Abstract
Myeloid malignancies are heterogenous disorders characterized by distinct molecular drivers but share convergence of oncogenic signaling pathways and propagation by ripe pro-inflammatory niches. Here, we establish a comprehensive transcriptional atlas across the spectrum of myeloproliferative neoplasms (MPN) and secondary acute myeloid leukemia (sAML) through RNA-sequencing of 158 primary samples encompassing CD34+ hematopoietic stem/progenitor cells and CD14+ monocytes. Supported by mass cytometry (CyTOF) profiling, we reveal aberrant networks of PI3K/AKT/mTOR signalling and NFκB-mediated hyper-inflammation. Combining ATAC-Seq, CUT&Tag, RNA-seq, and CyTOF, we demonstrate that targeting of ribosomal protein S6 kinase A1 (RSK1) suppresses NFκB activation and diminishes pro-inflammatory mediators including tumor necrosis factor (TNF) associated with MPN disease severity and transformation. We further evaluate a therapeutic approach utilizing a first-in-class RSK inhibitor, PMD-026, currently in Phase 2 development for breast cancer, for use in myeloid malignancies. Treatment with PMD-026 suppressed disease burden across seven syngeneic and patient-derived xenograft leukemia mouse models spanning the spectrum of driver and disease-modifying mutations. These findings uncover a therapeutic avenue for a conserved dependency across MPN and sAML.
Similar content being viewed by others
Introduction
Myeloproliferative neoplasms (MPNs), chronic myelomonocytic leukemia (CMML), and acute myeloid leukemia (AML) exist among a spectrum of myeloid malignancies. These clonal disorders are driven by canonical genetic alterations such as in JAK2, MPL, or CALR for MPNs and in NPM1, DNMT3A, and FLT3 for de novo AML, but also share mutated disease modifiers including in TP53, IDH1/2, NRAS, chromatin modifiers ASXL1, EZH2, and TET2, and spliceosome factors SRSF2 and SF3B11,2,3.
Current therapeutic approaches primarily targeting JAK2 (e.g., ruxolitinib) in MPNs demonstrate symptom control but are not curative4,5. The most dismal complication of chronic MPNs is transformation to secondary AML (sAML), which, other than allogenic stem cell transplant, has no reliable alternative therapy, as the emergence of malignant subclones is more aggressive and often no longer dependent on JAK-STAT signaling6,7. Recent technological and genomic advances in single-cell RNA and DNA sequencing have enabled greater resolution into the lens of dysfunctional clonal hematopoiesis8,9, but concerted efforts required of larger datasets that encompass the spectrum of the disease remain necessary to assign significance and context in malignant progression.
Furthermore, disease progression and transformation are multifactorial and involve the interplay of somatic mutation accumulation (and their functional transcriptional phenotype) propagated by a ripe pro-inflammatory niche10,11,12,13. Indeed, the presence of a pro-inflammatory cytokine/chemokine profile is a shared hallmark among myeloid malignancies often denoted by heightened expression of mediators such as TNF, IL-6, and IL-8 and hyperactive transduction through the NFκB pathway11,14,15. CD14 + monocytes are a potent and key population conducive to inflammatory dysfunction as they respond to microenvironmental agonists, stimulants, and cues through activation of the p65/RELA cascade and consequently mobilizing a reactive and cyclic transcriptional network16,17,18. As such, therapeutic strategies aimed to dampen this response may prove efficacious to drought in this fertile “soil”.
RSK1 (RPS6KA1; p90RSK; ribosomal protein S6 kinase A1) is an ERK-activated kinase with canonical function for propagating S6 signaling in addition to other effectors including NFκB and CREB19,20,21. We previously identified that the DUSP6-RSK1-S6 axis is an important driver of MPN transformation to sAML22. RSK1 was also indispensable for leukemic survival and its elevated expression in AML patients was associated with worse survival outcomes14. Current pre-clinical pan-RSK inhibitors targeting RSK1, such as FMK, LJI308, and BI-D1870, demonstrate efficacy in cell line models, however, their poor pharmacokinetic profiles limit further evaluation across in vivo and clinical studies23,24. Recently, an RSK1-4 kinase inhibitor, PMD-026, has been developed displaying promising pharmacokinetic profiles and potent on-target efficacy in patient-derived xenograft (PDX) breast cancer models25,26. This first-in-class oral RSK inhibitor is currently being evaluated in phase 1/1b/2 clinical trials (NCT04115306) in patients with metastatic breast cancer, targeting aberrant expression of RSK2. These observations provide a strong rationale for the evaluation of RSK1 as an exploitable driver in myeloid malignancies.
