls.
Cell Synchronization
Cells were seeded at a density of 1 × 106 / plate for western blotting and 3 × 106 for co-immunoprecipitation. See Supplementary Table 1 for detailed synchronization protocols. Thymidine was purchased from Sigma Aldrich.
Statistics and reproducibility
All experiments were performed at least twice and statistics were derived from experiments performed at least three times. Paired two-tailed Student’s t tests were used to determine P values (α = 0.05). The log-rank test was used to determine P values for survival curves. Data in bar graphs are represented as means ± SD or means ± SEM, as indicated in legends, and statistical significance was expressed as follows: *P < 0.05; **P < 0.005; ***P < 0.001; ns, not significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data supporting the findings of this study are available within the article and its supplementary information files. Source data are provided with this paper.
References
Green, D. R. Inflammasomes and other caspase-activation platforms. Cold Spring Harb. Perspect Biol. 14 https://doi.org/10.1101/cshperspect.a041061 (2022).
Man, S. M. & Kanneganti, T. D. Converging roles of caspases in inflammasome activation, cell death and innate immunity. Nat. Rev. Immunol. 16, 7–21 (2016).
Parrish, A. B., Freel, C. D. & Kornbluth, S. Cellular mechanisms controlling caspase activation and function. Cold Spring Harb. Perspect Biol. 5, https://doi.org/10.1101/cshperspect.a008672 (2013).
Tinel, A. & Tschopp, J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science 304, 843–846 (2004).
Ando, K. et al. PIDD death-domain phosphorylation by ATM controls prodeath versus prosurvival PIDDosome signaling. Mol. Cell 47, 681–693 (2012).
Ando, K. et al. NPM1 directs PIDDosome-dependent caspase-2 activation in the nucleolus. J. Cell Biol. 216, 1795–1810 (2017).
Dawar, S. et al. Caspase-2-mediated cell death is required for deleting aneuploid cells. Oncogene 36, 2704–2714 (2017).
Hiregange, D., Naick, H. & Rao, B. J. ATR signalling mediates the prosurvival function of phospho-NPM against PIDDosome mediated cell death. Cell Signal 71, 109602 (2020).
Li, Y. et al. A noncanonical IRAK4-IRAK1 pathway counters DNA damage-induced apoptosis independently of TLR/IL-1R signaling. Sci. Signal. 16, eadh3449 (2023).
Liu, P. H. et al. An IRAK1-PIN1 signalling axis drives intrinsic tumour resistance to radiation therapy. Nat. Cell Biol. 21, 203–213 (2019).
Shah, R. B. et al. FANCI functions as a repair/apoptosis switch in response to DNA crosslinks. Dev. Cell 56, 2207–2222.e2207 (2021).
Thompson, R. et al. An inhibitor of PIDDosome formation. Mol. Cell 58, 767–779 (2015).
Burigotto, M. et al. Centriolar distal appendages activate the centrosome-PIDDosome-p53 signalling axis via ANKRD26. EMBO J. 40, e104844 (2021).
Evans, L. T. et al. ANKRD26 recruits PIDD1 to centriolar distal appendages to activate the PIDDosome following centrosome amplification. EMBO J. 40, e105106 (2021).
Fava, L. L. et al. The PIDDosome activates p53 in response to supernumerary centrosomes. Genes Dev. 31, 34–45 (2017).
Sladky, V. C. et al. Centriole signaling restricts hepatocyte ploidy to maintain liver integrity. Genes Dev. 36, 843–856 (2022).
Sladky, V. C. et al. PIDDosome-induced p53-dependent ploidy restriction facilitates hepatocarcinogenesis. EMBO Rep. 21, e50893 (2020).
Burigotto, M. & Fava, L. L. The PIDDosome: centrosome guardian and backup on the DNA damage response. Mol. Cell Oncol. 8, 1893625 (2021).
Weiler, E. S., Szabo, T. G., Garcia-Carpio, I. & Villunger, A. PIDD1 in cell cycle control, sterile inflammation and cell death. Biochem Soc. Trans. 50, 813–824 (2022).
Sladky, V., Schuler, F., Fava, L. L. & Villunger, A. The resurrection of the PIDDosome - emerging roles in the DNA-damage response and centrosome surveillance. J. Cell Sci. 130, 3779–3787 (2017).
Jang, T. H. & Park, H. H. PIDD mediates and stabilizes the interaction between RAIDD and caspase-2 for the PIDDosome assembly. BMB Rep. 46, 471–476 (2013).
