Abstract
Phytopathogens often secrete effectors to enhance their infection of plants. In the case of Sclerotinia sclerotiorum, a necrotrophic phytopathogen, a secreted protein named SsPEIE1 (Sclerotinia sclerotiorum Plant Early Immunosuppressive Effector 1) plays a crucial role in its virulence. During the early stages of infection, SsPEIE1 is significantly up-regulated. Additionally, transgenic plants expressing SsPEIE1 exhibit increased susceptibility to different phytopathogens. Further investigations revealed that SsPEIE1 interacts with a plasma membrane protein known as hypersensitive induced reaction (HIR) that dampens immune responses. SsPEIE1 is required for S. sclerotiorum virulence on wild-type Arabidopsis but not on Arabidopsis hir4 mutants. Moreover, Arabidopsis hir2 and hir4 mutants exhibit suppressed pathogen-associated molecular pattern-triggered reactive oxygen species (ROS) bursts and salicylic acid (SA)-associated immune gene induction, all of which are phenocopied by the SsPEIE1 transgenic plants. We find that the oligomerization of AtHIR4 is essential for its role in mediating immunity, and that SsPEIE1 inhibits its oligomerization through competitively binding to AtHIR4. Remarkably, both Arabidopsis and rapeseed plants overexpress AtHIR4 display significantly increased resistance to S. sclerotiorum. In summary, these results demonstrate that SsPEIE1 inhibits AtHIR4 oligomerization-mediated immune responses by interacting with the key immune factor AtHIR4, thereby promoting S. sclerotiorum infection.
Similar content being viewed by others
Introduction
Necrotrophic pathogenic fungi are widely distributed and cause significant crop losses. Significant challenges posed by necrotrophs to crop production are expected to increase with climate change1,2. Sclerotinia sclerotiorum (Lib.) de Bary, a destructive ascomycete fungus with a wide host range, can infect over 700 plant species, including economically important crops such as Brassica napus, Glycine max, Helianthus annuus L., Beta vulgaris and Lactuca sativa3,4,5,6. S. sclerotiorum causes substantial yield losses and economic damage on many crops. Additionally, this fungus is very difficult to control due to the complexity of its disease cycle7,8. Studying the pathogenesis of S. sclerotiorum and its interactions with host plants is essential for developing innovative, environmentally friendly control strategies for Sclerotinia diseases.
Plants have evolved complex and diverse receptor proteins to interact with pathogens. These receptors are distributed in the cell membrane and cytoplasm, monitor the activity of pathogens, and generate various immune responses against pathogen invasion9,10. S. sclerotiorum is also inevitably recognized by plants through multiple pathways. For example, chitin in its cell wall is recognized by the receptor LYK5 on the cell membrane of Arabidopsis, transmitting immune signals downstream via the co-receptor kinase CERK111,12,13. In addition, various secreted proteins of this necrotrophic fungus can be recognized by receptors on plant membranes, triggering pattern-triggered immunity (PTI)14,15,16. The PTI signaling pathway rapidly transmits signals intracellularly and initiates early plant immune responses, including reactive oxygen species (ROS) burst, mitogen-activated protein kinase (MAPK) activation and increased transcription of disease-resistant related genes to defend against pathogen invasion17,18,19.
In contrast, S. sclerotiorum has long evolved a powerful arsenal to combat plants. Numerous studies have shown that the successful infection of plants by S. sclerotiorum is closely linked to the production of oxalic acid (OA) and cell wall degrading enzymes (CWDEs). OA plays multiple roles in the plant immune response, including creating a low pH environment for invasion, sequestering calcium, impairing the oxidative burst of the plant, inducing programmed cell death similar to apoptosis, and inhibiting autophagy20,21,22,23,24,25,26,27. Moreover, this fungus is induced by the host to secrete large amounts of CWDEs to break down the plant cell wall and digest plant cells, ultimately facilitating successful infection28,29,30. Equally important, small secretory proteins are also involved in the pathogenesis of S. sclerotiorum. Although many secreted proteins are predicted for S. sclerotiorum, only a few have been empirically investigated. For example, the secreted protein SsCaf1 is required for appressorium formation in the early stage of S. sclerotiorum infection31. Additionally, S. sclerotiorum finely regulates plant immunity or metabolism by secreting effectors into plant cells. For example, the small secretory protein SsSSVP1 interacts with QCR8 and alters its mitochondrial localization, thereby interfering with plant energy metabolism32. The secretory protein SsITL localizes in plant cell chloroplasts and interacts with CAS to suppress salicylic acid pathway-related immunity, promoting fungal infection33. SsCP1 interacts with plant PR1 in the apoplast to promote S. sclerotiorum infection, but it can also be recognized by plants to trigger defense responses via the salicylate (SA) signaling pathway16. Recently, it was reported that SCP, a small cysteine-rich secreted protein of S. sclerotiorum, can be recognized by Arabidopsis RLP30 and trigger plant immunity34. The apoplast is also an important battleground for S. sclerotiorum effectors. A recent study reported that S. sclerotiorum secretes a “rescuer” protein, SsPINE1, that binds to the PGIPs of the host to “rescue” the PGs, leading to successful infection35. These reports strongly suggest that the secretory proteins of S. sclerotiorum play crucial roles in the infection process. However, the strategies by which S. sclerotiorum secretory proteins overcome plant resistance remain largely unexplored, making it essential to investigate their functions to further understand the pathogenesis of S. sclerotiorum.
Hypersensitive induced reaction (HIR) proteins belong to the SPFH (stomatin/prohibitin/flotillin/HfK/C) superfamily, a diverse family of membrane proteins anchored in membrane micro-regions. These proteins contain a highly conserved SPFH structural domain and are involved in regulating plant growth and responses to biotic and abiotic stresses36,37,38. HIR genes are up-regulated upon attack by pathogens, including bacteria, fungi, and viruses39,40,41,42, and the accumulation of HIR proteins activates the host hypersensitive response (HR)39,43,44. In Nicotiana benthamiana, NbHIR3 can promote plant-based resistance through EDS1 and salicylic acid-dependent pathways42. Arabidopsis has four HIR genes (AtHIR1-4), with all AtHIR proteins enriched in membrane micro-regions of the plasma membrane, mediating resistance to Pseudomonas syringae pv. tomato DC300040. It has been reported that AtHIR1 may recruit plant immune-associated proteins such as H+-ATPase 2 (AHA2) at the plasma membrane to form a large complex in response to pathogen sensing45. Overexpression of HIR1 in N. benthamiana can trigger cell death, and this ability is associated with the self-interaction of HIR1 to form homo-oligomers46. Additionally, HIRs also positively regulate resistance to pathogens in wheat and rice41,42. These findings suggest that HIR proteins are important immunoregulatory proteins in plants, but the mechanisms by which they regulate plant disease resistance are not yet clear.
In this study, we identified a secreted protein important for S. sclerotiorum virulence and related to inhibition of early immunity in A. thaliana, named SsPEIE1 (Sclerotinia sclerotiorum Plant Early Immunosuppressive Effector 1). Through phenotypic analysis of S. sclerotiorum mutants, transgenic and mutant Arabidopsis, we found that SsPEIE1 is essential for the full virulence of S. sclerotiorum and that HIRs are virulence targets of SsPEIE1. We also clarified the important role of AtHIR4 oligomerization in resistance to S. sclerotiorum. SsPEIE1 competitively interacts with AtHIR4 to impair its oligomer formation, inhibiting plant disease resistance. This study contributes to our understanding of the virulence mechanisms and plant immune signaling networks in S. sclerotiorum, potentially aiding in the development of solutions to plant diseases caused by this devastating pathogen.
Results
Secretory protein SsPEIE1 plays an essential role in full virulence of S. sclerotiorum
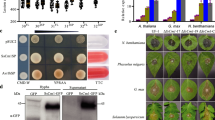
SsPEIE1 was identified from the transcriptome analysis as being substantially up-regulated during S. sclerotiorum infection47. Phylogenetic analysis reveals that homologs of SsPEIE1 are widespread among necrotrophic pathogenic fungi, including several significant plant pathogens (Fig. 1a), but there is no known functional domain within SsPEIE1. SsPEIE1 (https://www.broadinstitute.org/fungal-genome-initiative/sclerotinia-sclerotiorum-genome-project) encodes a secreted protein with 152 amino acids, and its N-terminal contains a predicted signal peptide (18-aa). Meanwhile, SsPEIE1 possesses four conserved cysteine residues (C38, C45, C64, and C86) (Fig. 1b and Supplementary Fig. 2a).
a Phylogenetic relationship of SsPEIE1 and its homologs from other fungi determined with the maximum-likelihood algorithm. Branch lengths are proportional to the average probability of change for characters on that branch. The phylogeny was constructed with Mega 6.0, using the neighbor-joining method. b SsPEIE1 is a secreted protein containing 152 amino acids including a signal peptide (SP) and four cysteine residues. c The invertase activity in TTC solution. TTC encountered sucrose breakdown products to produce triphenylformazan, which showed a red reaction to confirm that the functional signal peptide enables secretion of the sucrose-converting enzyme. d The secretory function of the SsPEIE1 signal peptide was verified by a yeast secretion trap screen assay. e Relative levels of SsPEIE1 transcript accumulation were determined by RT-qPCR using total RNA extracted from Arabidopsis plants inoculated with S. sclerotiorum in an infection time course (gray columns) or from fungal culture grown on PDA plates at 20 °C (black columns). Levels of β-tubulin transcripts of S. sclerotiorum were used to normalize different samples (n = 3 biologically repetitions). f, g Growth rates and colony morphology of wild-type (WT), ΔSsPEIE1-1, ΔSsPEIE1-2, ΔSsPEIE1-1-C1, and ΔSsPEIE1-1-C2 strains grown at 20 °C for 15 days on PDA, n = 3 biologically repetitions. h Virulence assay of S. sclerotiorum WT, ΔSsPEIE1 mutants ΔSsPEIE1-1 and ΔSsPEIE1-2, and complementary strains ΔSsPEIE1-1-C1 and ΔSsPEIE1-1-C2 on Col-0, photographed at 40 hpi. i Lesion areas by the cross-over method in (h), n = 8 biologically independent samples. j Relative biomass in (h) analyzed by qPCR (n = 3 biologically repetitions). Data represent means ± SD. Different letters on the same graph indicate statistical significance at p < 0.01 in one-way ANOVA. Source data are provided as a Source Data file.