Here, we show shared dependency of RSK1 across MPN, secondary AML, and chronic myelomonocytic leukemia, in propagating disease progression and transformation. Pharmacological targeting of RSK with PMD-026 suppresses oncogenic mTOR/AKT/S6 signaling and NFκB-mediated inflammation and demonstrates potent efficacy across myeloid mouse models reflecting a spectrum of driver and high-risk disease modifying mutations.
Results
The landscape of MPN/sAML CD34 + HSPCs
We first characterized the mutational landscape of 73 chronic-phase MPN and post-MPN sAML patients treated at Washington University School of Medicine. 39/73 patients underwent next-generation sequencing with a myeloid gene panel, and 34/73 underwent JAK2, CALR, and/or MPL testing by targeted sequencing (Fig. 1a and Supplementary Fig. 1a). All (26/26) sAML patients retained MPN driver mutations. Mutations observed in TET2, ASXL1, and TP53, in addition to spliceosome factors SRSF2, SF3B1, and ZRSR2 were consistent with the current mutational landscape of MPNs and sAML7. In comparing hotspot mutations in sAML to de novo leukemia cohorts, TCGA LAML and BeatAML2, there was increased prevalence of ASXL1 (23%, 2.5%, 10.3%), TP53 (23%, 8%, 9.2%), and SF3B1 (11.5%, 0.5%, 5.4%; Fig. 1b). In contrast, mutations in DNMT3A, FLT3, and NPM1 were rarely observed.
a Oncoprint of MPN/sAML samples in the WashU Cohort CD34 + RNA-seq. b Comparison of genetic alterations in WashU sAML (n = 34) and additional non-myeloproliferative neoplasm AML cohorts, TCGA (n = 200) and BeatAML2 (n = 893). c Schematic of the WashU Cohort CD34 + RNA-seq encompassing 90 primary samples representative of PV (n = 6), ET (n = 9), MF (n = 30), sAML (n = 34) and NBM (n = 11). d Principal component analysis of primary samples in the WashU Cohort CD34 + RNA-seq by gene expression. e Boxplots showing expression of key genes mediating oncogenic signaling through the mTOR/PI3K/AKT and receptor tyrosine kinase/RAS/RAF pathways across the WashU Cohort by disease. Statistics assessed by a two-tailed student’s t test. † denotes the comparison between MF and NBM. ‡ denotes the comparison between sAML and NBM. Sample numbers are denoted in panel (c). Boxplot showing median, upper, and lower quartiles with whiskers representing highest and lowest values excluding outliers, and potential outliers. f Schematic of mass cytometry analysis of CD34 + cells from the WashU Cohort encompassing 106 primary samples representative of PV (n = 17), ET (n = 5), MF (n = 34), sAML (n = 15) and normal healthy donors (n = 35). g Left: Heatmap of phosphorylated and total protein expression by mass cytometry. Values from patient samples were normalized to values from the normal control from each CyTOF run and are denoted as 90th percentile arcsinh ratios. Right: Boxplots showing phosphorylation of key protein by disease. Statistics assessed by a two-tailed student’s t test. † denotes the comparison between MF and NBM. ‡ denotes the comparison between sAML and NBM. Sample numbers are denoted in panel (f). Boxplot showing median, upper, and lower quartiles with whiskers representing highest and lowest values excluding outliers, and potential outliers.