Park, H. H. et al. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell 128, 533–546 (2007).
Telliez, J. B., Bean, K. M. & Lin, L. L. LRDD, a novel leucine rich repeat and death domain containing protein. Biochim Biophys. Acta 1478, 280–288 (2000).
Tinel, A. et al. Autoproteolysis of PIDD marks the bifurcation between pro-death caspase-2 and pro-survival NF-kappaB pathway. Embo J. 26, 197–208 (2007).
Tinel, A. et al. Regulation of PIDD auto-proteolysis and activity by the molecular chaperone Hsp90. Cell Death Differ. 18, 506–515 (2011).
Garcia-Carpio, I. et al. Extra centrosomes induce PIDD1-mediated inflammation and immunosurveillance. EMBO J. e113510, https://doi.org/10.15252/embj.2023113510 (2023).
Li, Y. et al. A non-canonical IRAK signaling pathway triggered by DNA damage. bioRxiv, https://doi.org/10.1101/2023.02.08.527716 (2023).
Terry, M. R. et al. Caspase-2 impacts lung tumorigenesis and chemotherapy response in vivo. Cell Death Differ. 22, 719–730 (2015).
Gareau, J. R. & Lima, C. D. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 11, 861–871 (2010).
Varejao, N., Lascorz, J., Li, Y. & Reverter, D. Molecular mechanisms in SUMO conjugation. Biochem Soc. Trans. 48, 123–135 (2020).
Flotho, A. & Melchior, F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev. Biochem 82, 357–385 (2013).
Jackson, S. P. & Durocher, D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell 49, 795–807 (2013).
Schwertman, P., Bekker-Jensen, S. & Mailand, N. Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat. Rev. Mol. Cell Biol. 17, 379–394 (2016).
Gibbs-Seymour, I. et al. Ubiquitin-SUMO circuitry controls activated fanconi anemia ID complex dosage in response to DNA damage. Mol. Cell 57, 150–164 (2015).
Wilkinson, K. A. & Henley, J. M. Mechanisms, regulation and consequences of protein SUMOylation. Biochem J. 428, 133–145 (2010).
Kunz, K., Piller, T. & Muller, S. SUMO-specific proteases and isopeptidases of the SENP family at a glance. J Cell Sci 131, https://doi.org/10.1242/jcs.211904 (2018).
Nayak, A. & Muller, S. SUMO-specific proteases/isopeptidases: SENPs and beyond. Genome Biol. 15, 422 (2014).
Li, P. et al. SUMO modification in apoptosis. J. Mol. Histol. 52, 1–10 (2021).
Li, S. et al. SUMOylation of MCL1 protein enhances its stability by regulating the ubiquitin-proteasome pathway. Cell Signal 73, 109686 (2020).
Hayashi, N., Shirakura, H., Uehara, T. & Nomura, Y. Relationship between SUMO-1 modification of caspase-7 and its nuclear localization in human neuronal cells. Neurosci. Lett. 397, 5–9 (2006).
Besnault-Mascard, L. et al. Caspase-8 sumoylation is associated with nuclear localization. Oncogene 24, 3268–3273 (2005).
Shirakura, H. et al. Caspase recruitment domain of procaspase-2 could be a target for SUMO-1 modification through Ubc9. Biochem Biophys. Res. Commun. 331, 1007–1015 (2005).
Bouchier-Hayes, L. et al. Characterization of cytoplasmic caspase-2 activation by induced proximity. Mol. Cell 35, 830–840 (2009).
Kim, Y. S., Keyser, S. G. & Schneekloth, J. S. Jr. Synthesis of 2’,3’,4’-trihydroxyflavone (2-D08), an inhibitor of protein sumoylation. Bioorg. Med Chem. Lett. 24, 1094–1097 (2014).
Lee, A., Zhu, Y., Sabo, Y. & Goff, S. P. Embryonic Cells Redistribute SUMO1 upon Forced SUMO1 Overexpression. mBio 10, https://doi.org/10.1128/mBio.01856-19 (2019).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Manzl, C. et al. Death of p53-defective cells triggered by forced mitotic entry in the presence of DNA damage is not uniquely dependent on Caspase-2 or the PIDDosome. Cell Death Dis. 4, e942 (2013).
Liu, B., Lois, L. M. & Reverter, D. Structural insights into SUMO E1-E2 interactions in Arabidopsis uncovers a distinctive platform for securing SUMO conjugation specificity across evolution. Biochem J. 476, 2127–2139 (2019).