To investigate whether the SsPEIE1 signal peptide exhibits normal secretory activity, we conducted experimental validation using the yeast secretion trap system48. YTK12 with pSUC2-Avr1bsp served as a positive control, while YTK12 and YTK12 with pSUC2 served as negative controls (Supplementary Fig. 2b). The transformed strains of YTK12 with pSUC2-SsPEIE1sp or pSUC2-SsPEIE1 were capable of degrading disaccharides to produce glucose, which results in the production of insoluble red-colored triphenylformazan from 2, 3, 5-triphenyltetrazolium chloride (TTC), allowing them to grow regularly on YPRAA media (Fig. 1c, d).
We used RT-qPCR to verify the expression of SsPEIE1 in S. sclerotiorum during infection. When inoculated onto leaves of A. thaliana (Col-0), the transcript level of SsPEIE1 rapidly increased peaking at a 68-fold increase at 6 h post-inoculation (hpi) (Fig. 1e). Notably, the transcript level of SsPEIE1 did not increase when S. sclerotiorum was grown on potato dextrose agar (PDA) medium (Fig. 1e), suggesting that the transcription of SsPEIE1 is specifically induced by host and this gene may play significant roles in S. sclerotiorum virulence. To further investigate the contribution of SsPEIE1 to S. sclerotiorum virulence, we obtained two SsPEIE1 deletion mutants (ΔSsPEIE1-1 and ΔSsPEIE1-2) through targeted gene replacement with a hygromycin resistance cassette (Supplementary Fig. 1a, b). We also generated complemented strains (ΔSsPEIE1-1-C1 and ΔSsPEIE1-1-C2) by introducing the SsPEIE1 wild-type (WT) allele into the deletion mutant using the ATMT method (Supplementary Fig. 1c).
The two ΔSsPEIE1 mutants showed similar growth rates, colony morphology, and OA production capacity to the wildtype strain 1980 (Fig. 1f, g and Supplementary Fig. 1d), indicating that SsPEIE1 is not required for normal growth and does not affect OA production. However, the ΔSsPEIE1 mutants caused significantly smaller disease lesions than the wildtype strain on Arabidopsis leaves (Fig. 1h, i). The ΔSsPEIE1 mutants also showed lower levels of relative Sclerotinia biomass compared to WT strain (Fig. 1j). Additionally, when we inoculated Brassica napus leaves with WT strain 1980 and the ΔSsPEIE1 mutants. Similarly, ΔSsPEIE1 mutants also exhibited a significant reduction in virulence (Supplementary Fig. 1e, f). Importantly, the complementation strains (ΔSsPEIE1-1-C1 and ΔSsPEIE1-1-C2) showed restored virulence, with lesion areas comparable to those of the wildtype strain (Fig. 1h–j). These results suggest that SsPEIE1 is a secreted protein required for the full virulence of S. sclerotiorum.
Transgenic Arabidopsis plants expressing SsPEIE1 have increased susceptibility to necrotrophic pathogens and suppressed immune responses
To investigate the roles of SsPEIE1 in plant-Sclerotinia interaction, stable transgenic plants constitutively expressing SsPEIE1 (35S:SsPEIE1-3×Flag) were generated in wildtype Arabidopsis (Col-0). The expression of SsPEIE1 in Arabidopsis was confirmed by western blot analysis (Supplementary Fig. 3a). SsPEIE1 transgenic lines (oxSsPEIE1−2, oxSsPEIE1−12, and oxSsPEIE1−22) exhibited smaller leaf areas compared to the WT Col-0 plants (Supplementary Fig. 3b–d).
We first investigated whether SsPEIE1 affects plant susceptibility to S. sclerotiorum infection. Compared to WT, SsPEIE1 transgenic plants exhibited significantly larger disease lesion areas and more relative Sclerotinia biomass (Fig. 2a–c). SsPEIE1 transgenic plants also showed higher levels of susceptibility to another necrotrophic fungal pathogen Botrytis cinerea (Fig. 2d–f). Furthermore, more dramatic increase in lesion area and relative Sclerotinia biomass was observed when ΔSsPEIE1 mutants were inoculated on SsPEIE1 transgenic plants compared to Col-0. The virulence of ΔSsPEIE1 mutants on SsPEIE1 transgenic plants increased to the level of WT S. sclerotiorum on WT Col-0 plants (Fig. 2g–i). These results further suggest that SsPEIE1 is an indispensable virulence factor for S. sclerotiorum and ectopic expression of SsPEIE1 in the host can adequately restore virulence of the ΔSsPEIE1 mutants to the WT level. These data indicate that SsPEIE1 may be involved in the plant-pathogen interaction by interfering with the plant immune response and reducing plant resistance.
a Virulence assay of S. sclerotiorum WT on Col-0 and SsPEIE1-3×Flag-expressing (35S:SsPEIE1) lines at 30 hpi. b Lesion areas by the cross over method in (a), n = 10 biologically repetitions. c Relative biomass in (a) analyzed by qPCR with equal area samples from infected sites used for DNA extraction (n = 3 biologically repetitions). d Virulence assay of B. cinerea wildtype B05.10 on Col-0 and SsPEIE1-3×Flag-expressing (35S:SsPEIE1) lines at 36 hpi. e Lesion area by the cross over method in (d), n = 6 biologically repetitions. f Relative biomass in (d) analyzed by qPCR with equal area samples from infected sites used for DNA extraction (n = 3 biologically repetitions). g Virulence assay of S. sclerotiorum WT and ΔSsPEIE1 mutants on Col-0 and 35S:SsPEIE1 Arabidopsis lines at 30 hpi. h Lesion area by the cross-over method in (g), n = 8 biologically repetitions. i Relative biomass in (g) analyzed by qPCR with equal area samples from infected sites used for DNA extraction (n = 3 biologically repetitions). j, k MAPK activation induced by chitin and flg22 in SsPEIE1 transgenic lines. l, m, n ROS burst in SsPEIE1 transgenic lines induced by chitin, Ssnlp20 and flg22, values represent means ± SEM (n = 12 biologically repetitions). o Induction of immune-related genes in SsPEIE1 transgenic lines. Four-week-old leaves were treated with 10 μg · mL−1 chitin for 1 h or 12 h. Data were normalized to AtUBQ5 expression in qPCR analysis (n = 3 biologically repetitions). Data represent means ± SD except for (l, m, and n). Different letters on the same graph indicate statistical significance at p < 0.01 in one-way ANOVA. Source data are provided as a Source Data file.
Consistent with these virulence phenotypes, SsPEIE1 transgenic plants exhibited a greatly reduced MAPK activation triggered by the PAMPs chitin or flg22 (Fig. 2j, k). Additionally, the ROS burst triggered by chitin, flg22, and Sclerotinia nlp20 was significantly compromised in SsPEIE1 transgenic plants (Fig. 2l–n). Furthermore, the immune marker genes FRK1, NHL10, and PR1 showed lower induction levels in SsPEIE1 transgenic plants upon chitin treatment (Fig. 2o). In summary, the plant early immune response in SsPEIE1 transgenic plants was strongly suppressed, further highlighting the importance of SsPEIE1 in the virulence of S. sclerotiorum.
SsPEIE1 interacts with Arabidopsis hypersensitive induced reaction 4 (AtHIR4) in the plasma membrane
Since SsPEIE1 is a secreted protein and inhibits the immune response in plants, we hypothesized that it might function as a fungal effector. We conducted a yeast two-hybrid (Y2H) screen of the Arabidopsis cDNA library. The cDNA screening identified a total of 106 positive clones. Sequencing these clones revealed 72 prey proteins (Supplementary Data 1). We validated the genes encoding proteins with a high number of clones and found that only six prey proteins interacted with SsPEIE1 (top six in Supplementary Data 1). Since SsPEIE1-GFP localizes to the cell membrane and cytoplasm in N. benthamiana (Fig. 3a), we focused on prey proteins with similar subcellular localization that also interacted with SsPEIE1. A. thaliana hypersensitive induced reaction 4 (AtHIR4) met both criteria (Supplementary Data 1 and Supplementary Fig. 5a).
a SsPEIE1-GFP was localized to the membrane and cytoplasm of N. benthamiana. The fluorescence of GFP was monitored at 3 days post-agroinfiltration using confocal laser scanning microscopy. N. benthamiana cells were treated with 0.5 M NaCl for 5 min to observe plasmolysis. Bars, 20 μm. b Yeast two-hybrid (Y2H) assays showing that SsPEIE1 interacted with AtHIR4. LW, SD–Leu/–Trp. LWHU+AbA+X-gal, SD–Ade/–His/–Leu/–Trp containing 125 ng · mL−1 Aureobasidin A (AbA) and 40 μg · mL−1 X-α-Gal. c SsPEIE1 and AtHIR4 showed strong interaction in the split-LUC and reverse direction split-LUC assay. SsPEIE1-nLUC and AtHIR4-cLUC or SsPEIE1-cLUC and AtHIR4-nLUC were co-expressed in N. benthamiana leaves and luciferase activity was assayed 2 days later. d Co-immunoprecipitation (Co-IP) and reverse direction Co-IP assays showed that SsPEIE1 was physically associated with AtHIR4. AtHIR4-FLAG and SsPEIE1-GFP or AtHIR4-GFP and SsPEIE1-FLAG fusion proteins were co-expressed in N. benthamiana leaves. Immunoprecipitation with anti-GFP agarose (GFP IP) was performed, and AtHIR4 was detected in the post-immunoprecipitation product with an anti-Flag antibody. The experiments were performed three times independently, and similar results were obtained. The red dots indicate the expected size of the associated proteins, respectively. e Subcellular localization of SsPEIE1 and AtHIR4 in N. benthamiana epidermal cells showed that both SsPEIE1-mCherry and AtHIR4-GFP were localized at the cell membrane. Source data are provided as a Source Data file.