From this cohort, we then investigated the underlying transcriptional landscape and performed bulk RNA-sequencing (RNA-seq) on sorted CD34 + hematopoietic stem/progenitor cells (HSPCs) from 45 chronic-phase MPN patients (6 polycythemia vera; PV, 9 essential thrombocythemia; ET, and 30 myelofibrosis; MF), 34 post-MPN sAML patients, and 11 healthy, normal bone marrow (NBM) donors to establish the WashU Cohort (Fig. 1c and Supplementary Data 1). Principal component analysis (PCA) revealed a gradient of disease progression from NBM/PV/ET to MF to sAML (Fig. 1d). Oncogenes that mediated PC trajectory included RICTOR and AKT3, and we observed further upregulated expression of other mTOR/PI3K/AKT (PIK3CA, PIK3C2A) and RAS/RAF (KRAS, BRAF) pathway effectors (Fig. 1e). Despite myeloid malignancies not commonly harboring hotspot mutations in these drivers compared to solid tumors, the increased transcript levels identified here may suggest similar pathway hyperactivity. In addition, despite a relatively low percentage of patients harboring TP53 mutations, MPN and sAML patients consistently demonstrated lower TP53 expression (Supplementary Fig. 1b). Consistent with our prior studies22, we also identified heightened FLT3 expression in sAML compared to MPN. PDK1 was also significantly elevated in sAML, which was also identified in de novo AML27. Further differential expressed gene (DEG) and gene set enrichment (GSE) analyses across the various disease subgroups revealed enrichment in JAK-STAT and TNF/NFκB signaling pathways observed in MPN/sAML HSPCs compared to NBM (Supplementary Fig. 1c–e and Supplementary Data 2). Lastly, we established an interactive browser that hosts this WashU RNA-seq data and allows for further exploration and DEG analysis.
To corroborate transcriptional enrichment of these oncogenic pathways, we performed signaling suspension mass cytometry (CyTOF) and compiled these results with our previous assays17,22 (Fig. 1f). In the CD34 + population, we observed consistent hyperactivation of signaling pathways including JAK-STAT, PI3K/AKT/mTOR/S6, NFκB, and MAPK across the spectrum of MPN and especially evident in sAML (Fig. 1g). These data reveal conserved dynamics governing malignant hematopoiesis and transformation, highlighting overexpression and hyperactivity of oncogene-driven signaling pathways that may inform rational therapeutic targeting.
RSK1 inhibition is efficacious in MPN
RSK family kinases are activated by RTKs and are at the convergence of the RAS/MEK/ERK and PI3K/AKT/mTOR pathways which thereby serve as central mediators of these hyperactive signaling pathways (Fig. 2a). We previously identified RPS6KA1 to be preferentially elevated in AML22 across 31 distinct cancer subtypes from the TCGA Pan-Cancer cohort and whose inhibition was most lethal following CRISPR–Cas9-mediated knockout across 804 pan-cancer cell lines in DepMap. As such, RSKs are potentially attractive therapeutic targets warranting further preclinical evaluation.
a Schematic of RSK1 as a key node along canonical oncogenic signaling pathways. b Pearson correlations of pRSK1 T573 and pRSK1 T359/S363 with other phosphorylated proteins (total n = 54 phosphorylated proteins) from RPPA data from 34 AML cell lines from CCLE. c KINOMEscan of PMD-026 across 398 kinases identifying specificity against RSK1-4. d Cell viability assay of HEL cells treated with RSK inhibitors BI-D1970 and PMD-026 at the indicated drug doses. Cells were treated for 5 days and with six biological replicates. Data are presented as mean values ± SD. e Immunoblot of HEL cells treated with 5 μM,10 μM PMD-026, or control for 48 h. f Cell viability assay of Ba/F3 cells ectopically expressing wild-type or mutant JAK2 and MPL treated with PMD-026 at the indicated drug doses. Cells were treated for 5 days and with six biological replicates. Data are presented as mean values ± SD. g Annexin V assay of HEL cells treated with 5 μM,10 μM PMD-026, or control for 48 h and 72 h. Cells were treated in triplicate (n = 3 independent experiments). Data are presented as mean values ± SD. h Cell cycle assay of HEL cells treated with 5 μM, 10 μM PMD-026, or control for 24 h and 48 h. Cells were treated in triplicate (n = 3 independent experiments). Data are presented as mean