Abstract
The biological relevance and dynamics of mRNA modifications have been extensively studied; however, whether rRNA modifications are dynamically regulated, and under which conditions, remains unclear. Here, we systematically characterize bacterial rRNA modifications upon exposure to diverse antibiotics using native RNA nanopore sequencing. To identify significant rRNA modification changes, we develop NanoConsensus, a novel pipeline that is robust across RNA modification types, stoichiometries and coverage, with very low false positive rates, outperforming all individual algorithms tested. We then apply NanoConsensus to characterize the rRNA modification landscape upon antibiotic exposure, finding that rRNA modification profiles are altered in the vicinity of A and P-sites of the ribosome, in an antibiotic-specific manner, possibly contributing to antibiotic resistance. Our work demonstrates that rRNA modification profiles can be rapidly altered in response to environmental exposures, and provides a robust workflow to study rRNA modification dynamics in any species, in a scalable and reproducible manner.
Similar content being viewed by others
Introduction
Increasing antibiotic resistance among pathogenic bacteria threatens healthcare and the efficacy of the majority of currently known antibiotics1. Most clinically used antibiotics inhibit bacterial growth by targeting protein synthesis2,3,4, often through direct binding to the bacterial ribosome, interfering with mRNA translation or blocking the formation of peptide bonds at the peptidyl transferase center. For example, aminoglycoside antibiotics such as kanamycin, streptomycin and neomycin, are potent and broad-spectrum antibacterials that were introduced in the clinic more than five decades ago, which interact with the 16S rRNA at the A site of the ribosome5. Unfortunately, their clinical efficacy is seriously threatened by multiple resistance mechanisms6. Currently, the most widely disseminated aminoglycoside resistance determinants are drug modification enzymes7, but 16S rRNA methyltransferases that modify the drug-binding site have recently emerged as a significant threat that can confer class-wide resistance to these drugs8,9. Thus, detailed studies of these emerging resistance mechanisms are urgently needed.
Bacterial rRNAs contain a large number of methylations that are placed by genomically encoded methyltransferases10. While these typically improve ribosome function under most conditions, when challenged with antibiotics the loss of specific modifications can confer low to moderate levels of antibiotic resistance11,12,13,14,15,16 (Supplementary Data S1). For example, bacterial strains lacking rsmA (rsmA_KO), responsible for dimethylation of adenine at position 1519 of the 16S rRNA (16S:m6,6A1519), show resistance to kasugamycin17,18, whereas bacterial strains lacking the rsmG methyltransferase (rsmG_KO), responsible for placing 7-methylguanosine in the 16S rRNA (16S:m7G527), show increased resistance to streptomycin19. To better comprehend the role that rRNA methylation dynamics plays in antibiotic resistance, and to decipher whether additional rRNA modifications might be contributing to increased antibiotic resistance, accurate methods to monitor and quantify rRNA modifications are sorely needed20,21,22.
Transcriptome-wide detection of RNA modifications has been typically achieved by coupling either antibody immunoprecipitation or chemical probing with next generation sequencing (NGS) technologies23,24,25,26,27,28,29,30,31,32,33,34,35. However, limited availability of selective antibodies and/or chemicals only allows for detection of ~5% of currently known RNA modifications21,36,37. Moreover, even when these reagents are available, these methodologies have high false positive rates38, are often not quantitative39 and are inconsistent when using different antibodies40, and can only detect one RNA modification type at a time. A promising alternative to NGS-based methods is the direct RNA nanopore sequencing (DRS) platform developed by Oxford Nanopore Technologies (ONT), which can detect diverse types of modified nucleotides in individual native RNA molecules41,42,43. In this platform, RNA molecules are translocated through the nanopores that are embedded in synthetic membranes coupled to an ammeter, causing changes in the ionic current, which are in turn used to identify the underlying nucleotide sequence using machine learning algorithms44,45,46, a process referred to as ‘base-calling’. RNA modifications can then be identified using two main approaches: (i) in the form of systematic base-calling ‘errors’47,48,49,50,51, or (ii) in the form of alterations in the current signal (i.e., altered current intensities, dwell times and/or trace)43,52,53,54,55,56,57. In recent years, a plethora of algorithms to detect RNA modifications in DRS datasets have been developed47,53,56,57,58,59; however, the overlap between predicted RNA modified sites by different algorithms is poor49,57, limiting our ability to extract meaningful biological conclusions from these datasets. Moreover, it is currently unclear how the performance of each algorithm varies depending on the RNA modification type, modification stoichiometry and sequencing depth, thus limiting the applicability of DRS for the detection of dynamically regulated RNA modifications in biological contexts45.
Here, we systematically benchmark diverse RNA modification detection softwares in DRS datasets, across diverse RNA modification types, stoichiometries and sequencing depths. We then propose a novel approach, NanoConsensus which uses as input the predictions of diverse RNA modification prediction softwares (EpiNano47, Nanopolish60, Tombo56 and Nanocompore57), re-scores them and weights them internally, and finally extracts a robust list of reproducible RNA modification sites that are differentially modified between two conditions (e.g., wild type versus knockout; antibiotic-treated versus untreated). Our results demonstrate that NanoConsensus is a robust strategy to detect multiple rRNA modification types simultaneously, outperforming all individual RNA modification softwares tested in this work, detecting RNA modification changes across diverse RNA modification types, with improved sensitivity and specificity across a wide range of modification stoichiometries and sequencing depths.
We then apply NanoConsensus to study rRNA modification dynamics on E. coli cultures from diverse genetic backgrounds (including strains lacking specific rRNA modification enzymes), which were subjected to either streptomycin (str) or (ii) kasugamycin (ksg) exposure, which bind to the A- and P-site of the ribosome, respectively (Fig. 1A). We find that upon antibiotic treatment, rRNA modification levels of a subset of sites are significantly decreased, in an antibiotic-dependent manner, and that this loss of rRNA modifications depends on the specific antibiotic employed. Notably, dysregulated rRNA modified sites are spatially located in the vicinity of the A site and the P-site of the ribosome. We show that this loss is not caused by the appearance of mutations in rRNA molecules, nor expression of alternative rRNA operons. Rather, we demonstrate that the loss of rRNA modifications is caused by the de novo appearance of a subpopulation of under-modified rRNA molecules that were not present in untreated E. coli cultures.
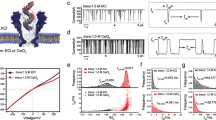
A 3D structure of the ribosome (gray) depicting the tRNA in the P-site (orange) and A-site (green). The residues surrounding the kasugamycin binding site are shown in blue, and are located near the anticodon region of the tRNA that is located at the P-site, whereas residues surrounding the streptomycin binding site are shown in pink, and are located near the anticodon region of the A-site tRNA. Surrounding residues were defined as those that have at least one of its atoms at less than 10 Å from the antibiotic. In the zoomed regions, modified nucleotides involved in antibiotic resistance are represented as brown surfaces, whereas the antibiotics are shown in black. PDB structure corresponds to 7K00. B Growth curves of E. coli WT (left) and E.coli rsmG KO (right) upon increasing concentrations of streptomycin. Data shown represents the average OD600 values of 2 independent biological replicates. Each biological replicate was calculated as the median of 3 technical replicates. Data was collected every 9 minutes, during a time course of 16 h. Source data are provided as a Source Data file. C Growth curves of E. coli WT (left) and E.coli rsmA KO (right) upon increasing kasugamycin concentrations. Data shown represents the average OD600 values of 2 independent biological replicates, and each biological replicate was calculated as the median of 3 technical replicates. Data was collected every 9 minutes, during a time course of 16 h. Source data are provided as a Source Data file. D IGV snapshots of E.coli WT and knockout strains illustrating loss of base-calling errors upon enzyme knockout (rsmG and rsmA). Positions with mismatch frequencies greater than 0.1 are colored, whereas positions with mismatch frequencies lower than 0.1 are shown in gray. E Scatterplots of the summed base-calling error frequencies (sum of insertion, deletion and mismatch frequencies) at each nucleotide position in the knockout strain, relative to WT. The rRNA modified sites that are lost upon knockout of the gene are shown in red (position 0); the neighboring positions (±4 nt) to the rRNA modification site (position 0) are shown in blue. Remaining positions are shown in gray.
Overall, our work reveals that rRNA modifications can be dynamically regulated upon antibiotic exposure, with altered rRNA modification patterns that are antibiotic-specific. Moreover, we demonstrate that NanoConsensus is a robust toolkit to study rRNA modification dynamics across varying stoichiometries and RNA modification types, with low false positive rates. To facilitate the use and applicability of NanoConsensus by future users, we integrate this pipeline into the MasterOfPores NextFlow workflow61, making the detection of differential RNA modifications DRS datasets simple, traceable and reproducible.
Results
Direct RNA nanopore sequencing can identify bacterial rRNA modifications implicated in antibiotic resistance mechanisms
Previous works have shown that bacteria have evolved an effective and elegant way of preventing drug binding to the ribosome by either adding or removing specific rRNA modifications at appropriate sites62,63. Yet, a systematic analysis of how antibiotic exposures affect the stoichiometry and dynamics of rRNA modifications is currently missing. The direct RNA nanopore sequencing (DRS) platform is well-suited to capture the dynamic changes in rRNA modified sites caused by environmental cues, such as antibiotics43,64. However, whether this is the case, and whether nanopore sequencing is sensitive enough to capture potential rRNA modification changes caused by the presence of antibiotics, remains unknown.
To address this question, we first examined whether E. coli mutant strains lacking specific rRNA methyltransferases would show increased antibiotic resistance phenotypes, as reported in the literature. To this end, we cultured E. coli strains lacking rsmG (responsible for placing 16S:m7G527) or rsmA (responsible for placing 16S:m6,6A1518,A1519) (Supplementary Data S1), and exposed them antibiotics at increasing concentrations (streptomycin or kasugamycin, for which their resistance had been reported, see Fig. 1A), and monitored their growth during 16 h. Results confirmed that rsmG and rsmA knockout strains showed increased resistance to streptomycin and kasugamycin, respectively (Fig. 1B, C), compared to wild type strains, in agreement with previous literature17,19.
Previous works using nanopore sequencing to identify RNA modifications have mainly focused their efforts on the detection of N6-methyladenosine (m6A)47,51,53,55,57,58,65 and pseudouridine (Ψ)43,66,67,68, and to a lesser extent inosine (I)69 and 2′O-methylation (Nm)43,70. However, whether DRS can detect other less frequent RNA modification types, such as those that have been previously implicated in antibiotic resistance mechanisms remains unclear. To this end, we sequenced total RNA from E. coli rsmG, rsmA and rsmF knockout strains (responsible for 16S:m7G527, 16S:m6,6A1518,A1519 and 16S:m5C1407, respectively), as well as from the parental E. coli wild type strain, using DRS (Fig. 1D, see also Fig. S1). EpiNano-Error47 was used to identify differentially modified rRNA sites by comparing the base-calling ‘errors’ of each knockout strain to those observed in the parental wild type strain, revealing that all 3 modifications examined (m7G, m6,6A and m5C), could be identified in the form of base-calling ‘errors’48,49 (Fig. 1D, E, see also Figure S1A). Indeed, we found that base-calling ‘errors’ decreased in the rsmG, rsmA and rsmF knockout strains (Fig. 1D, see also Fig. S1B), supporting that DRS can be used to identify rRNA modification types that are involved in antibiotic resistance mechanisms.