Targeted Y2H assays revealed a well-defined interaction between AtHIR4 and SsPEIE1 in yeast (Fig. 3b). We also performed a split-luciferase (split-LUC) assay to investigate the association of AtHIR4 with SsPEIE1. Results indicated that AtHIR4 fused with the C-terminal LUC produced a strong LUC signal when co-expressed with SsPEIE1-nLUC in N. benthamiana leaves, with similar results observed when the positions of AtHIR4 and SsPEIE1 were switched (Fig. 3c). These results were further confirmed by Co-IP assay in N. benthamiana leaves showing that GFP or FLAG-tagged SsPEIE1 could co-immunoprecipitate with FLAG or GFP-tagged AtHIR4 (Fig. 3d). Co-expression of SsPEIE1 and AtHIR4 in Arabidopsis protoplasts yielded similar results to those obtained from Co-IP experiments in N. benthamiana (Supplementary Fig. 5b). When SsPEIE1-mCherry and AtHIR4-GFP were co-expressed in N. benthamiana leaves, the mCherry and GFP signals overlapped perfectly, indicating that SsPEIE1 and AtHIR4 co-localize in the plasma membrane (Fig. 3e). These data demonstrate that SsPEIE1 physically interacts with AtHIR4 in plants.
AtHIR2 and AtHIR4 play essential roles in resistance to necrotrophic fungi
Previous research has shown that HIR proteins are located on the cell membrane and play a positive regulatory role in tomato resistance to Tomato leaf curl Yunnan virus (TLCYnV)46. To explore the role of AtHIR4 in regulating plant immune responses to S. sclerotiorum, we obtained a T-DNA insertion mutant Arabidopsis line, hir4, from Arashare and bred it to obtain a purified mutant (Supplementary Fig. 3e). Loss of function of AtHIR4 in Arabidopsis did not affect on plant morphology or growth (Supplementary Fig. 3f). However, the hir4 mutant exhibited significantly increased susceptibility to S. sclerotiorum and B. cinerea (Fig. 4a, d), with lesion areas and relative fungal biomass increased by ~50% (Fig. 4b, c, e, f), similar to the SsPEIE1 transgenic plants. Both the hir4 mutant and SsPEIE1 transgenic plants also showed impaired chitin-induced expression of early immune marker genes (Supplementary Fig. 6a). Additionally, the hir4 mutant and SsPEIE1 transgenic plants exhibited defects in resistance to the foliar hemi-biotrophic bacterial pathogen Pst DC3000 (Supplementary Fig. 6b, c). These data indicate that AtHIR4 is indispensable for plant resistance to both fungal and bacterial pathogens. Notably, the virulence of ΔSsPEIE1 mutant strains on hir4 mutant plants was restored (Fig. 4g–i). These results further suggested that SsPEIE1 inhibits the plant immune response by interacting with AtHIR4.
a Virulence assay of S. sclerotiorum WT on Col-0, hir4 mutant and oxSsPEIE1 lines at 30 hpi. b Lesion areas by the cross-over method in (a), n = 8 biologically independent samples. c Relative biomass in (a) analyzed by qPCR (n = 3 biologically repetitions). d Virulence assay of B. cinerea WT on Col-0, hir4 and oxSsPEIE1 lines at 36 hpi. e Lesion area by the cross-over method in (d), n = 12 biologically independent samples. f Relative biomass in (d) analyzed by qPCR (n = 3 biologically repetitions). g Virulence assay of S. sclerotiorum WT and ΔSsPEIE1 mutants on Col-0 and hir4 mutant at 36 hpi. h Lesion area by the cross over method in (g), n = 10 biologically independent samples. i Relative biomass in (g) analyzed by qPCR (n = 3 biologically repetitions). j Expression of four AtHIRs in A. thaliana at different time points after S. sclerotiorum inoculation (hyphal fragment spray) using qPCR (n = 3 biologically repetitions). AtUBQ5 was used as the reference. k Virulence assay of S. sclerotiorum WT on Col-0, hir2, hir4 and hir24 mutant at 30 hpi. l Lesion areas by the cross-over method (n ≥ 13 biologically repetitions). m Relative biomass in k analyzed by qPCR (n = 3 biologically repetitions). n Virulence assay of S. sclerotiorum WT and ΔSsPEIE1 mutants on Col-0 and hir24 mutant at 36 hpi. o Lesion area by the cross-over method (n = 12 biologically repetitions). p Relative biomass in (n) analyzed by qPCR (n = 3 biologically repetitions). q MAPK activation induced by chitin in Col-0 and hir2, hir4 and hir24 mutants. r ROS burst induced by chitin in Col-0 and hir2, hir4 and hir24 mutant, values represent means ± SEM (n = 12 biological replicates). Data represent means ± SD except for (r). Different letters on the same graph indicate statistical significance at p < 0.01 in one-way ANOVA. Source data are provided as a Source Data file.
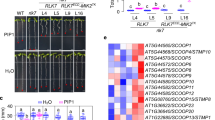
In addition to AtHIR4, A. thaliana contains three other AtHIR proteins: AtHIR1, AtHIR2, and AtHIR3. To determine whether SsPEIE1 interacts with any of the other HIRs, we performed Y2H and split luciferase assays. The results showed that SsPEIE1 also associates with AtHIR1, AtHIR2, and AtHIR3 (Supplementary Fig. 7a, b). The expression levels of the HIRs are rapidly induced by biotic stresses40,46. We examined the transcription patterns of AtHIR genes in Arabidopsis during S. sclerotiorum infection. The results indicated that the expression levels of AtHIRs significantly increased at the early stages of S. sclerotiorum infection (1 hpi), followed by a gradual decrease in AtHIR1 and AtHIR3 expression. In contrast, AtHIR2 and AtHIR4 expression levels decreased at 3 hpi and then significantly re-increased at 12 hpi (Fig. 4j). These results suggest that AtHIR genes are significantly induced in the early stages of S. sclerotiorum infection, with AtHIR2 and AtHIR4 possibly involved in the later immune response to resist S. sclerotiorum invasion in Arabidopsis.
To further clarify the biological significance of AtHIR2 and AtHIR4 in plant immunity, we edited AtHIR2 in hir4 mutant Arabidopsis to obtain the hir2/hir4 double mutant Arabidopsis (hir24) (Supplementary Fig. 3f–h). We then separately challenged hir2, hir4, and hir24 mutants with S. sclerotiorum. The hir2, hir4, and hir24 mutants exhibited faster disease development and higher biomass of S. sclerotiorum than the WT plants Col-0 (Fig. 4k–m), highlighting the important roles of AtHIR2 and AtHIR4 in regulating resistance to S. sclerotiorum. Additionally, the virulence of the ΔSsPEIE1 mutant strains on hir24 mutant plants was restored to the level of the WT strain (Fig. 4n–p). Consistent with the phenotypes of SsPEIE1 transgenic plants, the hir2, hir4, and hir24 mutants also exhibited impaired chitin-triggered MAPK activation and ROS burst, especially in the hir24 double mutant Arabidopsis (Fig. 4q, r). Furthermore, chitin-induced up-regulation of early immune marker genes was significantly reduced in hir2 or hir4 mutant lines (Supplementary Fig. 6d). These results support that both AtHIR2 and AtHIR4 are important for early plant immune responses and further clarify that the plant susceptibility induced by SsPEIE1 is achieved through its targeting of AtHIR2 and AtHIR4.
SsPEIE1 competitively binds to AtHIR4 and disrupts its oligomerization capacity and oligomerization-mediated disease resistance
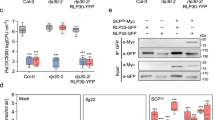
HIR proteins are indispensable for plant immunity, and recent studies have shown that interfering with the homo-oligomerization of HIR proteins significantly affects both HIR-mediated HR and disease resistance46,49. We found that AtHIR4 can form homo-oligomers in the Co-IP experiments, but this homo-oligomer formation is obviously reduced in the presence of SsPEIE1 (Fig. 3d). The same phenomenon was also observed when AtHIR4 and SsPEIE1 were co-expressed in N. benthamiana (Fig. 5a). These observations led us to hypothesize that AtHIR4 undergoes homo-oligomerization and that SsPEIE1 can interact with AtHIR4 to impair this process. To further confirm this hypothesis, we performed both in vivo and in vitro validation experiments.
a SsPEIE1 inhibits homo-oligomerization of AtHIR4. AtHIR4-Flag and SsPEIE1-GFP fusion proteins were co-expressed in N. benthamiana leaves. b Yeast tree-hybrid (Y3H) assay to confirm that SsPEIE1 competitively binds to AtHIR4. c In vitro traction assays demonstrated that SsPEIE1 competitively bind to AtHIR4 and that the AtHIR4 self-interaction weakened with increasing SsPEIE1 protein. AtHIR4-His or GST-AtHIR4, MBP and MBP-SsPEIE1 fusion proteins were dissolved in PBS (PH = 7.4), MBP and MBP-SsPEIE1 with different concentration gradients were incubated with AtHIR4-His and GST-AtHIR4 mixed proteins and immunoprecipitated with Glutathione Resin. d Co-immunoprecipitation (Co-IP) assays show that SsPEIE1 physically binds to AtHIRs in vivo to block the self-interaction of AtHIRs. AtHIRs-Flag, AtHIRs-Myc, and SsPEIE1-GFP fusion proteins were co-expressed in N. benthamiana leaves. e Co-immunoprecipitation (Co-IP) assay showed that the self-interaction of AtHIR4v157a was significantly weakened. f Co-immunoprecipitation (Co-IP) experiments showed that SsPEIE1 interacts with AtHIR4v157a. g Symptoms on N. benthamiana leaves transiently expressing different proteins 36 h after inoculation with wildtype S. sclerotiorum. h Western blot analysis for expression of AtHIR4 and AtHIR4v157a proteins in infiltrated N. benthamiana leaves. i Lesion area by the cross over method in (g), n = 6 biologically independent samples. j Relative biomass in (g) analyzed by qPCR with equal area samples from infected sites used for DNA extraction (n = 3 biologically repetitions). Data represent means ± SD. Different letters on the same graph indicate statistical significance at p < 0.05 in one-way ANOVA. The red dots indicate the expected size of the associated proteins, respectively. Source data are provided as a Source Data file.