values ± SD. i Top altered Hallmark pathways by enrichment analysis from top 500 DEGs from RNA-seq analysis of HEL cells treated with 10 μM PMD-026 compared to control for 24 h. Gene count represents the number of genes enriched across each pertinent gene set. Gene ratio represents the number of genes enriched/total number of genes in each gene set. j Colony counts from CD34 + colony assays. Sorted CD34 + cells from five patients were seeded into MethoCult H4034 in indicated doses of PMD-026 at micromolar concentration. Cells were plated in triplicate per condition (n = 3 biological replicates). Colonies were enumerated after 14 days. Data are presented as mean values ± SD. k Schematic of the CD34 + 784981 sAML pMF patient-derived xenograft mouse model. Two shRNAs targeting RPS6KA1 or control vector were ectopically expressed in isolated CD34 + cells after which transformed cells were transplanted into NSGS mice by intra-tibial injection into the bone marrow and followed for 12 weeks. l Percentage of human CD45 (hCD45) in the peripheral blood and bone marrow of transplanted mice ectopically expressing control (n = 4), shRPS6KA1 #1 (sh #1; n = 5), or shRPS6KA1 #2 (sh #2; n = 5) across multiple timepoints, and spleen and liver weights of mice at endpoint normalized by mouse weight. %hCD45 in PB statistics assessed by two-way ANOVA with Dunnett’s multiple comparisons test with control. %hCD45 in BM, and normalized spleen and liver weights statistics were assessed by one-way ANOVA with Dunnett’s multiple comparisons test with control. Data are presented as mean values +/− SD.
In addition to the elevated oncogenic signatures observed at the transcriptome and phosphoprotein level, we also observed an elevated Reactome RSK activation signature in sAML compared to normal bone marrow CD34 + cells (Supplementary Fig. 2a). To further establish an RSK1 signaling network, we analyzed 54 phosphorylated effectors from reverse phase protein array (RPPA) data of 34 AML cell lines from the Cancer Cell Line Encyclopedia28 (CCLE) and examined correlations with pRSK1 T359/S363 and pRSK1 T573 (Fig. 2b). We observed correlations of both pRSK1 markers with effectors along the mTOR cascade in p-mTOR, pS6, and p4E-BP1. RSK1 is known to be phosphorylated by ERK1/2 at T573 (correlations with pERK1/2, pMEK1/2); correlations of pRSK1 T359/S363 with SRC were also observed. These data highlight RSK1 as a central node coordinating oncogenic signal transduction in MPNs and AML.
BI-D1870 is one of the pre-clinical RSK1-4 inhibitors, which we have previously demonstrated to induce apoptosis and G2/M cell arrest in JAK2 V617F-mutant cell lines HEL and UKE-122. However, due to the in vivo limitations of BI-D1870, we utilized another small molecule compound targeting RSK family kinases, PMD-026, that harbors promising pharmacokinetic profiles and is formally under clinical evaluation for breast cancer. We performed a KINOMEscan across 398 kinases that demonstrated 99% specificity of PMD-026 for RSK1-4 with minimal inhibition of other kinases (Fig. 2c). A subsequent counter-screen was performed using an 11-concentration curve to establish IC50 values of 2 nM (RSK1), 0.7 nM (RSK2), 0.9 nM (RSK3) and 2 nM (RSK4) (Supplementary Fig. 2b). In short-term viability assays, PMD-026 demonstrated similar potency as BI-D1870 (Fig. 2d). PMD-026 also potently suppressed pRSK1, pS6, and p-mTOR (Fig. 2e), further establishing RSK1 as an important signaling propagator. Recent studies have identified splicing factor YB-1 (YBX1) to be an important mediator of JAK2 inhibitor persistence in MPNs and a dependency in AML29,30. YB-1 is also a direct substrate activated by RSK131, whose activity was inhibited by PMD-026 in HEL cells (Fig. 2e). Since PMD-026 is a pan RSK inhibitor, we employed lentiviral knockdown of RPS6KA1, in which genetic perturbation demonstrated similar signaling suppression as pharmacological inhibition (Supplementary Fig. 2c). Although suppressive effects on RSK2-4 could contribute to the observed anti-proliferative phenotype, these findings suggest on target RSK1 suppression by PMD-026. In support of this, other RSK family genes (RPS6KA2 (RSK3); RPS6KA2 (RSK2); RPS6KA6 (RSK4)) were not deemed essential in AML from DepMap32 (Supplementary Fig. 2d).