Poor reproducibility across RNA modification detection algorithms limits our ability to identify antibiotic-induced rRNA modification dynamics
Considering that m7G and m6,6A-deficient strains (rsmG and rsmA knockouts) displayed increased resistance to streptomycin and kasugamycin, respectively (Fig. 1B,C), we then wondered whether the wild type E. coli strain would dynamically modulate its rRNA modification levels of 16S:m7G527 and 16S:m6,6A1518,1519 upon antibiotic exposure. To address this question, we treated wild type E. coli cultures with either streptomycin (str) or kasugamycin (ksg) for either 1 h or 16 h, and sequenced treated and untreated samples using DRS (Supplementary Data S2). Differences in rRNA modification profiles were determined using EpiNano-Error, as well as with 3 additional softwares (Nanocompore57, Tombo56 and Nanopolish71) that employ pairwise comparison approaches to identify differential RNA modifications between two samples, and therefore, are not limited to detecting a single RNA modification type. We should note that Nanopolish does not identify RNA modifications per se; however, it can be used to ‘resquiggle’ the reads to then predict differences in current intensity at each position45. To ensure that unequal coverage along the transcript would not cause biases in the detection of differentially modified sites along the transcript and across algorithms, only full-length reads were kept for downstream analyses (see Methods and Supplementary Data S2). For every algorithm, we used its own scoring system to identify differentially modified sites (see Methods), which we refer to as ‘raw scores’.
Our results showed that each algorithm predicted many dynamic sites between the two conditions (antibiotic-treated versus untreated). However, the sites were poorly reproducible across biological replicates. Notably, we observed a poor overlap when comparing the predictions made by each algorithm (Fig. S2). In the case of Tombo, most of rRNA sites were in fact predicted as differentially modified, in agreement with previous works reporting that Tombo performance was poor due to very large proportions of false positives53,57.
Systematic benchmarking reveals that performance of nanopore-based RNA modification detection algorithms depends on modification type, sequencing coverage and stoichiometry
To investigate the reasons behind the poor reproducibility of RNA modification detection across softwares, we systematically analyzed how different features (sequencing coverage, RNA modification type and modification stoichiometry) might impact the performance of each algorithm. To this end, we analyzed DRS datasets from E. coli and S. cerevisiae wild type and mutant strains lacking distinct RNA modification types at known rRNA positions (see Supplementary Data S3).
Firstly, we examined whether different RNA modification types (Am, m5C, m7G, m6,6A, Um, Ψ) would be detected by each different software. We should note that RNA modifications appear in the form of altered base-calling ‘errors’ and/or current intensities in positions that are neighboring the modified site (i.e., positions −2,−1, +1 and/or +2 relative to the modified site)43,47,57. For this reason, we quantified the scores in the 5 nucleotides surrounding the differential modified site (5-mer), and then examined how each modification type was detected by each software, finding that the position within the 5-mer identified as ‘altered’ varied both with the RNA modification type and algorithm of choice (Fig. 2A). For example, EpiNano typically identified alterations in the base-calling signatures at the position 0 of the 5-mer, whereas current intensity-based methods, such as Tombo, Nanopolish or Nanocompore, frequently identified the alterations at the neighboring positions. Moreover, alterations in the signal were often seen at different positions of the 5-mer, even when comparing the same RNA modification type across different algorithms. For example, 2′-O-methyladenosine (Am) could be detected by all 4 softwares, but the altered signal was detected at distinct positions of the 5-mer depending on the algorithm. Altogether, these results suggest that in order to compare the predictions from individual softwares, raw scores from all positions of the 5-mer should be taken into consideration for downstream analyses, rather than comparing the nucleotide positions predicted by each software, as each software associated the modification signal to distinct nucleotide position(s).
A Dotplots of raw differential RNA modification scores along the modified 5-mer obtained by each software when detecting different RNA modification types. In the x-axis, 0 denotes the modified position. Position within the 5-mer of the ‘altered feature’ varies depending on the RNA modification type, but also depending on the software used to identify the modification changes. Data from three technical replicates from four different coverage levels is included. Box, first to last quartiles; whiskers, 1.5× interquartile range; center line, median. Source data are provided as a Source Data file. B Raw RNA modification scores along the E. coli 16S rRNA transcript for each individual software tested, obtained when comparing E. coli wild type and rsmF knockout strains. Analyses have been performed using either 100 (upper panel) or 500 randomly selected reads (lower panel). The modified site that is reported to be lost in the rsmF knockout strain (16S:m5C1407) is indicated with an asterisk (*). See also Figs. S3–S6. C Barplots of median differential modification raw scores obtained by each software in the modified 5-mer for each modification type present in the benchmarking. Raw scores are obtained by comparing the wild type and knockout strains, for each given site. Two randomly chosen rRNA modified sites that are not expected to change across any of the datasets have been included in the analyses as controls (16S:730 and 25S:2549). Error bars indicate median ± s.d. of three replicates. Source data are provided as a Source Data file. D Barplots of median raw scores in each modified 5-mer analyzed. Samples were analyzed using 1000 reads per sample, and with a diverse range of stoichiometry levels (see Methods). Error bars indicate median ± s.d. of three replicates. Source data are provided as a Source Data file.
Next, we examined how sequencing coverage affected the performance of each software. To this end, each DRS dataset (Supplementary Data S3) was downsampled into three independent subsets of 50, 100, 500 and 1500 reads (see Methods). Each wild type subset was then compared to a knockout subset of reads, and differential peaks were identified by each algorithm (Fig. 2B, see also Fig. S3–S6). Our results revealed that the identification of differentially modified sites was strongly dependent on sequencing coverage. Notably, the minimal coverage required to detect a differentially modified site varied both depending on the algorithm as well as on the modification type. For example, loss of m6,6A could be detected by Tombo when having a minimum coverage of 500 reads whereas Epinano was able to detect it with a coverage of 50 reads (Fig. 2C). Overall, our results demonstrate that coverage significantly affects the performance of all algorithms, that not all softwares efficiently detect all RNA modification types given a specific sequencing coverage, and that using 50 reads per position (which is the recommended threshold by several of the softwares) is often insufficient to detect RNA modifications, even when the site modified at high stoichiometry.
Finally, we assessed the impact of RNA modification levels (i.e., stoichiometry) in the performance of each algorithm. To this end, we artificially generated samples with decreasing modification stoichiometries (100, 75, 50, 40, 30, 20 and 10%) by mixing reads from wild type and knockout samples at different proportions, with the assumption that wild type samples are 100% modified at the site of interest, and knockout samples are 0% modified (see Methods). Subsamplings were performed in triplicate for each RNA modification type and stoichiometry level. We should note that all artificial mixes representing distinct RNA modification stoichiometries contained the same number of reads (n = 1000), to ensure that coverage would not be a confounder in the analysis. Our results revealed that stoichiometry had a major effect in the detection of RNA modifications (Fig. 2D), in agreement with previous works57,59. Notably, we observed that the effect of stoichiometry was strongly dependent on the RNA modification type. For example, Am was detected by most of softwares across all stoichiometries tested; by contrast, the majority of algorithms struggled to detect m5C, even at high stoichiometries (Fig. 2D).
NanoConsensus outperforms individual softwares at predicting RNA modifications
Our results showed that RNA modification type, stoichiometry levels and coverage impact the detection of differentially modified sites in DRS datasets (Fig. 2). Notably, our results also revealed that all softwares examined predicted a significant number of false positives (Fig. S2). Indeed, each software identified several ‘differentially modified’ sites when comparing wild type and knockout strains, even though only one RNA modified site is absent in the knockout strain, relative to the wild type (Fig. 2B, see also Figs. S3–S6).
We reasoned that the detection and replicability could be improved if predictions took into account 5-mer data instead of single nucleotide one. In addition, we reasoned that Z-score normalization would decrease the proportion of false positives identified by each software (see Methods). Indeed, we observed that Z-score rescaling significantly decreased the number of false positives, and decreased the minimum number of reads that were required to identify a site as differentially modified (Figs. S7–S11).
We then examined whether results using Z-score rescaled scores would improve the detection of differentially modified sites in E. coli cultures upon streptomycin (str) or kasugamycin (ksg) exposure (Figs. S12–S13). We found that all 4 softwares showed good overlap of predicted differential sites across biological replicates, within each software (38–69% replicability, depending on the software) (Fig. S14A). However, we still observed a poor overlap of predicted differentially modified sites across softwares (15–25%) (Fig. S14B) suggesting that a robust pipeline to detect differentially modified RNA sites from DRS datasets is greatly needed.
We reasoned that the detection could be further improved if the predictions from different algorithms would be combined in a consensual manner, as this should decrease the number of false positives while retaining the true differentially modified sites. To this end, we developed NanoConsensus (Fig. 3A), an algorithm that reports putative modified regions at per-transcript level, which takes as input the results generated by different RNA modification detection algorithms (EpiNano, Nanopolish, Tombo and Nanocompore), and generates as output a list of differentially modified sites that are robustly predicted across softwares and replicates. Briefly, NanoConsensus converts the outputs of each software into Z-scores, identifies putative modified sites by each software, and assigns a final NanoConsensus score to each position of the transcript. Because the alteration of the signal caused by a modification does not necessarily overlap across softwares (Fig. 2A, see also S7A), NanoConsensus expands each putative modified site predicted by each software into a 5-mer window, and predicts the overlap of 5-mer regions predicted by each software. Finally, all regions supported by at least two softwares and with a NanoConsensus score higher than a threshold (which is dependent on the background ‘noise’, see Methods) are reported as ‘differentially modified’ sites (Fig. 3A).
A Schematic overview of the NanoConsensus pipeline. B Per-position NanoConsensus scores along the 16 s and 18 s rRNA transcripts, for the battery of RNA modification types benchmarked in this work (m66A, Y, m5C, Am, Um and m7G), where the WT strain has been compared to a different knockout strain that lacks a specific rRNA modification (see Supplementary Data S1 for list of knockout strains used for each modification type). The known modified site that is lost is highlighted with an asterisk (*). Only one replicate is shown for each modification type, results obtained for additional replicates are shown in Fig. S15. C Barplot of NanoConsensus scores of the 5-mer regions centered in each modified site analyzed. Samples included in the analysis consist of 100% modified samples (WT), with varying read coverage levels, and are the same that were used to generate Fig. 2C. Error bars indicate median ± s.d. of three replicates. Source data are provided as a Source Data file. D Barplot of median NanoConsensus scores of the 5-mer regions centered in each modified site analyzed. Samples were analyzed with a range of stoichiometry levels (and fixed coverage of 1000 reads), which are the same samples that were used to generate Fig. 2D. Error bars indicate median ± s.d. of three replicates. Source data are provided as a Source Data file. E Radar plots of positive predictive values (PPV) for each software, RNA modification type and stoichiometry using either raw scores (upper track) or Z-scores (lower track). PPVs for raw scores were computed using the following threshold: for EpiNano, all sites reported as modified; for Nanopolish, sites whose Δmedian current intensity /median(Δmedian current intensity)>=5; for Tombo, sites whose p-value is lower than 0.01 after Benjamini–Hochberg correction; for Nanocompore, sites whose p-value is lower than 0.01 and with a GMM log odds ratio>0.5. PPVs for Z-scores were computed with a Z-Score threshold > 5. Source data are provided as a Source Data file. F Radar plots of positive predictive values (PPV) for NanoConsensus. PPV were computed using a Z-Score threshold > 5 and NanoConsensus score threshold > 5*(median across transcript). Source data are provided as a Source Data file.
Using this approach, we observed that NanoConsensus was able to robustly identify all modification types examined (Fig. 3B), in all independent biological replicates (n = 3) (Fig. S15). We then assessed Nanoconsensus’ performance in the detection of RNA modifications across diverse stoichiometry levels, coverage and modification types (Fig. 3C,D), using as input the same datasets used to assess the performance of each individual software (Fig. 2, see also Fig. S7 and Methods). To compare the performance of NanoConsensus to that of each individual software, Receiver Operating Characteristic (ROC) curves were built for each algorithm, modification type, coverage and stoichiometry, and the comparative performance was assessed by comparing the Area Under the Curve (AUC) values for each ROC curve. Our results showed that NanoConsensus was more robust in predicting RNA modifications across diverse modification types, stoichiometries and coverage (Figs. S16 and S17). Indeed, NanoConsensus globally outperformed all individual softwares in terms of AUC values (AUC = 0.959) when using as input all the subsets (RNA modification types, stoichiometries, and coverage) included in the benchmarking (Fig. S18).