First, the yeast three-hybrid (Y3H) assay showed that SsPEIE1 expression was repressed in the presence of methionine (Met), allowing yeast strains to grow on SD/-Leu/-Trp/-His medium, suggesting that AtHIR4 can self-interact. However, in the absence of Met, SsPEIE1 transcription was activated, and strains expressing SsPEIE1 failed to grow normally on SD/-Leu/-Trp/-His-Met medium, indicating that SsPEIE1 significantly inhibits AtHIR4 self-interaction (Fig. 5b). To determine whether SsPEIE1 directly affects AtHIR4 self-interactions, we conducted in vitro pull-down assays using MBP, MBP-SsPEIE1, GST-AtHIR4, and AtHIR4-His fusion recombinant proteins. MBP-SsPEIE1 was pulled down by GST-AtHIR4, and as the amount of MBP-SsPEIE1 increased, the amount of AtHIR4-His pulled down by GST-AtHIR4 gradually reduced. This not only indicated a direct interaction between AtHIR4 and SsPEIE1 in vitro but also confirmed that SsPEIE1 inhibits the self-association of AtHIR4 in a dose-dependent manner (Fig. 5c). Additionally, we performed Co-IP assays where AtHIRs-Flag, AtHIRs-Myc, and SsPEIE1-GFP or GFP were co-expressed in N. benthamiana leaves. The results showed that AtHIRs-Myc could be strongly immunoprecipitated with AtHIRs-Flag, confirming that AtHIR4 can self-interact to form homo-oligomers. However, the interaction between AtHIRs-Flag and AtHIRs-Myc was significantly weakened in the presence of SsPEIE1 (Fig. 5d). These findings indicate that SsPEIE1 competitively binds to AtHIR4, impairing its self-interaction.
To further clarify the role of AtHIR4 oligomerization in plant resistance to S. sclerotiorum and the biological significance of SsPEIE1 impairing AtHIR4 self-interaction, we predicted the key amino acid site for AtHIR4 homo-oligomer formation using Alphafold 2. Mutating the valine at position 157 to glycine (AtHIR4v157a) significantly reduced its oligomer formation ability (Supplementary Fig. 8b and Supplementary Fig. 9b, c). AtHIR4v157a exhibited reduced self-association compared to wildtype AtHIR4 in Co-IP experiments (Fig. 5e). Through Y2H assays, we confirmed that the valine at position 157 of AtHIR4, as well as homologous valines in other HIR family members, are essential for their self-interactions (Supplementary Fig. 9d). Additionally, the interaction between SsPEIE1 and AtHIR4v157a was weakened (Fig. 5f).
To elucidate the effect of AtHIR4 oligomerization on plant disease resistance, GFP, SsPEIE1, AtHIR4v157a, and AtHIR4 were expressed in N. benthamiana leaves and inoculated with S. sclerotiorum 36 h after infiltration (Fig. 5h and Supplementary Fig. 8b). The results showed that SsPEIE1 expression made N. benthamiana more susceptible to S. sclerotiorum, consistent with the inoculation of SsPEIE1 transgenic Arabidopsis (Fig. 2a). Expression of AtHIR4 significantly increased the resistance of N. benthamiana to S. sclerotiorum, but AtHIR4v157a lost the ability to enhance the plant resistance (Fig. 5g). The lesion area on AtHIR4v157a expressing leaves did not differ significantly from that on GFP-expressing leaves (Fig. 5i). The relative Sclerotinia biomass also showed a more consistent trend (Fig. 5j). These results illustrate that AtHIR4 oligomer formation is crucial for plant resistance to S. sclerotiorum, while SsPEIE1 promotes plant susceptibility by interacting with AtHIR4 to inhibit its homo-oligomer formation.
Constitutive over-expression of AtHIR4 enhances plant resistance
AtHIR4 is indispensable for plant immunity, and its absence leads to increased susceptibility to S. sclerotiorum. We further investigated the active role of AtHIR4 in plant disease resistance as a potential method for developing disease-resistant plants. To rapidly assess the function of AtHIR4, we generated transgenic Arabidopsis plants constitutively overexpressing AtHIR4 (Supplementary Fig. 10a). The 35S:AtHIR4-3×Flag Arabidopsis plants exhibited slightly smaller aboveground morphology and greener leaves compared with wildtype Col-0 at 4 weeks of growth (Supplementary Fig. 10b). At the same time, AtHIR4 overexpression significantly enhanced plant resistance to both S. sclerotiorum and B. cinerea (Fig. 6a–f). We also examined the intensity of the early immune response by treating AtHIR4-overexpressing Arabidopsis with chitin. The results showed that AtHIR4-overexpressing plants exhibited stronger ROS burst and activation of MAPKs compared to WT plants (Fig. 6g, h).
a Virulence assay of S. sclerotiorum WT on Col-0 and AtHIR4 overexpression transgenic Arabidopsis lines at 40 hpi. b Lesion area by the cross-over method in (a), n = 10 biologically independent samples. c Relative biomass in (a) analyzed by RT-qPCR with equal area samples from infected sites used for DNA extraction (n = 3 biologically repetitions). d Virulence assay of B. cinerea WT on Col-0 and AtHIR4 overexpression transgenic Arabidopsis lines at 48 hpi. e Lesion areas by the cross-over method in (d), n = 8 biologically independent samples. f Relative biomass in (d) analyzed by RT-qPCR with equal area samples from infected sites used for DNA extraction (n = 3 biologically repetitions). g chitin-induced MAPK activation was significantly enhanced in AtHIR4 overexpression lines. h chitin-induced ROS burst was increased in AtHIR4 overexpression lines, values represent means ± SE (n = 12 biologically repetitions). i Virulence assay of S. sclerotiorum WT on AtHIR4 transgenic rapeseed plants, empty vector transgenic rapeseed plants, and WT rapeseed plants (Y127) at 48 hpi. j Lesion areas by the cross-over method in (I) (n ≥ 10 biologically repetitions). k Relative biomass in (I) analyzed by qPCR with equal area samples from infected sites used for DNA extraction (n = 3 biologically repetitions). l Virulence assay of B. cinerea WT on AtHIR4 transgenic rapeseed plants, empty vector transgenic rapeseed plants, and WT rapeseed plants (Y127) at 48 hpi. m Lesion area by the cross-over method in (l), n = 6 biologically independent samples. n Relative biomass in (l) analyzed by qPCR with equal area samples from infected sites used for DNA extraction (n = 3 biologically repetitions). Data represent means ± SD except for (h). Different letters on the same graph indicate statistical significance at p < 0.01 in one-way ANOVA. Source data are provided as a Source Data file.
More significantly, we also created transgenic rapeseed plants expressing AtHIR4 by infecting hypocotyls with A. tumefaciens50 (Supplementary Fig. 10c). The growth phenotype of transgenic rapeseed was not noticeably different from that of WT plants (XiaoYun, Y127)51 and empty vector plants (Supplementary Fig. 10d and Supplementary Table 2). Consistent with AtHIR4-overexpressing Arabidopsis plants, transgenic rapeseed plants expressing AtHIR4 showed marked increase in resistance to S. sclerotiorum and B. cinerea (Fig. 6i–n). These results further indicate that AtHIR4 does play an important role in plant resistance and that its function is conserved among different plant species.
Discussion
As a necrotrophic phytopathogen, S. sclerotiorum possesses powerful weapons, including a large array of CWDEs and OA secretion, which contribute to its wide host range and strong virulence. While the mechanisms underlying plant resistance to biotrophic pathogens have been extensively studied, much less is known about the resistance mechanisms against necrotrophs52. Recent studies have predicted that numerous secreted proteins are involved in S. sclerotiorum infection5,53,54, and are crucial for its virulence. Some secreted proteins of S. sclerotiorum have been experimentally identified and characterized, with most reported as elicitors associated with cell death55,56,57. However, only a few effectors have been studied in depth. In this study, we demonstrated that the plant membrane-localized SsPEIE1, a core effector of S. sclerotiorum, targets multiple host HIR factors to suppress immunity. Notably, SsPEIE1 disrupts the integrity of the plant immune system by impairing the oligomerization of AtHIR4, leading to compromised host resistance (Fig. 5). Specifically, this fungal effector weakens critical immune responses of the host (Fig. 2). We found that the virulence of ΔSsPEIE1 mutants was significantly reduced but restored in SsPEIE1-complemented strains, confirming that SsPEIE1 is essential for the full virulence of S. sclerotiorum (Fig. 1h). Furthermore, the expression of SsPEIE1 in Arabidopsis restored the lost virulence of ΔSsPEIE1 mutants, further substantiating the key role of SsPEIE1 in manipulating host defense and facilitating pathogen infection. Additionally, a homologous protein of SsPEIE1 is present in many necrotrophic fungal pathogens, including B. cinerea. Importantly, our inoculation results revealed that SsPEIE1 transgenic Arabidopsis were significantly more susceptible to B. cinerea (Fig. 2d–f) than the WT Col-0, indicating that PEIE1 is a conserved effector in necrotrophic pathogenic fungi.
Previous research on the secreted proteins of S. sclerotiorum has largely focused on their role in inducing plant cell death, which can facilitate S. sclerotiorum colonization16,32,57. However, early plant immunity is also crucial for S. sclerotiorum pathogenesis, with ROS bursts in plants negatively affecting early invading S. sclerotiorum58,59. We discovered that SsPEIE1 interacts with AtHIR4 in plants, significantly inhibiting early immune responses to chitin and flg22 in Arabidopsis expressing SsPEIE1 (Figs. 2j–n and 3). This provides direct evidence that the S. sclerotiorum effector inhibits early immunity in plants. It was proposed that S. sclerotiorum may have a transient biotrophic phase during infection and secrete effectors to suppress plant immunity60. Indeed, S. sclerotiorum likely controls the onset of plant cell death at specific infection stages, thereby negatively impacting plant immunity beforehand61. Notably, hir4 mutants exhibited similar immunosuppression and susceptibility to necrotrophic fungal pathogens as SsPEIE1 transgenic Arabidopsis (Fig. 4a–f and Supplementary Fig. 6a), and the virulence of ΔSsPEIE1 mutants was obviously restored on hir4 and hir24 mutant Arabidopsis plants (Fig. 4g, h, i, n, o, p). Additionally, hir4 mutant was less responsive to flg22 and more susceptible to Pst DC3000, consistent with the phenotype of SsPEIE1 transgenic Arabidopsis (Supplementary Fig. 6b, c and Supplementary Fig. 11c, d). These findings provide strong genetic evidence that AtHIR4 is targeted by SsPEIE1 as a crucial positive regulator of plant immunity.