Pichler, A., Fatouros, C., Lee, H. & Eisenhardt, N. SUMO conjugation - a mechanistic view. Biomol. Concepts 8, 13–36 (2017).
Eisenhardt, N. et al. A new vertebrate SUMO enzyme family reveals insights into SUMO-chain assembly. Nat. Struct. Mol. Biol. 22, 959–967 (2015).
Zhang, T. et al. Crosstalk between SUMOylation and ubiquitylation controls DNA end resection by maintaining MRE11 homeostasis on chromatin. Nat. Commun. 13, 5133 (2022).
Hickey, C. M., Wilson, N. R. & Hochstrasser, M. Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 13, 755–766 (2012).
Logette, E. et al. PIDD orchestrates translesion DNA synthesis in response to UV irradiation. Cell Death Differ. 18, 1036–1045 (2011).
Haindl, M., Harasim, T., Eick, D. & Muller, S. The nucleolar SUMO-specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing. EMBO Rep. 9, 273–279 (2008).
Yun, C. et al. Nucleolar protein B23/nucleophosmin regulates the vertebrate SUMO pathway through SENP3 and SENP5 proteases. J. Cell Biol. 183, 589–595 (2008).
Lucking, U. et al. Damage incorporated: discovery of the potent, highly selective, orally available ATR inhibitor BAY 1895344 with favorable pharmacokinetic properties and promising efficacy in monotherapy and in combination treatments in preclinical tumor models. J. Med Chem. 63, 7293–7325 (2020).
Wengner, A. M. et al. The novel ATR inhibitor BAY 1895344 is efficacious as monotherapy and combined with DNA damage-inducing or repair-compromising therapies in preclinical cancer models. Mol. Cancer Ther. 19, 26–38 (2020).
Zhao, Q. et al. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 42, W325–W330 (2014).
Beauclair, G., Bridier-Nahmias, A., Zagury, J. F., Saib, A. & Zamborlini, A. JASSA: a comprehensive tool for prediction of SUMOylation sites and SIMs. Bioinformatics 31, 3483–3491 (2015).
Chang, P. C. et al. Kaposi’s sarcoma-associated herpesvirus (KSHV) encodes a SUMO E3 ligase that is SIM-dependent and SUMO-2/3-specific. J. Biol. Chem. 285, 5266–5273 (2010).
Song, J., Zhang, Z., Hu, W. & Chen, Y. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J. Biol. Chem. 280, 40122–40129 (2005).
Ayaydin, F. & Dasso, M. Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol. Biol. Cell 15, 5208–5218 (2004).
Liu, X. et al. Sumoylation of nucleophosmin/B23 regulates its subcellular localization, mediating cell proliferation and survival. Proc. Natl. Acad. Sci. USA 104, 9679–9684 (2007).
Matafora, V., D’Amato, A., Mori, S., Blasi, F. & Bachi, A. Proteomics analysis of nucleolar SUMO-1 target proteins upon proteasome inhibition. Mol. Cell Proteom. 8, 2243–2255 (2009).
Bouchier-Hayes, L. & Sidi, S. The nucleolus: a new home for the PIDDosome. Cell Cycle 16, 1562–1563 (2017).
Colussi, P. A., Harvey, N. L. & Kumar, S. Prodomain-dependent nuclear localization of the caspase-2 (Nedd2) precursor. A novel function for a caspase prodomain. J. Biol. Chem. 273, 24535–24542 (1998).
Mancini, M. et al. Caspase-2 is localized at the Golgi complex and cleaves golgin-160 during apoptosis. J. Cell Biol. 149, 603–612 (2000).
Zhivotovsky, B., Samali, A., Gahm, A. & Orrenius, S. Caspases: their intracellular localization and translocation during apoptosis. Cell Death Differ. 6, 644–651 (1999).
Hattersley, N., Shen, L., Jaffray, E. G. & Hay, R. T. The SUMO protease SENP6 is a direct regulator of PML nuclear bodies. Mol. Biol. Cell 22, 78–90 (2011).
Shen, T. H., Lin, H. K., Scaglioni, P. P., Yung, T. M. & Pandolfi, P. P. The mechanisms of PML-nuclear body formation. Mol. Cell 24, 331–339 (2006).
Psakhye, I. & Jentsch, S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151, 807–820 (2012).
Wu, C. S. et al. SUMOylation of ATRIP potentiates DNA damage signaling by boosting multiple protein interactions in the ATR pathway. Genes Dev. 28, 1472–1484 (2014).