Although NanoConsensus can robustly detect all RNA modification types examined in this work (Fig. 3B, see also S15), we should note that m5C modifications were among the worst performing of all modifications examined, as these could not be accurately identified at low stoichiometry levels (Fig. 3D, F). Despite these limitations, we found that NanoConsensus could robustly detect m5C modifications in datasets with ≥30% modification stoichiometry (Fig. S19). To further confirm the ability of NanoConsensus to detect m5C modifications, we examined its performance in independent mouse rRNA DRS datasets, by comparing wild type and NSUN5 KO mice, which are expected to lack 28S:m5C3438. The lack of modification in NSUN5 KO mice was confirmed by both RT-qPCR and bisulfite sequencing (Fig. S20, see also Methods). Our results confirm that NanoConsensus could accurately identify m5C in mouse rRNA molecules (Fig. S21), further supporting our observation that NanoConsensus is able to robustly identify m5C modifications in diverse sequence contexts, as long as the modification stoichiometry of the modified site is ≥30%.
Finally, we examined the number of false positives reported by each individual software, compared to NanoConsensus. To this end, we computed the positive predictive values (PPV) across diverse stoichiometries and RNA modification types, for each algorithm. The PPV reflects the proportion of true positives (TP) among the set of predicted ‘positive’ (P) differentially modified sites. Our results showed that the PPV was affected both by modification stoichiometry and coverage, and that it was also dependent on the RNA modification type (Fig. 3E, see also Figs. S22–S23). Notably, the PPV of each software was significantly improved after Z-score normalization, due to the decrease in false positives (Fig. S24). Globally, we found that Nanopolish showed the worst performance in terms of PPV, followed by EpiNano and Nanocompore. On the other hand, NanoConsensus showed the best performance in terms of PPV across all stoichiometries and modification types, implying that it reports fewer false positives, compared to individual softwares tested (Fig. 3F).
Aminoglycoside exposure leads to changes in the modification pattern of the 16S in the bacterial ribosome
We then used NanoConsensus to re-examine the question of how antibiotic exposures, such as kasugamycin or streptomycin, may impact the rRNA modification patterns of the bacterial ribosome. To this end, E. coli cultures were grown in LB until log phase, and were then exposed to streptomycin (str), kasugamycin (ksg) or left untreated (Fig. 4A). Cultures were collected 1 h or 16 h post-treatment, and the experiment was performed in independent biological replicates, on different days. DRS libraries from total RNA were built as previously described43 (see Methods), and the data was analyzed in a pairwise manner using NanoConsensus. This analysis revealed multiple regions (5 in 16 s rRNA, 1 in 23 s rRNA) that were ‘differentially modified’ in both replicates (Fig. 4B, labeled in red). Notably, all the identified regions located in the 16S rRNA overlapped with known rRNA-modified sites (Supplementary Data S5). In terms of directionality of the RNA modification change, all sites identified as differentially modified decreased in their modification stoichiometry upon antibiotic exposure (Fig. S25). We should note that increased duration of antibiotic treatment (1 h or 16 h) (Fig. 4B); did not significantly change the set of differentially modified sites identified in the case of kasugamycin, whereas in the case of streptomycin, some differentially rRNA modified sites were lost with extended exposures.
A Experimental design of the exposure of non-resistant E. coli to both streptomycin and kasugamycin. (Created in BioRender. Novoa, E. (2024) BioRender.com/b62y778). B NanoConsensus scores along the 16S rRNA (upper panel) and 23S rRNA (lower panel) of E. coli samples collected after 1 h or 16 h exposure to streptomycin (left panel) or kasugamycin (right panel). In gray, non-significant positions; in blue, regions identified by NanoConsensus in only one replicate; in red, regions identified in both replicates. Known modified sites found in replicable differentially modified regions identified by Nanoconsensus are also shown. Source data are provided as a Source Data file. C 3D structure of the bacterial ribosome (PDB: 7K00), highlighting the differentially modified rRNA residues upon streptomycin (upper panel) or kasugamycin (lower panel) exposure identified in this work. Differential rRNA modified residues are shown in red; in black, the antibiotic; in light green and orange, the tRNA molecule located in the A and P-sites, respectively.
Finally, we examined the 3D location of ‘differentially modified’ regions upon antibiotic treatment within the ribosome structure, and found that these regions were very close in the 3D space despite being far in the linear rRNA molecule (Fig. 4B). More specifically, these regions were all located within the A- and P-sites of the ribosome (Fig. 4C see also Supplementary Data S5), thus matching the binding site areas of streptomycin and kasugamycin, respectively (Fig. 4C). Thus, our results suggest that the alteration of a subset of rRNA modification might constitute an adaptive response of bacterial cells to diminish the binding affinity of kasugamycin and streptomycin in the P-site and A-site, respectively. Altogether, our results demonstrate that rRNA modification levels can be dynamically regulated upon environmental exposures, and that rRNA modification dysregulation might constitute a common molecular mechanism employed by bacterial species to increase their tolerance towards antibiotics binding the ribosome.
Mutations or differential rRNA usage do not explain the observed differences in DRS profiles
We and others have shown that differential RNA modifications in DRS datasets cause alterations in the current intensities and/or base-calling ‘error’ patterns45,72. However, changes in the nucleotide sequence (e.g., SNPs, alternative rRNA usage) can also lead to such alterations, and thus could constitute a confounder in the analysis, misguiding the detection of differential rRNA modifications.
To exclude such possibility, we examined whether the use of alternative E. coli rRNA genes could explain our results. E. coli has seven rRNA operons encoded in its genome, with small sequence differences among them73,74. Notably, previous works have shown that stress can lead to change in its operon usage75,76. To examine whether differential rRNA operon usage could explain our observations, we performed a multiple sequence alignment of the 7 annotated E. coli 16S rRNA sequences, finding that all 7 rRNA sequences were identical across operons within regions identified as ‘differentially modified’, (Fig. 5A) demonstrating that differential rRNA operon usage could not explain the observed changes in the DRS data.
A Multiple sequence alignment (MSA) of the seven rRNA operons sequences in all regions identified by NanoConsensus upon antibiotic exposure. B IGV tracks of nanopore cDNA reads aligning to the ribosome sequence in all regions identified by NanoConsensus upon antibiotic exposure. Positions with mismatch frequencies greater than 0.1 are colored, whereas positions with mismatch frequencies lower than 0.1 are shown in gray. C Differential expression analysis results between treated and untreated samples. In gray, non-significant genes; in green, genes with abs(log2FC) > 2 and; in red, genes with abs(log2FC) > 2 and p adjusted value >=0.05. P-values were calculated using the two-sided Walsh test and corrected with Benjamini–Hochberg approach. Source data are provided as a Source Data file. D GO term enrichment analysis using molecular process terms using as input all genes with abs(log2FC) > 2, which corresponded to n = 195 (upregulated) and n = 214 (downregulated) genes upon streptomycin treatment, and n = 197 (upregulated) and n = 203 (downregulated) genes upon kasugamycin treatment. In orange, terms related to downregulated genes upon antibiotic exposure and; in green, related to upregulated genes. P-values were obtained with the Fisher’s Exact Test, followed by calculating the False Discovery Rate (FDR). Only terms with a FDR < 0.05 were included in the final results. Source data are provided as a Source Data file. E Cumulative distribution of summed basecalling errors at per read level from both untreated (n = 10,953 reads), str-treated (n = 3689 reads) and ksg-treated (n = 14,376 reads) samples. In the upper-left corner, p-values between distributions calculated with the two-sided KS test are shown.
Another possible confounder in the rRNA modification analysis could be the presence of mutations that might arise at the genomic level upon bacterial divisions, which might be selected for if they confer a selective advantage upon antibiotic exposure77. To examine whether rRNA mutations could explain our observations, we sequenced antibiotic-treated and untreated samples using Nano3P-seq78, a cDNA-based nanopore sequencing method that efficiently captures both the coding and non-coding transcriptome, regardless of their tail composition (see Methods and Supplementary Data S4). Analysis of rRNA molecules sequencing using Nano3P-seq revealed no mutations at the rRNA regions identified as “differentially modified” by Nanoconsensus (Fig. 5B). Altogether, our analyses support that the changes seen in the DRS data are caused by alterations in the rRNA modification patterns, and not by changes occuring at the rRNA nucleotide sequence or caused by alternative use of rRNA operons.
De novo rRNA transcription occurs upon streptomycin exposure
Our results show that alterations in bacterial rRNA modification patterns can already be seen 1 h after antibiotic exposure (Fig. 5A, B), demonstrating that rRNA modifications can be rapidly regulated upon environmental exposures. This, in turn, opens new questions regarding how these differentially modified rRNA molecules are produced.
To decipher the mechanism(s) used by bacterial cells to alter their rRNA modification profiles, we first examined whether specific pathways might be altered upon antibiotic exposure. To this end, we sequenced ribodepleted RNA samples from untreated, ksg-treated and str-treated cultures (1 h post-antibiotic exposure) using Nano3P-seq, in biological duplicates. We performed differential expression analysis (Fig. 5C and Supplementary Data S6, see also Methods) and GO term enrichment analysis on the set of genes that had at least 4-fold changes in their expression levels (Fig. 5D), which revealed that both upon kasugamycin and streptomycin treatment, transmembrane transport-associated genes are downregulated, possibly to reduce the antibiotic uptake. In addition, we also found that de-novo ribosome synthesis genes were significantly upregulated upon streptomycin treatment, suggesting that the origin of lowly modified 16S rRNA molecules might arise from newly synthesized 16S rRNA molecules. However, we cannot exclude other possibilities, such as the existence of eraser enzymes, that can be responsible for the appearance of lowly modified 16S rRNA molecules.
To test this hypothesis, we examined the cumulative distribution of per-read ‘errors’ (which can be used as a proxy for the relative number of RNA modifications per molecule) from str-treated, ksg-treated and untreated full-length DRS reads (see Methods). Our analysis revealed that a population of lowly modified reads (i.e., summed errors <4) was present in both str- and ksg-treated samples, but was largely absent in untreated samples, suggesting that lowly-modified rRNA reads appear in response to antibiotic treatments (Fig. 5E). Similar results were obtained using PCA analysis of per-read rRNA modification profiles, which also showed that most of the lowly-modified rRNA reads were mainly appearing upon antibiotic exposure (Fig. S26).
Excluding rsmA and rsmG, the lack of individual methyltransferases does not lead to an increased resistance to streptomycin nor kasugamycin
Previous works have reported that the lack of rsmA (responsible of placing m6,6A in 16S:1518-1519), and rsmG (responsible of m7G in 16S:527) leads to increased resistance to kasugamycin and streptomycin, respectively. In our work, we find that modification levels of both 16S:m6,6A1518-1519 and 16S:m7G527 are significantly decreased upon antibiotic exposure, suggesting an adaptive response of bacteria in their rRNA modification profiles to increase their resistance to antibiotics. Notably, we also found that additional rRNA modified sites, apart from these two, were significantly decreased in their modification levels upon antibiotic exposure (Fig. 4B). Therefore, we wondered whether the lack of methyltransferases responsible for placing rRNA modifications that we find dysregulated upon antibiotic treatment, in addition to rsmA and rsmG, might lead to increased antibiotic resistance.