HIR proteins have been extensively studied as positive regulators of plant HR in Solanaceae39,62. HIR proteins in the Solanaceae family induce HR in N. benthamiana, and the expression of CaHIR1 and OsHIR1 in Arabidopsis significantly enhances resistance, although excessive immune responses can lead to Arabidopsis dwarfing43,44. In contrast, reduced HR and increased accumulation of PVX-CP protein were observed in N. benthamiana silencing HIR146. Current studies suggest that HIR proteins likely play active roles in virus and bacterial infections through the regulation of the salicylic acid pathway40,42,46. However, the role of HIR in fungal resistance remains unclear. We found that the transcripts of AtHIR2 and AtHIR4 were significantly up-regulated during S. sclerotiorum infection (Fig. 4j), suggesting that AtHIR2 and AtHIR4 might play crucial roles in resistance to S. sclerotiorum. We further evaluated the effects of AtHIR2 and AtHIR4 on Arabidopsis resistance, finding that hir2, hir4, and hir24 mutants were more susceptible to various pathogens, particularly S. sclerotiorum. (Fig. 4k–m and Supplementary Fig. 11a, b). Our study reveals that AtHIR2 and AtHIR4 in Brassicaceae are indispensable for resistance to fungal pathogens. Furthermore, overexpression of AtHIR4 in Arabidopsis resulted in high levels of resistance to necrotrophic fungi and the bacterial pathogen Pst DC3000 (Supplementary Fig. 11e, f). Concurrently, hir2, hir4, and hir24 mutants exhibited reduced early immune responses to treatments with PAMPs from different pathogen sources (Fig. 4q, r and Supplementary Fig. 11c, d), whereas Arabidopsis overexpressing AtHIR4 showed significantly enhanced early immune responses (Fig. 6g, h and Supplementary Fig. 11g, h). These biochemical changes in resistance are consistent with the genetic experiment results. Additionally, HIR1 has been reported to play a role in antiviral processes46. The related results provide ample evidence that HIR proteins are broad-spectrum and vital plant disease resistance proteins, playing critical roles in resistance to a wide range of pathogens.
To elucidate the significance of the interaction between SsPEIE1 and HIR proteins, we co-expressed SsPEIE1 and AtHIR4 both in vivo and in vitro and found that SsPEIE1 inhibits the oligomerization of AtHIR4 by competitively binding to it (Fig. 5a–d). Previous studies have shown that MdHIR can self-interact to form homo-oligomers, which is essential for its pathogen resistance capability49. Similarly, the TLCYnV C4 protein interacts with HIR1 and promotes the interaction of NbLRR1 with HIR1, thereby interfering with HIR1 oligomerization and leading to HIR1 degradation46. Our study further clarified the correlation between AtHIR4 oligomerization and resistance, demonstrating that impaired homo-oligomer formation of the AtHIR4v157a variant significantly reduces disease resistance (Fig. 5e, g, i, j). These findings elucidate the interaction mechanism between SsPEIE1 and AtHIRs, highlighting that AtHIR4 oligomerization is essential for resistance. Additionally, HIR proteins are widely present in various S. sclerotiorum hosts, and we verified that SsPEIE1 can interact with CaHIR1 and SlHIR1 in Solanaceae plants (Supplementary Fig. 7a). This suggests that SsPEIE1 likely employs a similar mechanism to inhibit the immune function of HIR proteins in other hosts, consistent with the broad host range of S. sclerotiorum.
The defense-related hormone salicylic acid (SA) plays significant roles in local and systemic resistance in plants63, activating specific immune pathways and the expression of thousands of genes, including key components of plant resistance64. Our results demonstrated that the up-regulated expression of chitin-induced SA pathway-related gene was significantly suppressed in hir2, hir4 and SsPEIE1 transgenic Arabidopsis (Fig. 2o and Supplementary Fig. 6d). Furthermore, we found that chitin-induced MAPKs phosphorylation and ROS bust were negatively affected in hir2, hir4, and hir24 mutant Arabidopsis (Fig. 4q, r), consistent with the phenotype of SsPEIE1 transgenic Arabidopsis (Fig. 2j, l). These immune responses are critical for SA pathway-mediated plant resistance and SA has been reported to play an active role in resistance to S. sclerotiorum19,33. Importantly, both Arabidopsis and rapeseed overexpressing AtHIR4 showed enhanced resistance to S. sclerotiorum and B. cinerea (Fig. 6a, d, i, l). Our study uncovered the critical roles of AtHIR2 and AtHIR4 in Arabidopsis resistance to fungal pathogens, indicating that their resistance function likely depends on the SA pathway. HIR-activated immune responses are closely related to SA, and NbHIR3-mediated HR requires EDS1 for SA response42. Elevated PR1 transcripts were observed in oxCaHIR1 and oxOsHIR1 transgenic Arabidopsis, resistance to Pst DC3000 was significantly increased39,44. Therefore, we hypothesize that overexpression of AtHIR4 enhances the immune response through the SA pathway. Our current understanding of HIRs is limited, and HIR-mediated plant disease resistance may involve complex pathways, potentially including the jasmonic acid or ethylene pathways. However, we have not yet identified key proteins interacting with HIRs in these pathways, and the mechanism of HIR-mediated disease resistance requires further exploration. Overall, our studies comprehensively illustrate the critical role of SsPEIE1 as a virulence factor of S. sclerotiorum. We found that SsPEIE1 disrupts the oligomerization of AtHIR4 by competitively binding to it, thereby inhibiting plant immunity. Consequently, SsPEIE1 creates a favorable environment within hostile plant for early infection and successful colonization by S. sclerotiorum. The enhanced resistance to S. sclerotiorum exhibited by oxAtHIR4 transgenic rapeseed also highlights the potential value of HIR proteins for further investigation. This unique interaction mode between necrotrophic pathogenic fungi and plants offers fresh insights into the dynamic relationship between necrotrophic pathogens and their host plants.
Methods
Fungal and bacterial strains, plant materials, and growth conditions
The S. sclerotiorum WT strain 1980 was cultured on PDA plates at 20 °C and stored on PDA at 4 °C. Gene deletion mutants and their complementation mutants were cultured on PDA plates amended with 100 μg · mL−1 hygromycin B or 100 μg · mL−1 G418 (Sigma-Aldrich) (Supplementary Table 2).
Pseudomonas syringae pv. tomato strain Pst DC3000 was grown in King’s B (KB) medium containing 50 mg · mL−1 rifampicin at 28 °C. Agrobacterium tumefaciens strain GV3101 carrying different constructs was incubated in LB medium with 50 mg · mL−1 rifampicin and respective antibiotics at 28 °C. All A. thaliana plants used in this study were of the Columbia-0 (Col-0) genetic background. Arabidopsis hir1, hir2, hir3, and hir4 mutants were obtained from AraShare (https://www.arashare.cn/index/). Arabidopsiss lines were grown in growth chamber soil at 22 °C, 75 mE−2 · s−1 (T5 LED tube light, 4000 K). The light/dark photoperiod was 12 h and relative humidity was 40%–60%. N. benthamiana plants were grown in jiffy pots in a growth chamber, under the same conditions as described above. Leaves of 4-week-old N. benthamiana plants were used for Agrobacterium-mediated transient expression (Supplementary Table 2).
Protein bioinformatics analysis
The SsPEIE1 protein sequence was downloaded from the NCBI GenBank database. SIGNALP 4.0 and SIGNALP 4.1 were used for signal peptide prediction65,66, DeepTMHMM (https://dtu.biolib.com/DeepTMHMM) was used for prediction of trans-membrane helices. Protein homology modeling was performed under the AlphaFold2 server with normal modeling mode67, and the PDB file produced was edited by PYMOL software. Sequence alignment was performed in the genome database using the BLASTP program to obtain homologous sequences of SsPEIE1 in other species. Multiple comparisons of amino acid sequences were generated by the DNAMAN program. Phylogenetic analysis was performed to reconstruct the phylogenetic tree using MEGA X using the maximum likelihood method.
Plasmid construction and generation of transgenic plants
The coding sequence of SsPEIE1 was amplified from S. sclerotiorum cDNA using primers containing homologous fragments of the pCNF3 vector. The PCR-amplified fragment was then cloned into CaMV 35S promoter-driven binary expression vectors pCNF3 with 3 × FLAG and pTF101 with GFP tags fused at the C terminus for assays in N. benthamiana and Agrobacterium-mediated floral dipping of Arabidopsis. The coding sequence of AtHIR genes was amplified from Arabidopsis Col-0 cDNA and cloned into the pCNF3 vector for expression in N. benthamiana and Agrobacterium-mediated floral dipping of Arabidopsis, and AtHIR genes were subcloned into a binary expression vector driven by the CaMV 35S promoter. The AtHIR4 mutant AtHIR4v157a was subcloned into the pCNF3, pCAMBIA 1300 (Luc) and pCAMBIA 2300 (2×myc) vectors for expression in N. benthamiana using homologous recombination (Vazyme, Cat. C113-02) (Supplementary Table 1).
Agrobacterium tumefaciens GV3101 carrying pCNF3-SsPEIE1-3×FLAG or pCNF3-AtHIR4-3×FLAG was cultured overnight in LB liquid medium containing 50 mg · mL−1 rifampicin and 50 mg · mL−1 kanamycin. After centrifugation at 3000 × g for 5 min, the bacterial cells were suspended in Agrobacterium infiltration buffer containing 5% sucrose and 0.04% (v/v) Silwet L-77 to an optical density (OD) 600 = 0.8. Arabidopsis buds were thoroughly immersed in the bacterial suspension, and the plants were kept moisturized for 8 h after immersion. Plants were then maintained at 22 °C and 45% relative humidity with a 16 h light/8 h dark photoperiod, conditions for seed harvesting.
Transgenic Arabidopsis was screened with 1/2 MS containing 50 mg · mL−1 kanamycin and further confirmed by immunoblotting using anti-FLAG antibody (Sigma-Aldrich F1804).