Alghoul, E. et al. Compartmentalization of the SUMO/RNF4 pathway by SLX4 drives DNA repair. Mol. Cell 83, 1640–1658 e1649 (2023).
Choi, S. G. et al. SUMO-modified FADD recruits cytosolic Drp1 and caspase-10 to mitochondria for regulated necrosis. Mol. Cell Biol. 37, https://doi.org/10.1128/MCB.00254-16 (2017).
Dong, D. et al. Inflammasome activity is controlled by ZBTB16-dependent SUMOylation of ASC. Nat. Commun. 14, 8465 (2023).
Barry, R. et al. SUMO-mediated regulation of NLRP3 modulates inflammasome activity. Nat. Commun. 9, 3001 (2018).
Qin, Y. et al. TRIM28 SUMOylates and stabilizes NLRP3 to facilitate inflammasome activation. Nat. Commun. 12, 4794 (2021).
Shao, L. et al. SUMO1 SUMOylates and SENP3 deSUMOylates NLRP3 to orchestrate the inflammasome activation. FASEB J. 34, 1497–1515 (2020).
Hietakangas, V. et al. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci. USA 103, 45–50 (2006).
Mohideen, F. et al. A molecular basis for phosphorylation-dependent SUMO conjugation by the E2 UBC9. Nat. Struct. Mol. Biol. 16, 945–952 (2009).
Vu, E. H., Kraus, R. J. & Mertz, J. E. Phosphorylation-dependent sumoylation of estrogen-related receptor alpha1. Biochemistry 46, 9795–9804 (2007).
Ptak, C. et al. Phosphorylation-dependent mitotic SUMOylation drives nuclear envelope-chromatin interactions. J. Cell Biol. 220, https://doi.org/10.1083/jcb.202103036 (2021).
Braun, V. Z. et al. Extra centrosomes delay DNA damage-driven tumorigenesis. Sci. Adv. 10, eadk0564 (2024).
Manzl, C. et al. PIDDosome-independent tumor suppression by Caspase-2. Cell Death Differ. 10.1038/cdd.2012.54 (2012).
Peintner, L. et al. The tumor-modulatory effects of Caspase-2 and Pidd1 do not require the scaffold protein Raidd. Cell Death Differ. 22, 1803–1811 (2015).
Mendes, A. V., Grou, C. P., Azevedo, J. E. & Pinto, M. P. Evaluation of the activity and substrate specificity of the human SENP family of SUMO proteases. Biochim Biophys. Acta 1863, 139–147 (2016).
Zunino, R., Schauss, A., Rippstein, P., Andrade-Navarro, M. & McBride, H. M. The SUMO protease SENP5 is required to maintain mitochondrial morphology and function. J. Cell Sci. 120, 1178–1188 (2007).
Yang, Y. et al. Small-molecule inhibitors targeting protein SUMOylation as novel anticancer compounds. Mol. Pharm. 94, 885–894 (2018).
Wei, J. et al. Recent research and development of inhibitors targeting sentrin-specific protease 1 for the treatment of cancers. Eur. J. Med Chem. 241, 114650 (2022).
Bunz, F. et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282, 1497–1501 (1998).
Manzl, C. et al. Caspase-2 activation in the absence of PIDDosome formation. J. Cell Biol. 185, 291–303 (2009).
Sidi, S. et al. Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2, and caspase-3. Cell 133, 864–877 (2008).
Janssens, S., Tinel, A., Lippens, S. & Tschopp, J. PIDD mediates NF-kappaB activation in response to DNA damage. Cell 123, 1079–1092 (2005).
Acknowledgements
We thank Christopher Lima and Aneel Aggarwal for helpful discussions and Stephen Goff, Andrew Holland, Weibin Wang and Lisa Bouchier-Hayes for sharing critical reagents. S.S. was supported by grants from NIH/NIGMS (RO1GM135301), NIH/NCI (R01CA178162), Pershing Square Sohn Cancer Research Alliance, New York Community Trust and Searle Scholars Program.
Author information
Authors and Affiliations
Contributions
R.B.S. performed the experiments with contributions from Y.L. (generation of PIDD1 K/R mutants) and technical assistance from H.Y. and E.K.; S.S. designed and supervised the study, and wrote the manuscript with R.B.S. All authors discussed the results and participated in the manuscript preparation and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shah, R.B., Li, Y., Yu, H. et al. Stepwise phosphorylation and SUMOylation of PIDD1 drive PIDDosome assembly in response to DNA repair failure. Nat Commun 15, 9195 (2024). https://doi.org/10.1038/s41467-024-53412-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-53412-0