To this end, we cultured seven E. coli knock-out strains (listed in Supplementary Data S1), including rsmA and rsmG, that we identified as responsible for placing rRNA modifications that are differentially methylated upon streptomycin and/or kasugamycin exposure (Fig. 4B). Each strain was cultured under a broad range of antibiotic (kasugamycin or streptomycin) concentrations, and their growth was monitored for 16 h (Figs. 6A and S27). To assess whether the lack of a given methyltransferase enzyme led to increased antibiotic resistance, we compared the growth of each knockout to that of the wild type strain by subtracting the AUC values of the growth curves of each strain and antibiotic concentration relative to the same conditions in the wild type strain. Our results showed that the lack of rsmA and rsmG led to increased antibiotic resistance (Fig. 6B), as expected. However, the lack of other methyltransferases examined did not lead to increased antibiotic resistance, despite being responsible for placing rRNA modifications at sites that we find differentially modified upon antibiotic exposure. The possible synergistic effect of removing several of these enzymes simultaneously remains unknown.
A Median growth curves from four biological replicates of different E. coli strains (wild type, rsmA knockout, rsmG knockout and rsmE knockout) across a range of antibiotic (streptomycin, upper panels; kasugamycin, lower panels) concentrations. Data for each biological replicate was computed as the median of three technical replicates. Growth curve data was fitted to a logistic curve. See also Figure S27 for growth curves in additional E. coli strains tested (rsmB, rsmD, rsmH and rsmJ knockout strains). B Barplots depicting Median Area Under the Curve (AUC) difference in growth curves between wild type and knockout strains (KO-WT), for every KO strain and across all antibiotic concentrations tested. In bold, the strains that are reported to have an increased resistance to each specific antibiotic. Error bars indicate standard deviation of four biological replicates. Source data are provided as a Source Data file.
Discussion
A plethora of computational methods for detecting RNA modifications in nanopore direct RNA sequencing (DRS) datasets have been developed in recent years43,48,49,50,52,53,54,55,56,57,58. These methods typically predict RNA modifications either through the analysis of the raw signal features (e.g., current intensity and/or dwell time)43,52,53,54,55,56,57,58 or in the form of differential base-calling ‘errors’47,48,49,50,51. Previous works have compared the performance of some softwares in detecting m6A modifications in DRS datasets, finding a relatively poor overlap between the predictions across softwares53,57,58,79,80. However, their comparative performance in detecting other RNA modification types (e.g., Ψ, Nm, ac4C) has so far not been assessed. Thus, it is unclear which software(s) should be used to study rRNA modification dynamics.
To tackle this question, here we benchmarked four softwares that can in principle detect diverse RNA modification types in DRS datasets, and assessed their ability in detecting diverse RNA modification types (i.e., Ψ, Am, Um, m6,6A, m7G and m5C) across diverse stoichiometries and sequencing coverage ranges. Our results showed that softwares relying on signal intensity features were better than those using basecalling errors at detecting some RNA modification types (e.g., m5C), but the opposite was seen for other RNA modification types (e.g., m6,6A) (Fig. 2C). Notably, we also found that in cases where RNA modifications showed a ‘spread of signal’ along several nucleotides, algorithms often identified different positions as the ‘differentially modified site’, partly explaining the poor overlap of RNA modification predictions across softwares (Fig. S2). In addition, our analyses showed that both coverage and stoichiometry levels had a strong impact in the performance of the algorithms (Fig. 2), lowering both their sensitivity and specificity (Figs. S3–S6 and S16). To overcome these limitations, we designed and implemented NanoConsensus, a workflow that identifies differentially modified RNA sites between two samples using as input the predictions from 4 different softwares, allowing for sliding-window overlaps to identify common predictions across softwares. We found that NanoConsensus outperformed all 4 individual softwares, and was robust across a range of RNA modification types, stoichiometry and coverage levels (Fig. 3E-F). To facilitate the use of this tool, we have made Nanoconsensus available as Nextflow module81, and have integrated it within the MasterOfPores61 Nextflow workflow, to ensure reproducibility, scalability and traceability of the analyses performed. We should note that similar ‘consensus’ approaches have also been successfully employed to improve the detection of DNA modifications in nanopore sequencing data82.
Once the performance of Nanoconsensus was benchmarked and optimized, we applied it to investigate whether bacterial rRNA modification profiles were altered upon antibiotic exposure, and determine whether variation in rRNA modification levels might constitute a natural mechanism that bacteria use to increase their resistance upon antibiotic exposure. To this end, we sequenced total RNA from streptomycin-treated, kasugamycin-treated and untreated E. coli cultures using nanopore DRS, and compared the rRNA modification profiles of treated and untreated cultures. We found that antibiotic exposure significantly altered the 16S rRNA modification profiles of the bacterial ribosome (Fig. 4B). Notably, differentially modified rRNA sites were largely located within the vicinity of the A and P-sites (Fig. 4C and Supplementary Data S5), which correspond to the binding regions of the streptomycin and kasugamycin, respectively. We should note that one site was identified in 23S rRNA, which did not overlap with any annotated rRNA modified site, and was located 91 Å and 105.4 Å away from streptomycin and kasugamycin binding sites, respectively, and thus, was considered as a false positive for the remaining downstream analyses.
The fact that rRNA modifications are altered precisely in the vicinity of the antibiotic binding site suggests that this phenomenon is a specialized response from bacteria to the specific antibiotic in question, possibly as a means to decrease the binding affinity of the antibiotic to the bacterial ribosome. In the case of streptomycin, it has been previously reported that the antibiotic interacts with positions 16S:530 in the loop19,83, and more specifically with 16S:m7G527. Therefore, the lack of this modification, possibly also enhanced by the loss of other neighboring modifications, likely causes a decrease in the binding affinity of the antibiotic. In the case of kasugamycin, it is known that both m6,6A residues (16S:m6,6A1518 and 16S:m6,6A1519) are not in direct contact with the antibiotic, which might explain why the absence of these modifications confers only low-level resistance84.
Previous works using DRS have reported differential RNA modification patterns upon heat and oxidative stress in snRNAs, snoRNAs and mRNAs, but not in rRNAs43,64. Contrary to previous observations, our results point to rRNA modifications being dynamically regulated upon environmental conditions, at least in the case of antibiotic exposure(s). An alternative explanation for the loss of the 16S rRNA modifications observed upon antibiotic exposure would be that antibiotics themselves interfere with the RNA modification machinery and/or alter the recognition of the sites during rRNA maturation.
Indeed, the selective loss of certain bacterial rRNA modifications upon antibiotic exposures, in an antibiotic-specific manner, opens new questions regarding what might be the mechanism(s) that bacteria use to achieve the outcome of differential rRNA modification. To answer this question, we performed single molecule analysis of rRNA modification patterns (Figs. 5E and S26) as well as differential expression analysis of treated and untreated cultures using Nano3P-seq (Fig. 5C and Supplementary Data S4). These efforts revealed that antibiotic-exposed bacteria have increased de novo expression of under-modified rRNA molecules, which ultimately leads to significantly altered rRNA modification profiles. It remains unclear, however, how antibiotics lead to altered rRNA maturation processes and to the production of under-modified rRNA molecules.
Future work is needed to confirm whether the observed changes in rRNA modification patterns are caused by de novo generation of under-modified rRNA molecules that appear upon antibiotic exposure, or other possible models, such as that the presence of antibiotics causes defects in ribosome assembly, which leads to the generation of an under-modified rRNA population. In this regard, it has been previously shown that kasugamycin exposure leads to the formation of 61S ribosomes85, although these ribosomes showed unaltered 16S:m66 A1518 and 16S:m66A1519 modification levels in previous studies, which differs from our findings.
In addition to the mechanism that leads to the appearance of under-modified rRNA populations, an exciting open question that is prompted by this work is whether differentially modified rRNA subpopulations that are naturally generated will be incorporated into translating ribosomes and used for protein synthesis, for example by performing sucrose gradient fractionation coupled to nanopore total RNA sequencing (to capture rRNA modifications) and mRNA sequencing (to capture selective translation of subsets of transcripts). Establishing whether these unmodified ribosomes are part of the translating pool of ribosomes, and whether they might be involved in preferential translation of selected subsets of mRNAs as a means to achieve antibiotic resistance, are still open questions that need to be addressed in the future.
Various resistance mechanisms, including enzymatic detoxification, target alteration (rRNAs and ribosomal proteins) and reduced accumulation (impermeability and efflux) have been shown to be involved in bacterial resistance to protein synthesis inhibitors86. Notably, rRNA alteration has recently gained increased interest due to several recent studies showing that some chloramphenicol-florfenicol resistant (Cfr) bacteria carried a gene encoding for a mutant version of an rRNA methyltransferase87,88. Here we systematically examined whether the loss of individual methyltransferases, responsible for placing rRNA modifications that we identified as differentially modified upon antibiotic exposure (Supplementary Data S5), might alter the resistance of bacteria towards the antibiotic in question. We found that only the loss of rsmG and rsmA, but not other methyltransferase enzymes examined, led to a resistance phenotype (Fig. 6A, B, see also Fig. S27). It is possible that the lack of these additional enzymes is not per se enough to lead to increased resistance, but may have a synergistic effect when lost simultaneously with rsmG and/or rsmA. In this regard, recent studies have reported that some Mycobacterium tuberculosis antibiotic-resistant strains carry multiple mutations in several methyltransferase genes89 that are ortholog to the genes identified by Nanoconsensus, suggesting that the lack of these additional methyltransferases might constitute a selective advantage towards antibiotic resistance.
While our work establishes a robust framework to identify differential rRNA modification patterns across conditions using DRS (Fig. 2), we should note that Nanoconsensus has some limitations, as well as room for future improvement. Firstly, it does not provide information regarding the directionality (i.e., increase or decrease) of the rRNA modification changes between two conditions, although this limitation can be partly alleviated by using the Epinano results that are generated as part of the Nanoconsensus pipeline (see Fig. S25). Secondly, NanoConsensus results are qualitative or at best semi-quantitative, and therefore, to obtain quantitative information on the differentially modified identified, other softwares such as nanoRMS43 should be used downstream in the analysis. Thirdly, Nanoconsensus lacks single nucleotide resolution; consequently, if several RNA modifications are located within the differentially modified regions identified, it is not possible to determine which specific rRNA modification is changing. Fourthly, NanoConsensus requires a minimum modification stoichiometry of 10–30% (depending on the RNA modification) to detect a differentially modified site, limiting its applicability to RNA molecules with high modification stoichiometries, such as rRNAs or tRNAs. Despite these limitations, we demonstrate that Nanoconsensus can be used to reveal antibiotic-dependent rRNA modification changes, contributing to the understanding of how bacteria tune their ribosomes as a response to environmental conditions. Notably, this knowledge may be applied in the future to improve the design of antibiotic chemical structures such that they have improved binding efficiencies towards both the fully-modified and under-modified ribosomal A and P-site regions.
Methods
Bacterial strains and culturing
E. coli strains used in this study (rsmA:JW0050, rsmG:JW3718 and rsmF:JW5301, see Supplementary Data S1 for additional details) were obtained from the Keio Knockout Collection90, including the reference wild type strain (BW25113). Knockout strains have a kanamycin cassette replacing the depleted gene. Strains were plated in LB-agar plates (E. coli BW25113) or in LB-agar plates supplemented with 25 μg/mL kanamycin (in the case of E. coli knockout strains).