Gene knockout and complementation of S. sclerotiorum
Gene knockout mutants of the SsPEIE1 gene in S. sclerotiorum were generated using the split-marker technique68. The knockout strategy is illustrated in Supplementary Fig. 1a. Two fragments of ~1000 bp each, SsPEIE1−5′ and SsPEIE1−3′, flanking the gene were amplified from genomic DNA by PCR reactions with primers P1/P2 (both containing SalI sites) and P3/P4 (both containing XbaI sites) using KOD DNA Polymerase (TOYOBO) (Supplementary Data 2). These PCR products were cloned into the Sal I and XbaI sites in the pUCH18 vector containing the hygromycin-resistant cassette, respectively16. The pUCH18-SsPEIE1−5′ and pUCH18-SsPEIE1−3′ constructs were obtained after successful cloning (Supplementary Table 1). The fusion sequences SsPEIE1−5′ and SsPEIE1−3′ with two truncated hygromycin-resistant genes, SsPEIE1−5′-HY and YG-SsPEIE1−3′, were mass amplified with primers P1/HY and YG/P4, respectively (Supplementary Data 2). The purified SsPEIE1−5′-HY and YG-SsPEIE1−3′ DNA fragments (10 μg or more each) were mixed in equimolar amounts and used for transformation to generate SsPEIE1 knockout mutants.
To obtain complementary strains, we amplified the full length of the SsPEIE1 gene, including its promoter sequence, from WT genomic DNA to obtain a 5′-XhoI- promoter SsPEIE1-SsPEIE1-KpnI-3′ fragment, which was then cloned into the pCETNS vector containing the geneticin resistance expression cassette (Supplementary Data 2 and Supplementary Table 1). The promoter SsPEIE1-SsPEIE1 fragment fused with the geneticin resistance expression cassette was subsequently amplified from the vector using KOD FX DNA Polymerase (TOYOBO), purified and used in at least 10 μg quantities to transform the protoplasts of the SsPEIE1-knockout strain.
For the preparation and transformation of S. sclerotiorum protoplasts, mycelial balls grown in PDB for 36 h were lysed with lysing enzymes (10 mg/mL) from Trichoderma harzianum (Sigma Cat. L1412) to obtain fresh protoplasts16. Fragments of SsPEIE1−5′-HY and YG-SsPEIE1−3′ were transferred into the protoplasts of WT strains covered with RM medium containing 200 μg · mL−1 hygromycin B. Putative transformants with hygromycin B resistance were obtained after 5–7 days. After verifying the correct substitution site according to the knockout strategy schematic, the transformants were induced to produce sexual state ascospores, which were then screened for hygromycin resistance to obtain pure syngeneic knockout mutant strains. After obtaining a pure SsPEIE1 gene knockout mutant and transferring the promoterSsPEIE1-SsPEIE1-PtrpC-NptII-TtrpC fragments into its protoplast, complemented transformant strains were obtained by geneticin resistance screening of the recovered mycelium. Subsequently, genomic DNA and total RNA were extracted from the putative back-complemented transformants, and reverse transcription of the RNA was performed to obtain cDNA. The DNA and cDNA of the putative transformants were amplified by PCR to verify the presence of the SsPEIE1 gene in the positive transformants (Supplementary Data 2). Gel electrophoresis, vector cloning, and sequencing were performed using standard procedures69.
Determination of the biological characteristics of S. sclerotiorum transformants
Transformant strains of S. sclerotiorum (ΔSsPEIE1 mutant and complementary strains) were characterized for growth rate, colony morphology, acid production capacity, and virulence. The WT, knockout, and complementary strains were inoculated in the center of PDA plates for 3 days at 20 °C to measure the growth rate and were incubated continuously for 14 days to record their colony morphology by photograph. To assay the ability of OA production, the WT strain, deletion mutants, and back-complemented strains were inoculated into the center of PDA plates containing 0.005% (w/v) bromophenol blue dye and grown at 20 °C for 36 h. Their acid production ability was qualitatively characterized by visual observation.
Fungal and bacterial inoculation assay
For the fungal inoculation assay, agar discs (2 mm in diameter) were punched from the actively growing edge of fungal 2 × SY plates (SY medium: 0.5% (w/v) sucrose and yeast extract, 1% (w/v) agar and inoculated onto the leaves of 4–5-week-old Arabidopsis plants, which were then incubated at 22 °C. Six to twelve biological replicates per treatment, taken from six Arabidopsis plants. Fungal virulence was assessed macroscopically by measuring the long and short axes of the lesion with a caliper, and molecularly by measuring the ratio of S. sclerotiorum DNA to host plant DNA in infected leaves. Disease lesion area was calculated based on the elliptical area formula. Relative fungal pathogen biomass was determined by the ratio of pathogen DNA to host plant DNA70. In each group of Arabidopsis leaves, after measuring the lesion area, equal area samples were taken from the infected sites using a 1.5-cm-diameter punch (or in the case of rapeseed leaves, with a 2.5-cm side square). The DNA of the samples was extracted and analyzed for the relative content of fungal pathogens and plant DNA by qPCR. Each treatment included three replicates (each replicate contained 2–4 diseased leaves), and each experiment was performed three times. Primers used are provided in Supplementary Data 2.
For bacterial inoculation assay, Pst DC3000 was grown overnight in KB medium supplemented with the appropriate antibiotics. After centrifugation, the bacterial cells were resuspended with 10 mM MgCl2 to a desired density. Leaves of 4-week-old Arabidopsis plants were soaked in the bacterial suspension and 2 days later the leaves were collected to detect the bacterial population. 12–24 leaves were divided into 6–12 replicates and then ground in 100 μL of 10 mM MgCl2 and serially diluted onto TSA medium containing the appropriate antibiotics. Colony-forming units (CFUs) were counted after 3 days of incubation at 28 °C.
ROS production analysis and MAPK activation assay
The third or fourth pair of true leaves from 4-week-old soil-grown Arabidopsis plants were cut into leaf discs (5 mm in diameter) and further cut into strips. The leaf strips were floated in 96-well plates with 100 µL ddH2O and shaken gently overnight to eliminate the wounding effect. For the assay, ddH2O was replaced with 100 µL reaction solution containing 50 µM L-012 (Wako CAS:143556-24-5), 10 µg · mL−1 horseradish peroxidase (Sigma Cat. P6782), and appropriate elicitors (10 μg · mL−1 chitin, 1 μM Ssnlp20 or 100 nM flg22). Measurements were taken immediately after the addition of the reaction solution using a Multimode Reader Platform (Tecan Austria GmbH, SPARK 10 M), with ROS values representing the relative light units of different plants.
Arabidopsis seedlings grown on 1/2 MS plates for 10 days were transferred to 1 mL sterilized ddH2O, allowed to recover overnight, and then treated with the indicated concentrations of flg22 or chitin for 0, 5, 15, and 30 min. Total protein from the samples was extracted using protein extraction buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, 10% glycerol, and 1% Triton X-100), and the samples were incubated at 95 °C for 10 min. The supernatant was collected after centrifugation at 10,000 × g for 2 min, and the protein samples containing 1 × SDS buffer were loaded onto 10% (v/v) SDS-PAGE gels and immunoblotted with anti-PERK1/2 antibody to detect pMPK3, pMPK4, and pMPK6 (CST Cat. 9101S).
Secretion trap screen assay
In this study, the predicted signal peptide fragment of the SsPEIE1 and SsPEIE1sp gene was fused to the N-terminus of the secretion-defective invertase gene (suc2) in the vector pSUC2 and then transformed into the yeast strain YTK12. Candidate yeast transformants were screened and cultured on medium lacking tryptophan (CMD-W) and YPRAA (10 g · L−1 yeast extract, 20 g · L−1 peptone, 20 g · L−1 raffinose, 2 mg · L−1 antimycin A, and 2% agar) medium, and strains with secretion activity were able to grow on YPRAA. TTC was used to assay the secretion sucrase activity of the candidate yeast transformants. The candidate yeast transformants were incubated in 10% sucrose solution at 30 °C for 35 min, then the supernatant was centrifuged and the final concentration of 0.1% TTC reagent was added and left at room temperature for 5 min to observe the color change in the test tube. A positive reaction changed from colorless to dark red, with the Avr1bsp transformant, YTK12-pUSC2, and YTK12 strains used as positive and negative controls, respectively.
Yeast two-hybrid and three-hybrid assays
For the Y2H assay, the coding sequences of the genes to be tested for interaction (without signal peptides) were PCR amplified and cloned into pGBKT7 and pGADT7 to generate bait and prey vectors, respectively, based on the Matchmaker Gold Yeast Two-Hybrid System for GAL4. The bait and prey plasmids were co-transformed into yeast strain Y2H Gold according to the manufacturer’s instructions. Transformed yeast cells were grown on synthetic dropout (SD)/-Trp-Leu plates for 3–4 days and single-colony cells were transferred to 2 mL of liquid SD/-Trp-Leu medium and cultured for 24 h. The cells were collected by centrifugation, adjusted to a concentration of 106 cells · mL−1 with sterile water, and 2 μL of yeast suspension was assayed for growth on SD/-Trp-Leu-His-Ade plates containing 5-bromo-4-chloro-3-indolyl α-D-galactopyranoside (X-α-gal).
For the Y3H assay, AtHIR4 and SsPEIE1 were cloned into the pBridge plasmid to obtain pBridge-AtHIR4-SsPEIE1(-Met)-BD, and AtHIR4 was cloned into pGADT7 to obtain AtHIR4-AD (Supplementary Table 1). Co-expression of pBridge-AtHIR4-SsPEIE1(-Met)-BD with AtHIR4-AD was performed in yeast strain Y2H Gold, with pBridge-AtHIR4-SsPEIE1(-Met)-BD and pGADT7 as a negative control, and pBridge-AtHIR4-MCS(-Met)-BD and AtHIR4-AD as a positive control. Yeast growth was analyzed on SD/-Leu/-Trp/-His medium with or without methionine (Met). The pBridge plasmid contains two promoters: the yeast constitutive expression promoter ADH1 and the methionine deficiency-inducible promoter MET25. In the absence of methionine, the yeast strain containing the pBridge-AtHIR4-SsPEIE1(-Met)-BD plasmid is induced to express SsPEIE1, while SsPEIE1 expression is repressed in the presence of methionine. Based on these principles, the effect of SsPEIE1 on AtHIR4 self-interaction was further clarified.