Growth curves of antibiotic-resistant E. coli strains
For each strain, E. coli starter cultures were grown O/N in 4 mL tubes at 37 °C and 180 rpm. Aliquots from the overnight cultures were added into a final volume of 200 μl in 96-well plates with an initial OD600 of 0.01. Different antibiotic concentrations (streptomycin: 8,16, 32, 64 and 128 μl/mL; kasugamycin: 62, 125, 250, 500 and 1000 μl/mL) were tested in both non-resistant and resistant strains. E.coli rsmF KO strain was not tested for kanamycin resistance as its KO was generated through the insertion of a kanamycin resistance cassette90. Bacteria was grown at 37 °C and OD600 was measured every 10 min with a TECAN M200 Plate Reader. We should note that the rsmF knockout strain was not included in the growth curve experiments as this mutant is expected to show increased resistance to kanamycin, and all Keio collection mutants contain a kanamycin resistance cassette replacing the gene that has been removed90. All experiments were performed in biological replicates (n = 4) for each strain and condition, and each strain and condition was measured at least in two independent plates and in two different days.
Bacterial exposure to antibiotics
E. coli BW25113 starter cultures were grown O/N in 4 mL of LB at 37 °C and 180 rpm. Three 500 mL LB cultures were then inoculated with the starter culture, to an initial OD600 ≈ 0.01. OD600 was monitored every hour with a spectrophotometer until all cultures reached OD600 ≈ 0.4 (early log phase). Then, the first 500 mL culture was supplemented with streptomycin 10 mg/mL (final concentration = 16 μl/mL) (Sigma-Aldrich, #S6501-5G), the second 500 mL culture was supplemented with kasugamycin 40 mg/mL (final concentration = 250 μl/mL) (Merck, #32354-100MG) and the third culture was supplemented with the equivalent volume of water (no antibiotic). OD600 was measured at time points 0 h, 1 h and 16 h after antibiotic addition (or no antibiotic). 50 mL from each erlenmeyer were taken at each of the three time points, and were immediately centrifuged in a pre-chilled benchtop centrifuge at 16,000 g 4 °C for 10 min. Supernatant was discarded, and pellets were stored at −80 °C until further use. This experiment was repeated with three independent biological replicates, which were cultured in different days.
E. coli total RNA extraction
Frozen pellets were thawed on ice, followed by the addition of 400uL of Trizol (Life Technologies, #15596018). After five minutes incubation at room temperature, 90uL of chloroform (Vidra Foc, #C2432) was added and mixed thoroughly by inversion. Samples were centrifuged for 15 min at 16,000 g at 4 °C. Supernatant was kept and an equal volume of 70% ethanol was added in a new eppendorf tube. Afterwards, small RNAs were removed with the Qiagen RNeasy Mini Kit following manufacturer’s recommendations (Qiagen, #74104). Samples were DNAse-treated using Turbo DNAse (Life Technologies, #AM2239) for 10 min at 37 °C followed by a final clean-up with Qiagen RNeasy MinElute Kit (Qiagen, #74204) using default manufacturer’s protocol (which keeps RNAs > 200nt) to remove excess of small RNAs that are typically present in E. coli total RNA extracts. RNA integrity was assessed using TapeStation (Fig. S28) and quantified using Nanodrop.
Direct RNA nanopore library preparation and sequencing
DNAse-treated E.coli total RNA (600 ng per sample) was in vitro polyadenylated using E. coli Poly(A) polymerase (New England Biolabs, #M0276L) using manufacturer’s recommendations with minor changes (the polyA tailing reaction was carried out for 10 minutes at 37 °C, instead of 30). Poly(A)-tailed total RNA was then prepared for sequencing using the direct RNA sequencing kit (SQK-RNA002), following the protocol guidelines (version: DRCE_9079_v2_revM_14Aug2019), with minor changes: RNA was linearized using SuperScript IV (Thermo Fisher Scientific, #18090010) and incubated for 15 min at 55 °C followed by a heat inactivation step (80 °C for 10 min), or linearized using Maxima enzyme (Life Technologies, #EP0751) and incubated for 30 minutes at 60 °C followed by a heat inactivation step of 5 min at 85 °C (see Supplementary Data S2 and S3).
Base-calling, demultiplexing and mapping of yeast and bacterial direct RNA sequencing runs
Raw fast5 reads from yeast and bacterial samples (Supplementary Data S2 and S3) were analyzed using the MasterOfPores (MoP) version 2 Nextflow workflow61. Briefly, the mop_preprocess module was used to demultiplex the FAST5 reads using DeePlexiCon with default parameters91. Demuxed FAST5 were then base-called using Guppy 3.1.5 (Oxford Nanopore Technologies) with the model rna_r9.4.1_70bps_hac, and aligned using graphmap v 0.5.2 with -v 1 -K fastq parameters to the the E. coli rRNA reference transcriptome, which comprised the 5S rRNA, the 16S rRNA and the 23S rRNA sequences (available in Github: NanoConsensus/ref/Escherichia_coli.rRNA.fa).
Detection of RNA modifications using diverse algorithms and implementation into the MasterOfPores Nextflow workflow
To detect differential RNA modifications across strains, each control condition (e.g., E. coli wild type BW25113) was compared in a pairwise manner to each of the knockout strains (e.g., to the E. coli knockout strains JW5301, JW0050 and JW3718, respectively) (Supplementary Data S1). For each pairwise comparison, four RNA modification detection algorithms were run in parallel: (i) EpiNano49, (ii) Nanopolish71, (iii) Tombo56 and (iv) Nanocompore57. EpiNano (version 1.1.1) was run with default parameters to extract per-site and per-kmer base-calling errors from every sample. Nanopolish (version 0.11.1) was used to resquiggle the basecalled fast5 using nanopolish eventalign (--samples --print-read-names --scale-events). Then, current intensity per position of the resquiggled reads and the coverage was extracted with a in-house python script, available in GitHub (https://github.com/biocorecrg/MOP2/blob/main/mop_mod/bin/mean_per_pos_v3.py)61. Tombo (v1.5) was first used to resquiggle the reads using tombo resquiggle (-rna option). The function tombo detect_modifications level_sample_compare with --store-p-value was then used to perform RNA modification detection. Finally, p-values at position level together with coverage data were retrieved with tombo text_output browser_files. Nanocompore (version 1.0.0) was run through the command nanocompore sampcomp (--sequence_context 2 --downsample_high_coverage 10000 --pvalue_thr 1 --logit --comparison_methods GMM,KS,MW,TT) (Fig. S29).
Improving the speed and reducing computational and time requirements of module mop_mod from MasterOfPores Nextflow workflow
Nanopolish eventalign output results are stored in csv files, which we found were very slow to parse, increasing the computational time required to perform all the downstream analyses, thus limiting its applicability transcriptome-wide. To increase the speed of the analysis, two in-house scripts based on the Apache Parquet format92 were implemented in Python3 (available in GitHub: https://github.com/biocorecrg/MoP3/mop_mod/bin/mean_per_pos.py and https://github.com/biocorecrg/MoP3/mop_mod/bin/Merging_processed_nanopolish_data.py). On the other hand, Tombo output files are given in the form of wig and bedgraph files. Therefore, to increase the efficiency of file parsing, we converted Tombo output files into bigWig files and processed them at per-transcript level using the pyBigWig package93. These scripts have been implemented in the MasterOfPores version 2.0 NextFlow workflow81, which is available in GitHub (https://github.com/biocorecrg/MoP2).
Prediction of RNA modifications using NanoConsensus
NanoConsensus reports putative modified regions at per-transcript level using as input the outputs obtained from each of the 4 different RNA modification detection algorithms previously described (EpiNano, Nanopolish, Tombo and Nanocompore).
In a first step, NanoConsensus converts per-position reported values by each software into normalized Z-Scores across each transcript. These scores are then used to select candidate RNA modified positions for each independent software, which must be higher than the provided user-defined threshold (default value = 5) (Fig. S30).Then, candidate positions for each individual software previously identified are extended into 5-mers. Flexible overlapping is then performed to identify overlapping k-mers across softwares. The regions supported by two or more softwares are saved as putative modified sites. In a second step, NanoConsensus re-scales Z-score values between 0 and 1 across each software’s set of values, to make the score comparable across transcripts (otherwise Z-scores are affected by the coverage of each transcript). Rescaled Z-scores are then converted to a NanoConsensus score, which is equal to the median of the rescaled Z-scores across all softwares (NanoConsensus score = median (Z-Score Epinano, Z-score Nanopolish, Z-Score Tombo, Z-Score Nanocompore). Thus, every position across the transcript has an assigned NanoConsensus score. Finally, NanoConsensus identifies the differentially modified sites as those having a Nanoconsensus score greater than 5 (default settings) times the median per-transcript NanoConsensus score. Thus, the threshold varies depending on the background signal of the transcript, i.e., if the data is “noisier” (often the case when coverage is low) the Nanoconsensus score threshold to identify a site as “differentially modified” within that given transcript will be higher. We should note that this threshold can be modified by the user. All benchmarking results included in this work used threshold=5 (default) throughout all datasets and RNA modification types. To capture modest variations in stoichiometry, the user might prefer to decrease the threshold to 3.5-4.
NanoConsensus will produce the following files as outputs: (i) a flat file with all raw results obtained by all softwares, (ii) a flat file with the putative modified sites according to NanoConsensus; (iii) BED tracks that can be loaded into IGV to visualize the results in a user-friendly manner; and (iv) a PDF file showing both the performance of each algorithm, represented in the form of Z-scores, and NanoConsensus scores along the analyzed transcripts. All code has been integrated into the mop_consensus module (RNA modification detection module) that is part of the MasterOfPores version 2 (MoP2) Nextflow workflow (https://github.com/biocorecrg/MoP2)81.
Benchmarking RNA modification predictions across datasets with known stoichiometry and coverage
For each modification type included in the benchmarking (Am. Um, Y, m7G, m5C and m66A), reads from wild type E. coli were assumed to be 100% modified, and the respective knockout rRNA reads were assumed to be 0% modified for that position (Supplementary Data S3), respectively. For each dataset analyzed, subsampled datasets were generated using one of the two approaches: (i) subsamples contained a fixed amount of reads or; (ii) samples contained a fixed number of modified reads (both scripts available at GitHub: https://github.com/novoalab/NanoConsensus/scripts/Downsampling). All benchmarking datasets only included full-length reads, defined as reads whose initial mapping position was 50 or lower and their final position was equal or larger than 1525 and 1779, for 16S and 18S transcripts respectively (available at GitHub: https://github.com/novoalab/NanoConsensus/scripts/Full_Length/Extract_FullLength_IDs.sh).
To compare datasets with different modification stoichiometry levels, we performed two complementary analyses using the two above-mentioned subsampling approaches. In the first subsampling approach, each subsampled dataset comprised the same number of reads (n = 1000). Then, to achieve different stoichiometry levels (100, 75, 50, 40, 30, 20 and 10%), a specific proportion of randomly selected modified and unmodified full-length reads were mixed. Triplicates per modification type and stoichiometry were built. In the second subsampling approach, the subsampled dataset were composed of triplicates that have a fixed number of modified reads. Then, for each stoichiometry level, an increasing amount of unmodified reads were added sequentially. As a result, the population of modified reads was kept constant whereas the total number of reads varied across the samples from the same replicate. Additionally, four full datasets with a different number of modified reads in the 100% modified sample (50, 100, 500 and 1500 reads) were generated. All samples were compared against 1000 full-length reads from their respective knock-out strain using mop_mod with default parameters (see previous section).
The ability of NanoConsensus to identify other modification types, such as m2G and m3U, was examined by comparing wild-type data and knock-out strains lacking these specific modifications, in a similar fashion to other RNA modification types included in the benchmarking (m66A, Y, m5C, Am, Um and m7G) (Figs. S15, S31, see also Supplementary Data S1). We should note that m4Cm was not included in this analysis as it is not a modification placed by a single enzyme, but rather by two sequential enzymes, and the double knockout strain that would be required was not commercially available. Thus, we did not demonstrate whether Nanoconsensus can detect m4Cm modifications.