Co-IP assays
For co-IP in N. benthamiana and Arabidopsis protoplasts, the cDNAs sequences of SsPEIE1 and AtHIR4 were constructed into binary expression vectors and transferred into A. tumefaciens strain GV3101 by electroporation, respectively. The proteins were co-expressed in leaves of N. benthamiana by Agrobacterium-mediated transient expression for 36 h. Alternatively, Arabidopsis protoplasts were transfected with indicated plasmids and incubated for 12 h. Subsequently, samples were then collected and lysed after vortexing in Co-IP Buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, 2 mM DTT, 10% glycerol, 0.5% Triton X-100, and protease inhibitor cocktail). Prior to co-IP, 30 μL of total protein was collected, and 10 μL of 4 × SDS Loading Buffer (200 mM Tris-HCl, pH 6.8; 40% glycerol; 0.04% bromophenol blue; 8% SDS; 5% β-mercaptoethanol; 4 mM DTT) was added and treated at 98 °C for 5 min to serve as input control, followed by the addition of anti-GFP agarose beads (Chromotek Cat. gta-20) to the total protein and immunoprecipitation at 4 °C for 3 h. The agarose beads were collected and washed with washing buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, and 0.1% Triton X-100) three times, and the supernatant was removed and 1 × SDS Loading Buffer was added (40 μL), then the beads were treated at 98 °C for 5 min before performing western blot assay. Immunoblotting analysis of immunoprecipitated and input proteins was performed with anti-GFP (1: 2000, GenScript) or anti-Flag antibodies (1: 2000, Sigma-Aldrich F1804).
In vitro pull-down assay
AtHIR4-His or GST-AtHIR4, MBP, and MBP-SsPEIE1 fusion proteins were expressed in Escherichia coli strain BL21. The obtained prokaryotically expressed proteins were dissolved in PBS (pH 7.4), MBP and MBP-SsPEIE1 at different concentration gradients were incubated with AtHIR4-His and GST-AtHIR4 mixed proteins at 4 °C for 1–2 h, followed by incubation with Glutathione Resin (GenScript, Cat. No. L00206) for 1 h at 4 °C to enrich GST-AtHIR4. The beads were collected and washed three times with PBS (5 mL). Proteins were detected with anti-His (1: 5000, abmart), anti-MBP (1: 5000, abmart) and anti-GST (1: 5000, abmart) antibodies by immunoblotting.
RNA isolation, cDNA synthesis, and RT-qPCR analysis
Plant and fungal samples were ground to a powder in liquid nitrogen, total RNA was extracted using tizol, and DNA was removed using RNase-free recombinant DNase I (Takara 2270A). To detect the expression pattern of SsPEIE1, the wildtype strain was cultured on PDA for 36 h, mycelium was collected and then transferred to new PDA plates or inoculated onto leaves of rape. Leaves of 4-week-old Col-0 plants sprayed with S. sclerotiorum mycelial suspension were collected at 0, 1, 3, 12, 24, and 48 h for nucleic acid extraction. Mycelia were harvested at 0 h, 1.5 h, 3 h, 6 h, 12 h, and 24 h and 1, 2, 3, 5, and 7 days to extract nucleic acids. The concentration of total RNA was quantified using a spectrophotometer (Thermo Fisher Scientific), and first-strand cDNA was synthesized using Easy Script One-Step gDNA Removal and cDNA Synthesis SuperMix (Transgen AE311-02). RT-PCR was performed using a CFX96 Real-Time PCR Detection System and TransStart Green qPCR SuperMix (Transgen AQ101-01). RNA samples for each real-time PCR were normalized with the β-tubulin gene Sstub1 of S. sclerotiorum and the ubiquitin 5 gene AtUBQ5 for Arabidopsis, respectively. For each gene, real-time PCR assays were repeated at least twice, with three biological replicates each time. Primers used are provided in Supplementary Data 2.
Accession numbers
Sequence data in this article can be found in the S. sclerotiorum Database or the Arabidopsis Information Resource under the following accession numbers: SsPEIE1 (Ss1G_00849), Sstub1 (Ss1G_04652), ATHIR1 (AT1G69840), ATHIR2 (AT3G01290), ATHIR3 (AT5G51570), ATHIR4 (AT5G62740), SLHIR1 (LOC101245344), CAHIR1 (LOC107862499), ATUBQ5 (AT3G62250), FRK1 (AT2G19190), NHL10 (AT2G35980), WRKY30 (AT5G24110), PR1 (AT2G14610), PHI1 (AT2G21870). The Arabidopsis mutant numbers are: hir1 (SALK_088328C), hir2 (SALK_124393C), hir3 (SALK_104547C), and hir4 (WiscDsLox489-492B7).
Statistics and reproducibility
Data from growth rate assay, inoculation assay, leaf area assay, and qPCR assay were expressed as mean ± standard deviation (SD), and data from ROS burst assay were expressed as mean ± standard error of mean (SEM). Statistical analyses were performed using one-way ANOVA, and graphs were generated by GraphPad Prism 8.0 software.
In this study, representative experimental results, such as growth rate assay, inoculation assays, ROS assay, MAPKs phosphorylation assays, fluorescence observations, and any western blot analyses, were independently repeated three times, producing similar results, with the most representative one being shown. All experimental observations were carried out without pre-selection of groups. The plants and strains used in each independent repeated experiment were from the same batch of planting or activation, and were randomly assigned to the control group and the experimental group. No other forms of randomization were relevant to this study.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All of our raw data including full uncropped images in the manuscript are provided in figshare (https://doi.org/10.6084/m9.figshare.27178839). The authors declare that the other data supporting the findings of this study are available from the corresponding author upon request.
References
Ghozlan, M. H., El-Argawy, E., Tokgöz, S., Lakshman, D. K. & Mitra, A. Plant defense against necrotrophic pathogens. Am. J. Plant Sci. 11, 2122–2138 (2020).
Liao, C. J., Hailemariam, S., Sharon, A. & Mengiste, T. Pathogenic strategies and immune mechanisms to necrotrophs: differences and similarities to biotrophs and hemibiotrophs. Curr. Opin. Plant Biol. 69, 102291 (2022).
Boland, G. J. & Hall, R. Index of plant hosts of Sclerotinia sclerotiorum. Can. J. Plant Pathol. 16, 93108 (1994).
Bolton, M. D., Thomma, B. P. H. J. & Nelson, B. D. Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 7, 1–16 (2006).
Derbyshire, M. et al. The complete genome sequence of the phytopathogenic fungus Sclerotinia sclerotiorum reveals insights into the genome architecture of broad host range pathogens. Genome Biol. Evol. 9, 593–618 (2017).
Chitrampalam, P., Figuli, P. J. & Matheron, M. E. Biocontrol of lettuce drop caused by Sclerotinia sclerotiorum and S. minor in desert agroecosystems. Plant Dis. 92, 1625–1634 (2008).
Zhou, F., Zhang, X., Li, J. & Zhu, F. Dimethachlon resistance in Sclerotinia sclerotiorum in China. Plant Dis. 98, 1221–1226 (2014).
Hou, Y. P. et al. Molecular and biological characterization of Sclerotinia sclerotiorum resistant to the anilinopyrimidine fungicide cyprodinil. Pestic. Biochem. Physiol. 146, 80–89 (2018).
Zhou, J. M. & Zhang, Y. Plant immunity: danger perception and signaling. Cell 181, 978–989 (2020).
Ngou, B. P. M., Jones, J. D. G. & Ding, P. Plant immune networks. Trends Plant Sci. 27, 255–273 (2022).
Cao, Y. et al. The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 3, e03766 (2014).
Miya, A. et al. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 104, 19613–19618 (2007).
Liu, T. et al. Chitin-induced dimerization activates a plant immune receptor. Science 336, 1160–1164 (2012).
Zhang, W. et al. Arabidopsis receptor-like protein30 and receptor-like kinase suppressor of BIR1-1/EVERSHED mediate innate immunity to necrotrophic fungi. Plant Cell 25, 4227–4241 (2013).
Song, T. et al. The N-terminus of an ustilaginoidea virens Ser-Thr-rich glycosylphosphatidylinositol-anchored protein elicits plant immunity as a MAMP. Nat. Commun. 12, 2451 (2021).
Yang, G. et al. A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytol. 217, 739–755 (2018).
Yu, X., Feng, B., He, P. & Shan, L. From chaos to harmony: responses and signaling upon microbial pattern recognition. Annu. Rev. Phytopathol. 55, 109–137 (2017).
Lewis, L. A. et al. Transcriptional dynamics driving MAMP-triggered immunity and pathogen effector-mediated immunosuppression in Arabidopsis leaves following infection with Pseudomonas syringae pv tomato DC3000. Plant Cell 27, 3038–3064 (2015).
Li, B., Meng, X., Shan, L. & He, P. Transcriptional regulation of pattern-triggered immunity in plants. Cell Host Microbe 19, 641–650 (2016).
Marciano, P., Di Lenna, P. & Magro, P. Oxalic acid, cell wall-degrading enzymes and pH in pathogenesis and their significance in the virulence of two Sclerotinia sclerotiorum isolates on sunflower. Physiol. Plant Pathol. 22, 339–345 (1983).
Cessna, S. G., Sears, V. E., Dickman, M. B. & Low, P. S. Oxalic acid, a pathogenicity factor for Sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. Plant Cell 12, 2191–2199 (2000).
Guimarães, R. L. & Stotz, H. U. Oxalate production by Sclerotinia sclerotiorum deregulates guard cells during infection. Plant Physiol. 136, 3703–3711 (2004).
Kim, K. S., Min, J. Y. & Dickman, M. B. Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol. Plant-Microbe Interact. 21, 605–612 (2008).
Williams, B., Kabbage, M., Kim, H. J., Britt, R. & Dickman, M. B. Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog. 7, e1002107 (2011).
Heller, A. & Witt-Geiges, T. Oxalic acid has an additional, detoxifying function in Sclerotinia sclerotiorum pathogenesis. PLoS ONE 8, e72292 (2013).
Kabbage, M., Williams, B. & Dickman, M. B. Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLoS Pathog. 9, e1003287 (2013).
Xu, L., Xiang, M., White, D. & Chen, W. pH dependency of sclerotial development and pathogenicity revealed by using genetically defined oxalate-minus mutants of Sclerotinia sclerotiorum. Environ. Microbiol. 17, 2896–2909 (2015).