Generation of NSUN5 KO mice
Experimental procedures with mice were conducted following protocols that were evaluated and approved by the Biosafety and Bioethics Committee at the University of Salamanca (Protocol #269) and the Competent Authority of Castilla y León. All mice were housed at the Animal Research Core Facility at the University of Salamanca, in ventilated filter cages under Specific Pathogen Free (SPF) conditions, with food and water available ad libitum. Mice were kept at a constant temperature (set up at 21 °C) and humidity with a 12-hour light/dark cycle.
B6N(Cg)-Nsun5tm2b(EUCOMM)Wtsi/J mice were purchased from The Jackson Laboratory (Fig. S20A). To get Nsun5−/− mice and their wild type littermates, mice were crossed in heterozygosity. A PCR-based strategy was developed to distinguish the wild-type and Nsun5 mutant alleles. The primers are as follows: 5′-TGCAGCTCAGCAATAAGCA-3′, 5′-GCAGTTCCTAGCACCGTGT-3′, 5′-ACACGGGGTTTGCAGATG-3′ and 5′-CGGTCGCTACCATTACCAGT-3′. Nsun5 +/+ mice yielded a single band of 240 bp, while Nsun5−/− mice yielded a single band of 428 bp (Fig. S20B). Nsun5 loss in Nsun5−/− mice was validated by RT-qPCR using TaqMan Probe Mm00520549_m1 (Thermo Fisher Scientific). Expression of the housekeeping gene Gapdh used to normalize was detected by TaqMan Probe Mm99999915_g1 was used to normalize Nsun5 expression (Fig. S20C).
RNA extraction from NSUN5 KO mice
RNA was extracted from limbs of post-natal day 2 wild type and Nsun5−/− mice using QIAzol lysis reagent (Qiagen) according to the manufacturer’s instructions. After isopropanol precipitation, TURBO DNAse I (Invitrogen) treatment was performed to remove any remaining DNA contamination. RNA integrity was analyzed using 2100 Bioanalyzer and RNA 6000 Nano chip (Agilent).
Detection of m5C in RNA from NSUN5 KO mice using PCR-bisulfite
DNAse-treated RNA was treated with sodium bisulfite using Methylamp RNA Bisulfite Conversion Kit (Epigentek) following manufacturer’s instructions. After the sodium bisulfite treatment for 5 minutes at 70 °C and 90 minutes at 60 °C, treated RNA was in-column desalted and deaminated. Converted RNA was used as template for cDNA analysis using Maxima H-minus cDNA synthesis MasterMix (Thermo Fisher Scientific) following manufacturer’s recommendations. cDNA was then used as template for PCR amplification using the following primers: 5′- AATGAAGTGTGGGTAAATGG-3′ and 5′- AAATAAAAACAATAAAAATCTCAT-3′. PCR product was ligated into the pCR 2.1 TOPO vector (Invitrogen), transformed into chemically-competent DH5a E. coli and sequenced by Sanger sequencing using the following primer: 5′- GTAAAACGACGGCCAG-3′. Sequences were aligned using MEGA11 software and compared to mouse reference 28S rRNA (NR_003279.1) (Fig. S20D).
Direct RNA nanopore library preparation and sequencing of total RNA from WT and Nsun5 -/- M. musculus samples
DNAse-treated M. musculus total RNA (600 ng per sample) was in vitro polyadenylated using E. coli Poly(A) polymerase (New England Biolabs, #M0276L) using manufacturer’s recommendations with minor changes (polyA tailing reaction was carried out for 10 minutes at 37 °C, instead of 30 min). Poly(A)-tailed total RNA was then prepared for sequencing using the direct RNA sequencing kit (SQK-RNA002), following the protocol guidelines (version: DRCE_9079_v2_revM_14Aug2019), with minor changes: RNA was linearized using Maxima enzyme (Life Technologies, #EP0751) and incubated for 30 minutes at 60 °C followed by a heat inactivation step of 5 min at 85 °C (see Supplementary Data S3).
Detection of RNA modifications from bacterial samples exposed to antibiotics
Bacterial DRS sequencing runs (Supplementary Data S2) were filtered to keep only full-length reads using in-house scripts (all available at GitHub: https://github.com/novoalab/NanoConsensus/scripts/Full_Length), which were defined as reads that covered the full transcript, leaving up to a maximum of 50 nt uncovered in the 5′ and/or 3′ ends. In other words, 16S rRNA reads that were kept covered at least from position 50 to 1525, and reads that aligned to the 23S rRNA covered at least from position 50 to 2894. Replicates per condition and time point were included in the analysis (Fig. S32). Pairwise comparisons of antibiotic-treated and untreated samples were analysed using the MasterOfPores (MoP2) module mop_mod, with default parameters, followed by NanoConsensus (also integrated in MoP2), with the following parameters across all transcripts: --MZS_thr 3.75 --NC_thr 4. The directionality of the changes in stoichiometry at the identified ‘differentially modified’ rRNA sites was assessed by comparing the sum of basecalling errors of treated and untreated samples using EpiNano (version 1.1.1). We should note that Nanoconsensus can be run comparing treated samples to untreated (t = 0 h) (Fig. S33A), but the signal-to-noise ratio improves if matched untreated time points are used (t = 1 h or t = 16 h, respectively) (Fig. S33B). Running time of these algorithms from two of the comparisons included in this study can be found in Supplementary Data S7.
Clustering of full length reads based on basecalling errors
To identify differentially modified populations of RNA molecules upon antibiotic exposure, per-read basecalling errors (mismatch, insertion and deletion frequency) were retrieved using EpiNano (v 1.0)49, which reports per-read base-calling errors. For each read, basecalling errors of the 5-mer region centered in each ‘differentially modified’ site identified by NanoConsensus were extracted, and used for downstream analyses. For each read, the total summed error per-read was calculated. The per-read sum of ‘errors’ provides an approximate measure of the modification levels of a given read in those selected regions, relative to the rest of reads. Reads were binned based on their total summed score, and the fraction of reads belonging to each bin, for each treated sample (str-treated, ksg-treated, untreated) was calculated.
Growth curves of bacterial strains lacking individual methyltransferases
E. coli starter cultures of multiple strains (BW25113, rsmA KO, rsmG KO, rsmD KO, rsmB KO, rsmJ KO, rsmH KO and rsmE KO) were grown overnight in 10 mL tubes at 37 °C 180 rpm, in biological duplicates. Overnight cultures were added into a final volume of 200 μl in 96-well plates with an initial OD600 of 0.01. Different antibiotic concentrations (streptomycin: 8, 16, 32, 64 and 128 μl/mL; kasugamycin: 62, 125, 250, 500 and 1000 μl/mL) were tested for each strain. For each strain and antibiotic concentration, three technical replicates were generated and the experiment was performed with two biological replicates, in two independent days. Bacteria were grown at 37 °C and OD600 was measured every 10 minutes with a TECAN M200 Plate Reader. Area Under the Curve (AUC) calculations were performed in R using the growthcurver package94.
Bacterial ribodepletion
Ribodepletion was performed using riboPOOL oligos (siTOOLs, cat #055) following the manufacturer’s protocol (version: riboPOOL Protocol_v1-8). Briefly, 5ug of total RNA was mixed together with 1uL resuspended riboPOOL oligos, 5 uL hybridization buffer (10 mM Tris-HCl pH 7.5, 1 mM EDTA, 2 M NaCl) and 0.5 uL SUPERase•In RNase Inhibitor (Thermo Fisher, AM2694) followed by a 10 min incubation at 68 °C and slow cool down to 37 °C for hybridization. In parallel, Dynabeads MyOne Streptavidin C1 (Thermo Fisher, #65001) beads were resuspended, and 80uL of the resuspended beads were transferred into a tube and placed on a magnetic rack. Then, the supernatant was removed and 100 uL of bead resuspension buffer (0.1 M NaOH, 0.05 M NaCl) was added to the sample. Beads were then resuspended in the buffer by agitation, the tube was placed again on a magnetic rack, and the supernatant was aspirated. This step was performed twice. Beads were then resuspended in 100 uL of washing buffer (0.1 M NaCl) and placed again onto the magnet to remove the supernatant. The beads were then resuspended in 160 uL of depletion buffer (10 mM Tris-HCl pH 7.5, 1 mM EDTA, 1 M NaCl). This mixture was then divided into two tubes of 80uL, which were used sequentially. 20 uL of hybridized riboPOOL oligos and total RNA was spun down briefly and then transferred into a tube with 80uL of beads in the depletion buffer. Then, it was mixed by pipetting carefully and incubated at 37 °C for 15 min, followed by a 50 °C incubation for 5 min. The reaction was placed on the magnetic rack and the supernatant was transferred into the second tube of beads, from which the supernatant was removed before its use. The solution was mixed and incubated at 37 °C for 15 min, followed by a 50 °C incubation for 5 min. After briefly spinning down the droplets, the mix was placed on a magnet for 2 min and the supernatant was transferred into a different tube. Finally, long RNA species (>200 bp) were separated from short ones (<200 nt) using RNA Clean & Concentrator-5 kit (Zymo, R1013).
End-capture nanopore cDNA sequencing (Nano3P-seq)
E. coli total RNA was sequenced using Nano3P-seq78, which is a cDNA library preparation variation that relies on the SQK-DCS109 direct cDNA nanopore sequencing kit. Notably, Nano3P-seq does not require the presence of polyA tails, which are typically absent in bacterial mRNAs, to initiate the library (whereas the standard direct cDNA nanopore sequencing library requires the presence of polyA tail). The protocol was executed as previously described (https://www.protocols.io/view/nano3p-seq-protocol-3byl4j292lo5/v1). Briefly, 50 ng of ribodepleted total RNA per sample was mixed with 1 µL pre-annealed RNA-DNA oligos (RNA oligo: 5′ rGrArArGrArUrArGrArGrCrGrArCrArGrGrCrArArGrUrGrArUrCrGrGrArArG/3SpC3/3′; DNA oligo: 5′/5Phos/CTTCCGATCACTTGCCTGTCGCTCTATCTTCN 3′), 1uL 100 mM DTT, 4 µL 5X TGIRT buffer, 1uL RNasin Ribonuclease Inhibitor (Promega, N2511), 1uL TGIRT (InGex) and nuclease-free water (NFW) up to 19uL.This reverse transcription mix was first incubated at RT for 30 minutes before 1uL 10 mM dNTP mix (NEB, N0447S) was added. Then, the solution was brought to 60 °C for 60 min, and inactivated by heating at 75 °C for 15 min before moving to ice. Afterwards, RNAse Cocktail (Thermo Scientific, AM2286) was added followed by an incubation at 37 °C for 10 minutes and a clean up step using 0.8X AMPure XP Beads (Agencourt, A63881). Then, 1 µL 100 µM of the DNA oligo (5′ GAAGATAGAGCGACAGGCAAGTGATCGGAAGA 3′), complementary to the initial one, was added to 15uL solution containing cDNA with 2.25 µL 0.1 M Tris pH 7.5, 2.25 µL 0.5 M NaCl and 2 µL NFW. The mix was incubated at 94 °C for 1 min and the temperature was ramped down to 25 °C (−0.1 °C/s). Later, 2.5uL from a single native barcode and 25 µL Blunt/TA Ligase Mix (NEB, M0367S) were added, followed by a 10 min incubation at RT and a clean up step with 0.5X AMPure XP beads and eluted with 16uL NFW. The concentration of each sample was quantified using Qubit ssDNA HS assay (Thermo Scientific, Q10212). Individual samples were pooled together with a final concentration that did not exceed 200 fmol and a final volume of 65uL. All the subsequent steps used reagents from the direct cDNA nanopore sequencing kit (ONT, SQK-DCS109). The pooled sample was mixed with 5uL Adapter Mix (AMI), 20uL 5X NEB Quick Ligation buffer (NEB, B6058S) and 10uL Quick T4 DNA ligase (NEB, M0202M) and then incubated for 10 min at RT. Finally, the sample was cleaned with 0.5X AMPure XP beads (Beckman Coulter, A63881), using washing buffer (WSB) and elution buffer (EB). The library was mixed with sequencing buffer (SQB) and loading Beads (LB) prior to loading it onto a primed R9.4.1 MinION flowcell. Biological triplicates were run on independent MinION flowcells, and prepared on different days.
Base-calling, demultiplexing and mapping of Nano3P-seq datasets
FAST5 reads sequenced with the Nano3Pseq protocol were analyzed using the MasterOfPores version 2 (MoP2) Nextflow workflow61 (Supplementary Data S4). Firstly, the mop_preprocess module was used to basecall and demultiplex all FAST5 using Guppy 4.0 (Oxford Nanopore Technologies) with the DNA basecalling model dna_r9.4.1_70bps_hac. Reads whose barcode could not be identified, went through a second round of demultiplexing using readucks (version 0.0.3)95 with parameters --limit_barcodes_to 1 2 3 4 5 6 --adapter_threshold 73 --threshold 50. Demuxed reads were then aligned to the E.coli BW251113 genome (NCBI ID: CP009273.1) using minimap2 v.2.17 with -ax spliced -k14 -uf parameters. Both per-gene counts and per-transcript counts were generated. Briefly, per-gene counts were obtained by htseq-count (version 0.13.5)96 with the option --stranded reverse as well as using the --nonunique all option to account for reads spanning more than one feature (ie: bacterial operons). On the other hand, to obtain per-transcript estimates, bacterial reads were aligned to the reference transcriptome that was built from the reference genome annotation (NCBI ID: CP009273.1, see code in GitHub: https://github.com/novoalab/NanoConsensus/Nano3Pseq_analysis/) using minimap2 v.2.17 with -ax map-ont -k14 parameters. Per-transcript abundance estimates were obtained from the aligned reads using salmon quant (version 1.9.0)97 with the parameters --ont -l SR. Both per-gene counts (using either –stranded and per-transcript estimates were used as input for differential expression analysis (Fig. 5C, see also Fig. S34).
Differential expression and GO term enrichment analysis
Differential expression analysis was performed both on per-gene and per-transcript counts of untreated samples compared to antibiotic-treated samples (streptomycin or kasugamycin) using DeSeq2 v.1.34.098. Genes with an absolute log2(Fold Change) greater or equal than 2 and an adjusted p-value lower or equal than 0.05 were considered as differentially expressed. Results were visualized using the R package EnhancedVolcano v.1.12.099 (Fig. 5C and Fig. S34). Scripts used for differential expression analysis are available in GitHub: https://github.com/novoalab/NanoConsensus/scripts/Differential_Expression). Genes with absolute log2(Fold Change) greater or equal than 2 were used as input for a GO term enrichment analysis, which was performed using Panther (release 17.0)100. All genes included in the differential expression analysis were included as the reference list for this analysis. The annotation data set used was GO biological process complete, the statistical test was the Fisher’s Exact Test, followed by calculating the False Discovery Rate (FDR). Only terms with a FDR < 0.05 were included in the final results (Fig. 5D).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The FAST5 data generated in this study have been deposited in the European National Archive (ENA) database under accession codes PRJEB42568 [https://www.ebi.ac.uk/ena/browser/view/PRJEB42568] (E. coli total RNA DRS samples from different strains), PRJEB48806 (E. coli total RNA DRS samples exposed to antibiotics), PRJEB58640 [https://www.ebi.ac.uk/ena/browser/view/PRJEB58640] (E. coli Nano3P-seq ribodepleted total RNA samples) and PRJEB78295 [https://www.ebi.ac.uk/ena/browser/view/PRJEB78295] (NSUN5 KO datasets). Base-called FAST5 files from yeast total RNA correspond to ENA under accession PRJEB37798101. A description of all samples used in this work with their corresponding ENA numbers can be found in Supplementary Data S2-S4. Source data are provided with this paper.
Code availability
NanoConsensus code and documentation is publicly available in GitHub (https://github.com/novoalab/NanoConsensus) and in Zenodo (https://doi.org/10.5281/zenodo.5805806) Moreover, to facilitate its implementation by future users, NanoConsensus has been integrated as a module into the MasterOfPores61 Nextflow102 workflow for the analysis of direct RNA nanopore sequencing data (https://github.com/biocorecrg/MoP2), under version 2.0 of MasterOfPores.
Change history
09 January 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41467-025-55846-6
References
Crofts, T. S., Gasparrini, A. J. & Dantas, G. Next-generation approaches to understand and combat the antibiotic resistome. Nat. Rev. Microbiol. 15, 422–434 (2017).
Arenz, S. & Wilson, D. N. Bacterial protein synthesis as a target for antibiotic inhibition. Cold Spring Harb. Perspect. Med. 6, a025361 (2016).
Sutcliffe, J. A. Antibiotics in development targeting protein synthesis. Ann. N. Y. Acad. Sci. 1241, 122–152 (2011).
Ennis, H. L. in The Quarterly Review of Biology (eds Gale, E. F., Cundliffe, E., Reynolds, P. E., Richmond, M. H. & Waring, M. J.). 336 (The University of Chicago Press, 1974).
Shandrick, S. et al. Monitoring molecular recognition of the ribosomal decoding site. Angew. Chem. Int. Ed. Engl. 43, 3177–3182 (2004).
Garneau-Tsodikova, S. & Labby, K. J. Mechanisms of resistance to aminoglycoside antibiotics: overview and perspectives. Medchemcomm 7, 11–27 (2016).
Ramirez, M. S. & Tolmasky, M. E. Aminoglycoside modifying enzymes. Drug Resist Updat. 13, 151–171 (2010).
Jackson, J., Chen, C. & Buising, K. Aminoglycosides: how should we use them in the 21st century? Curr. Opin. Infect. Dis. 26, 516 (2013).
Conn, G. L., Savic, M. & Macmaster, R. Antibiotic resistance in bacteria through modification of nucleosides in 16S ribosomal RNA. DNA and RNA Modification Enzymes: Comparative Structure, Mechanism, Functions, Cellular Interactions and Evolution. Landes Bioscience, Austin, TX 524–536 (2009).
Sergiev, P. V., Aleksashin, N. A., Chugunova, A. A., Polikanov, Y. S. & Dontsova, O. A. Structural and evolutionary insights into ribosomal RNA methylation. Nat. Chem. Biol. 14, 226–235 (2018).
Sigmund, C. D., Ettayebi, M. & Morgan, E. A. Antibiotic resistance mutations in 16S and 23S ribosomal RNA genes of Escherichia coli. Nucleic Acids Res. 12, 4653–4664 (1984).
Ofengand, J. & Del Campo, M. Modified nucleosides of Escherichia coli ribosomal RNA. EcoSal Plus 1, (2004).
Kehrenberg, C., Schwarz, S., Jacobsen, L., Hansen, L. H. & Vester, B. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol. Microbiol. 57 1064–1073 (2005).
Toh, S.-M. & Mankin, A. S. An indigenous posttranscriptional modification in the ribosomal peptidyl transferase center confers resistance to an array of protein synthesis inhibitors. J. Mol. Biol. 380, 593–597 (2008).
Weisblum, B. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39, 577–585 (1995).
Lai, C. J. & Weisblum, B. Altered methylation of ribosomal RNA in an erythromycin-resistant strain of Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 68, 856–860 (1971).
Sparling, P. F. Kasugamycin resistance: 30S ribosomal mutation with an unusual location on the Escherichia coli chromosome. Science 167, 56–58 (1970).
Vila-Sanjurjo, A., Squires, C. L. & Dahlberg, A. E. Isolation of kasugamycin resistant mutants in the 16 S ribosomal RNA of Escherichia coli. J. Mol. Biol. 293, 1–8 (1999).
Okamoto, S. et al. Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol. Microbiol. 63, 1096–1106 (2007).
Helm, M. & Motorin, Y. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet. 18, 275–291 (2017).
Jonkhout, N. et al. The RNA modification landscape in human disease. RNA 23, 1754–1769 (2017).
Alfonzo, J. D. et al. A call for direct sequencing of full-length RNAs to identify all modifications. Nat. Genet. 53, 1113–1116 (2021).
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Dominissini, D. et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 530, 441–446 (2016).
Carlile, T. M. et al. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515, 143–146 (2014).
Sas-Chen, A. et al. Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping. Nature https://doi.org/10.1038/s41586-020-2418-2 (2020).
Schwartz, S. et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162 (2014).
Marchand, V. et al. AlkAniline-Seq: profiling of m7 G and m3 C RNA modifications at single nucleotide resolution. Angew. Chem. Int. Ed. Engl. 57, 16785–16790 (2018).
Marchand, V., Blanloeil-Oillo, F., Helm, M. & Motorin, Y. Illumina-based RiboMethSeq approach for mapping of 2′-O-Me residues in RNA. Nucleic Acids Res. 44, e135–e135 (2016).
Dai, Q. et al. Nm-seq maps 2′-O-methylation sites in human mRNA with base precision. Nat. Methods 14, 695–698 (2017).
Delatte, B. et al. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science 351, 282–285 (2016).
Lovejoy, A. F., Riordan, D. P. & Brown, P. O. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One 9, e110799 (2014).
Li, X. et al. Base-resolution mapping reveals distinct m1A methylome in nuclear- and mitochondrial-encoded transcripts. Mol. Cell 68, 993–1005.e9 (2017).
Edelheit, S., Schwartz, S., Mumbach, M. R., Wurtzel, O. & Sorek, R. Transcriptome-Wide Mapping of 5-methylcytidine RNA Modifications in Bacteria, Archaea, and Yeast Reveals m5C within Archaeal mRNAs. https://doi.org/10.1371/journal.pgen.1003602 (2013).
Suzuki, T., Ueda, H., Okada, S. & Sakurai, M. Transcriptome-wide identification of adenosine-to-inosine editing using the ICE-seq method. Nat. Protoc. 10, 715–732 (2015).
Motorin, Y. & Helm, M. Methods for RNA modification mapping using deep sequencing: established and new emerging technologies. Genes 10, 35 (2019).
Novoa, E. M., Mason, C. E. & Mattick, J. S. Charting the unknown epitranscriptome. Nat. Rev. Mol. Cell Biol. 18, 339–340 (2017).
Anreiter, I., Mir, Q., Simpson, J. T., Janga, S. C. & Soller, M. New twists in detecting mRNA modification dynamics. Trends Biotechnol. 39, 72–89 (2021).
Zheng, H.-X., Zhang, X.-S. & Sui, N. Advances in the profiling of N6-methyladenosine (m6A) modifications. Biotechnol. Adv. 45, 107656 (2020).
Grozhik, A. V. et al. Antibody cross-reactivity accounts for widespread appearance of m1A in 5′UTRs. Nat. Commun. 10, 5126 (2019).
Garalde, D. R. et al. Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 15, 201–206 (2018).
Smith, A. M., Jain, M., Mulroney, L., Garalde, D. R. & Akeson, M. Reading canonical and modified nucleobases in 16S ribosomal RNA using nanopore native RNA sequencing. PLoS One 14, e0216709 (2019).
Begik, O. et al. Quantitative profiling of pseudouridylation dynamics in native RNAs with nanopore sequencing. Nat. Biotechnol. https://doi.org/10.1038/s41587-021-00915-6 (2021).
Wick, R. R., Judd, L. M. & Holt, K. E. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 20, 129 (2019).