Riou, C., Freyssinet, G. & Fevre, M. Production of cell wall-degrading enzymes by the phytopathogenic fungus Sclerotinia sclerotiorum. Appl. Environ. Microbiol. 57, 1478 (1991).
Issam, S. M. et al. A β-Glucosidase from Sclerotinia sclerotiorum biochemical characterization and use in oligosaccharide synthesis. Appl. Biochem. Biotechnol. 112, 63 (2004).
Ellouze, O. E., Loukil, S. & Marzouki, M. N. Cloning and molecular characterization of a new fungal xylanase gene from Sclerotinia sclerotiorum S2. BMB Rep. 44, 653–658 (2011).
Xiao, X. et al. Novel secretory protein Ss-Caf1 of the plant-pathogenic fungus Sclerotinia sclerotiorum is required for host penetration and normal sclerotial development. Mol. Plant-Microbe Interact. 27, 40–55 (2014).
Lyu, X. et al. A small secreted virulence-related protein is essential for the necrotrophic interactions of Sclerotinia sclerotiorum with its host plants. PLoS Pathog. 12, e1005435 (2016).
Tang, L. et al. An effector of a necrotrophic fungal pathogen targets the calcium-sensing receptor in chloroplasts to inhibit host resistance. Mol. Plant Pathol. 21, 686–701 (2020).
Yang, Y. et al. Convergent evolution of plant pattern recognition receptors sensing cysteine-rich patterns from three microbial kingdoms. Nat. Commun. 14, 3621 (2023).
Wei, W. et al. A fungal extracellular effector inactivates plant polygalacturonase-inhibiting protein. Nat. Commun. 13, 2213 (2022).
Nadimpalli, R., Yalpani, N., Johal, G. S. & Simmons, C. R. Prohibitins, stomatins, and plant disease response genes compose a protein superfamily that controls cell proliferation, ion channel regulation, and death. J. Biol. Chem. 275, 29579–29586 (2000).
Rostoks, N., Schmierer, D., Kudrna, D. & Kleinhofs, A. Barley putative hypersensitive induced reaction genes: genetic mapping, sequence analyses and differential expression in disease lesion mimic mutants. Theor. Appl. Genet. 107, 1094–1101 (2003).
Ma, C. et al. Structural insights into the membrane microdomain organization by SPFH family proteins. Cell Res. 32, 176–189 (2022).
Jung, H. W. & Hwang, B. K. The leucine-rich repeat (LRR) protein, CaLRR1, interacts with the hypersensitive induced reaction (HIR) protein, CaHIR1, and suppresses cell death induced by the CaHIR1 protein. Mol. Plant Pathol. 8, 503–514 (2007).
Qi, Y. et al. Physical association of Arabidopsis hypersensitive induced reaction proteins (HIRs) with the immune receptor RPS2. J. Biol. Chem. 286, 31297–31307 (2011).
Duan, Y. et al. Wheat hypersensitive-induced reaction genes TaHIR1 and TaHIR3 are involved in response to stripe rust fungus infection and abiotic stresses. Plant Cell Rep. 32, 273–283 (2012).
Li, S. et al. The hypersensitive induced reaction 3 (HIR3) gene contributes to plant basal resistance via an EDS1 and salicylic acid-dependent pathway. Plant J. 98, 783–797 (2019).
Jung, H. W. et al. Distinct roles of the pepper hypersensitive induced reaction protein gene CaHIR1 in disease and osmotic stress, as determined by comparative transcriptome and proteome analyses. Planta 227, 409–425 (2007).
Zhou, L. et al. Rice hypersensitive induced reaction protein 1 (OsHIR1) associates with plasma membrane and triggers hypersensitive cell death. BMC Plant Biol. 10, 290 (2010).
Lv, X. et al. Membrane microdomains and the cytoskeleton constrain AtHIR1 dynamics and facilitate the formation of an AtHIR1-associated immune complex. Plant J. 90, 3–16 (2017).
Mei, Y., Ma, Z., Wang, Y. & Zhou, X. Geminivirus C4 antagonizes the HIR1-mediated hypersensitive response by inhibiting the HIR1 self-interaction and promoting degradation of the protein. New Phytol. 225, 1311–1326 (2020).
Lyu, X. et al. Comparative genomic and transcriptional analyses of the carbohydrate-active enzymes and secretomes of phytopathogenic fungi reveal their significant roles during infection and development. Sci. Rep. 5, 15565 (2015).
Lee, S. J., Kim, B. D. & Rose, J. Identification of eukaryotic secreted and cell surface proteins using the yeast secretion trap screen. Nat. Protoc. 1, 2439 (2006).
Jing, Y. et al. The apple FERONIA receptor‐like kinase MdMRLK2 negatively regulates Valsa canker resistance by suppressing defence responses and hypersensitive reaction. Mol. Plant Pathol. 23, 1170–1186 (2022).
Dai, C. et al. An efficient Agrobacterium-mediated transformation method using hypocotyl as explants for Brassica napus. Mol. Breed. 40, 96 (2020).
Wang, P. et al. Xiaoyun, a model accession for functional genomics research in Brassica napus. Plant Commun. 5, 100727 (2024).
Spanu, P. D. & Panstruga, R. Editorial: biotrophic plant-microbe interactions. Front. Plant Sci. 8, 192 (2017).
Richardson, P. M. et al. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 7, e1002230 (2011).
Guyon, K., Balagué, C., Roby, D. & Raffaele, S. Secretome analysis reveals effector candidates associated with broad host range necrotrophy in the fungal plant pathogen Sclerotinia sclerotiorum. BMC Genom. 15, 336 (2014).
Seifbarghi, S. et al. Receptor-like kinases BAK1 and SOBIR1 are required for necrotizing activity of a novel group of Sclerotinia sclerotiorum necrosis-inducing effectors. Front. Plant Sci. 11, 1021 (2020).
Newman, T. E. et al. The broad host range pathogen Sclerotinia sclerotiorum produces multiple effector proteins that induce host cell death intracellularly. Mol. Plant Pathol. 24, 866–881 (2023).
Yang, C. et al. SsNEP2 contributes to the virulence of Sclerotinia sclerotiorum. Pathogens 11, 446 (2022).
Arfaoui, A., El Hadrami, A. & Daayf, F. Pre-treatment of soybean plants with calcium stimulates ROS responses and mitigates infection by Sclerotinia sclerotiorum. Plant Physiol. Biochem. 122, 121–128 (2018).
Beracochea, V. C. et al. Sunflower germin-like protein HaGLP1 promotes ROS accumulation and enhances protection against fungal pathogens in transgenic Arabidopsis thaliana. Plant Cell Rep. 34, 1717–1733 (2015).
Kabbage, M., Yarden, O. & Dickman, M. B. Pathogenic attributes of Sclerotinia sclerotiorum: switching from a biotrophic to necrotrophic lifestyle. Plant Sci. 233, 53–60 (2015).
Stajich, J. E., Andrew, M., Barua, R., Short, S. M. & Kohn, L. M. Evidence for a common toolbox based on necrotrophy in a fungal lineage spanning necrotrophs, biotrophs, endophytes, host generalists and specialists. PLoS ONE 7, e29943 (2012).
Choi, H. W., Kim, Y. J. & Hwang, B. K. The hypersensitive induced reaction and leucine-rich repeat proteins regulate plant cell death associated with disease and plant immunity. Mol. Plant-Microbe Interact. 24, 68–78 (2011).
Pieterse, C. M. J., Leon-Reyes, A., Van der Ent, S. & Van Wees, S. C. M. Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316 (2009).
Hillmer, R. A. et al. The highly buffered Arabidopsis immune signaling network conceals the functions of its components. PLoS Genet. 13, e1006639 (2017).
Bendtsen, J. D., Nielsen, H., Heijne, G. & Brunak, S. Improved prediction of signal peptides: SignalP 3.0. Mol. Biol. 340, 783–795 (2004).
Petersen, T. N., Brunak, S., Heijne, G. & Nielsen, H. SIGNALP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 (2011).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Catlett, N. L., Lee, B. N., Yoder, O. C. & Turgeon, B. Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genet. Rep. 50, 9–11 (2003).
Sambrook, J. E., Fritsch, E. F. M. & Maniatis, T. E. Molecular Cloning: A Laboratory Manual (Could Spring Habor, 1989).
Li, H. et al. Pathogen protein modularity enables elaborate mimicry of a host phosphatase. Cell 186, 3196–3207 (2023).
Acknowledgements
We thank Yangrong Cao of Huazhong Agricultural University for provision of pBridge vector. We also acknowledge Dengfeng Hong of Huazhong Agricultural University for providing rapeseed Xiao Yun and genetic transformation method. This research was supported by the National Nature Science Foundation of China (32172368, 32130087), the earmarked fund of China Agriculture Research System (CARS-12), and the Huazhong Agricultural University Scientific and Technological Self-innovation Foundation (2021ZKPY014), and US Department of Agriculture Agricultural Research Service Sclerotinia Initiative. The funders were not involved in study design, data collection and analysis, decision to publish, or manuscript preparation.
Author information
Authors and Affiliations
Contributions
X.L. and J.C. designed experiments, analyzed the data, and wrote the manuscript; H.Z. performed gene complement, identifying, and partial vector construction; M.Y. conducted rapeseed transformation; P.L. constructed the relevant vectors and contributed to experimental design; D.J., J.X., Y.F., B.L., X.Y., T.C. and Y.L. designed the research and provided critical feedback; W.C. contributed to revising the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Yangdou Wei, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, X., Zhao, H., Yuan, M. et al. An effector essential for virulence of necrotrophic fungi targets plant HIRs to inhibit host immunity. Nat Commun 15, 9391 (2024). https://doi.org/10.1038/s41467-024-53725-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-53725-0
This article is cited by
-
Revealing the early pathogenic mechanisms of Agroatehlia rolfsii in Arachis hypogaea through RNA-seq and identification of ArNIS1-like effectors
BMC Plant Biology (2026)
-
The elicitation of early defence responses and improvement of disease resistance in oil palm by extracellular proteins from Ganoderma boninense
European Journal of Plant Pathology (2025)
-
A Model System for Understanding Host–Pathogen Relationships Through In Vitro Host Mimicry
European Journal of Plant Pathology (2025)