Furlan, M. et al. Computational methods for RNA modification detection from nanopore direct RNA sequencing data. RNA Biol. 18, 31–40 (2021).
Wan, Y. K., Hendra, C., Pratanwanich, P. N. & Göke, J. Beyond sequencing: machine learning algorithms extract biology hidden in Nanopore signal data. Trends Genet. https://doi.org/10.1016/j.tig.2021.09.001 (2021).
Liu, H., Begik, O. & Novoa, E. M. EpiNano: detection of mA RNA modifications using oxford nanopore direct RNA sequencing. Methods Mol. Biol. 2298, 31–52 (2021).
Jenjaroenpun, P. et al. Decoding the epitranscriptional landscape from native RNA sequences. Nucleic Acids Res. 49, e7 (2021).
Liu, H., et al. Accurate detection of m6A RNA modifications in native RNA sequences. Nat. Commun. 10, 4079 (2019)
Parker, M. T. et al. Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m6A modification. Elife 9, e49658 (2020).
Price, A. M. et al. Direct RNA sequencing reveals m 6 A modifications on adenovirus RNA are necessary for efficient splicing. Nat. Commun. 11, 1–17 (2020).
Parker, M. T., Barton, G. J. & Simpson, G. G. Yanocomp: robust prediction of m6A modifications in individual nanopore direct RNA reads. bioRxiv 2021.06.15.448494 https://doi.org/10.1101/2021.06.15.448494 (2021).
Pratanwanich, P. N. et al. Identification of differential RNA modifications from nanopore direct RNA sequencing with xPore. Nat. Biotechnol. https://doi.org/10.1038/s41587-021-00949-w (2021).
Ueda, H. nanoDoc: RNA modification detection using Nanopore raw reads with Deep One-Class Classification. bioRxiv (2020).
Gao, Y. et al. Quantitative profiling of N6-methyladenosine at single-base resolution in stem-differentiating xylem of Populus trichocarpa using Nanopore direct RNA sequencing. Genome Biol. 22, 22 (2021).
Stoiber, M. et al. De novo Identification of DNA Modifications Enabled by Genome-Guided Nanopore Signal Processing. Cold Spring Harbor Laboratory 094672 https://doi.org/10.1101/094672 (2017).
Leger, A., Amaral, P. P., Pandolfini, L. & Capitanchik, C. RNA modifications detection by comparative Nanopore direct RNA sequencing. BioRxiv (2019).
Hendra, C. et al. Detection of m6A from direct RNA sequencing using a Multiple Instance Learning framework. bioRxiv 2021.09.20.461055 https://doi.org/10.1101/2021.09.20.461055 (2021).
Acera Mateos, P. et al. Identification of m6A and m5C RNA modifications at single-molecule resolution from Nanopore sequencing. bioRxiv 2022.03.14.484124 https://doi.org/10.1101/2022.03.14.484124 (2022).
Simpson, J. T. et al. Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods 14, 407–410 (2017).
Cozzuto, L. et al. MasterOfPores: a workflow for the analysis of Oxford nanopore direct RNA sequencing datasets. Front. Genet. 11, 211 (2020).
Blair, J. M. A., Webber, M. A., Baylay, A. J., Ogbolu, D. O. & Piddock, L. J. V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51 (2015).
Inoue, K., Basu, S. & Inouye, M. Dissection of 16S rRNA methyltransferase (KsgA) function in Escherichia coli. J. Bacteriol. 189, 8510–8518 (2007).
Bailey, A. D. et al. Concerted modification of nucleotides at functional centers of the ribosome revealed by single-molecule RNA modification profiling. Elife 11, e76562 (2022).
Lorenz, D. A., Sathe, S., Einstein, J. M. & Yeo, G. W. Direct RNA sequencing enables m6A detection in endogenous transcript isoforms at base-specific resolution. RNA 26, 19–28 (2020).
Huang, S. et al. Interferon inducible pseudouridine modification in human mRNA by quantitative nanopore profiling. Genome Biol. 22, 330 (2021).
Tavakoli, S. et al. Semi-quantitative detection of pseudouridine modifications and type I/II hypermodifications in human mRNAs using direct long-read sequencing. Nat. Commun. 14, 334 (2023).
Hassan, D., Acevedo, D., Daulatabad, S. V., Mir, Q. & Janga, S. C. Penguin: a tool for predicting pseudouridine sites in direct RNA nanopore sequencing data. Methods 203, 478–487 (2022).
Nguyen, T. A. et al. Direct identification of A-to-I editing sites with nanopore native RNA sequencing. Nat. Methods 19, 833–844 (2022).
Stephenson, W. et al. Direct detection of RNA modifications and structure using single-molecule nanopore sequencing. Cell Genom 2, 100097 (2022).
Loman, N. J., Quick, J. & Simpson, J. T. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods 12, 733–735 (2015).
Begik, O., Mattick, J. S. & Novoa, E. M. Exploring the epitranscriptome by native RNA sequencing. RNA https://doi.org/10.1261/rna.079404.122 (2022).
Acinas, S. G., Marcelino, L. A., Klepac-Ceraj, V. & Polz, M. F. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 186, 2629–2635 (2004).
Hillebrand, A., Wurm, R., Menzel, A. & Wagner, R. The seven E. coli ribosomal RNA operon upstream regulatory regions differ in structure and transcription factor binding efficiencies. Biol. Chem. 386, 523–534 (2005).
Kurylo, C. M. et al. Endogenous rRNA sequence variation can regulate stress response gene expression and phenotype. Cell Rep. 25, 236–248.e6 (2018).
Condon, C., Philips, J., Fu, Z. Y., Squires, C. & Squires, C. L. Comparison of the expression of the seven ribosomal RNA operons in Escherichia coli. EMBO J. 11, 4175–4185 (1992).
Watson, M., Liu, J.-W. & Ollis, D. Directed evolution of trimethoprim resistance in Escherichia coli. FEBS J. 274, 2661–2671 (2007).
Begik, O. et al. Nano3P-seq: transcriptome-wide analysis of gene expression and tail dynamics using end-capture nanopore cDNA sequencing. bioRxiv 2021.09.22.461331 https://doi.org/10.1101/2021.09.22.461331 (2022).
Zhong, Z.-D. et al. Systematic comparison of tools used for m6A mapping from nanopore direct RNA sequencing. Nat. Commun. 14, 1906 (2023).
Maestri, S. et al. Benchmarking of computational methods for m6A profiling with Nanopore direct RNA sequencing. Brief. Bioinform. 25, bbae001 (2024).
Cozzuto, L., Delgado-Tejedor, A., Hermoso Pulido, T., Novoa, E. M. & Ponomarenko, J. Nanopore direct RNA sequencing data processing and analysis using MasterOfPores. Methods Mol. Biol. 2624, 185–205 (2023).
Yuen, Z. W.-S. et al. Systematic benchmarking of tools for CpG methylation detection from nanopore sequencing. Nat. Commun. 12, 3438 (2021).
Carter, A. P. et al. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407, 340–348 (2000).
Schuwirth, B. S. et al. Structural analysis of kasugamycin inhibition of translation. Nat. Struct. Mol. Biol. 13, 879–886 (2006).
Kaberdina, A. C., Szaflarski, W., Nierhaus, K. H. & Moll, I. An unexpected type of ribosomes induced by kasugamycin: a look into ancestral times of protein synthesis? Mol. Cell 33, 227–236 (2009).
Doi, Y., Wachino, J. & Arakawa, Y. Aminoglycoside Resistance: The Emergence of Acquired 16S Ribosomal RNA Methyltransferases. Infect. Dis. Clin. North Am. 30, 523–537 (2016).
Vester, B. The cfr and cfr-like multiple resistance genes. Res. Microbiol. 169, 61–66 (2018).
Toh, S.-M. et al. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol. Microbiol. 64, 1506–1514 (2007).
Bijpuria, S., Sharma, R. & Taneja, B. Loss of U1498 methylation in 16S rRNA by RsmE methyltransferase associates its role with aminoglycoside resistance in mycobacteria. J. Glob. Antimicrob. Resist 23, 359–369 (2020).
Baba, T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006).
Smith, M. A. et al. Molecular barcoding of native RNAs using nanopore sequencing and deep learning. Genome Res. 30, 1345–1353 (2020).
Vohra, D. Apache Parquet. in Practical Hadoop Ecosystem 325–335 (Apress, 2016).
Ryan, D., Grüning, B. & Ramirez, F. pyBigWig 0.2.4. https://doi.org/10.5281/zenodo.45238 (2016).
Sprouffske, K. & Wagner, A. Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinforma. 17, 1–4 (2016).
GitHub - artic-network/readucks: Nanopore read de-multiplexer. GitHub https://github.com/artic-network/readucks.
Anders, S., Pyl, P. T. & Huber, W. HTSeq - A Python framework to work with high-throughput sequencing data. bioRxiv https://doi.org/10.1101/002824 (2014) .
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Mi, H., Muruganujan, A., Ebert, D., Huang, X. & Thomas, P. D. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, D419–D426 (2019).
Begik, O. et al. Quantitative profiling of pseudouridylation dynamics in native RNAs with nanopore sequencing. Nat. Biotechnol. 39, 1278–1291 (2021).
Di Tommaso, P. et al. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 35, 316–319 (2017).
Acknowledgements
We thank all the members of the Novoa lab for their insightful discussions. We thank Prof. Xavier Barril for his discussions and help with Pymol to obtain the structural alignments of PDB structures of ribosomes from different species containing diverse antibiotics. AD-T is supported by an FPI Severo-Ochoa fellowship by the Spanish Ministry of Economy, Industry and Competitiveness (MEIC). The work of S.B. work was supported by the Spanish Ministry of Science and Innovation MCIN/AEI/10.13039/501100011033 and by “ERDF A way of making Europe” (PID2019-111692RB-I00 and PID2022-139598OB-I00), and The Scientific Foundation AECC AECC (LABAE19040BLAN). J.L-L. was supported by the AECC (PRDSA19002LÓPE) and MIU (FPU19/01190). This work was supported by funds from the Spanish Ministry of Economy, Industry and Competitiveness (MEIC) (PID2021-128193NB-100 to EMN) and the European Research Council (ERC-StG-2021 No 101042103 to EMN). We acknowledge the support of the MEIC to the EMBL partnership, Centro de Excelencia Severo Ochoa and CERCA Program / Generalitat de Catalunya.
Author information
Authors and Affiliations
Contributions
A.D.-T. performed most wet lab experiments and data analyses described in this work, including the development, implementation and benchmarking of NanoConsensus. R.M. helped with the E. coli growth curve experiments. O.B. helped with preparing and sequencing some of the direct RNA libraries used in this work. L.C. implemented NanoConsensus into a Nextflow module and into the MasterOfPores general workflow. J.P. supervised the implementation of NanoConsensus into MasterOfPores. S.B. and J.L. generated NSUN5 KO mice and validated its phenotype. E.M.N. conceived and supervised the work. A.D.-T. built the figures. A.D.-T. and E.M.N. wrote the manuscript, with contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
E.M.N. is a member of the Scientific Advisory Board of IMMAGINA Biotech. E.M.N. has received travel and accommodation expenses to speak at Oxford Nanopore Technologies conferences. A.D.-T. and O.B. have received travel bursaries from ONT to present their work in conferences. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Delgado-Tejedor, A., Medina, R., Begik, O. et al. Native RNA nanopore sequencing reveals antibiotic-induced loss of rRNA modifications in the A- and P-sites. Nat Commun 15, 10054 (2024). https://doi.org/10.1038/s41467-024-54368-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-54368-x
This article is cited by
-
Direct detection of 8-oxo-dG using nanopore sequencing
Nature Communications (2025)
-
Nano3P-seq: charting the coding and noncoding transcriptome at single-molecule resolution
Nature Protocols (2025)