To further assess whether cells harboring MPN drivers would be preferentially sensitive to RSK inhibition, we established gain-of-function in vitro models by ectopically expressing wild-type (WT) and mutant JAK2 and MPL in Ba/F3 cells. Transduced cells demonstrated IL-3-independent proliferation and were preferentially sensitive to PMD-026 relative to parental Ba/F3 cells (Fig. 2f). Since PMD-026 does not mechanistically inhibit JAK2 or MPL, selectivity of proliferation inhibition between WT or mutant-transduced cells was not observed. However, hyperactivation of S6 signaling was evident upon transduction with these oncogenes, and acute PMD-026 treatment exhibited rapid suppression of S6 phosphorylation but not of STAT3 (Supplementary Fig. 2e) further highlighting both on-target effector suppression and preferential dependency of the RSK/S6 pathway.
Additional further profiling of PMD-026 demonstrated time-dependent induction of apoptosis and necroptosis (Fig. 2g), and cell cycle arrest in S and G2/M phases (Fig. 2h), consistent with BI-D1870 treatment22. We then performed RNA-seq and GSEA on HEL cells treated with PMD-026, which also revealed suppression of G2/M checkpoint and mitotic spindle pathways, and E2F targets at the transcriptional level (Fig. 2i and Supplementary Data 3). Amongst these top 500 DEGs included AURKA, PLK1, CCND1, CCND2, and MYC, in line with the observed cell cycle arrest phenotype. GSEA analysis also revealed alterations in mTORC1 signaling, which is consistent with our immunoblots. As these pathways can also promote malignant transformation, we sought to determine the capacity of RSK inhibition to revert sAML disease to a precursor/ancestral state. DEG analysis of CD34 + cells from the WashU Cohort revealed 1810 candidates with greater than 1 log2 fold change in mRNA expression in sAML compared to MF HSPCs, which we define as an sAML disease transformation signature (Supplementary Fig. 2f and Supplementary Data 2). This signature was significantly suppressed by both PMD-026 and BI-D1780 treatment (Supplementary Fig. 2g).
We next assessed the effects of RSK1 perturbation utilizing primary samples. Dose-dependent inhibition was observed in ex vivo CD34 + colony assays, where PMD-026 inhibited MF and sAML patient samples (Fig. 2j). We also transduced sorted CD34 + cells from post-MF sAML patient 784981 with two independent shRNAs targeting RPS6KA1 and performed colony formation assays. RPS6KA1 perturbation reduced the number and size of colonies in sAML but not in normal CD34 + cells (Supplementary Fig. 3a, b). Furthermore, we characterized the hematopoietic impact of targeting RSK family kinases across normal cord blood CD34 + cells. In colony assays, there were no significant alteration of total number of colonies following RPS6KA1, RPS6KA3, or RPS6KA2 knockdown (Supplementary Fig. 3c, d), and RPS6KA6 (RSK4) was lowly expressed in CD34 + cells (Supplementary Fig. 3e). These data demonstrate that RSK family genes may play distinct roles in hematopoietic lineage determination and importantly, targeting of RSK1 leads to significant anti-leukemic effects in disease relative to healthy tissue.
Lastly, we established a patient-derived xenograft (PDX) model in which CD34 + cells from sAML 784981 following RPS6KA1 knock-down were engrafted in NSGS mice by intra-tibial injection into the bone marrow. Animals were then monitored for 12 weeks to assess impact on disease burden (Fig. 2k). Remarkably, RPS6KA1 knockdown led to near complete elimination of human CD45-positive (hCD45 +) cells in both the peripheral blood and bone marrow and resulted in a trend of spleen and liver weight reduction (Fig. 2l). This essentiality recapitulates the vital dependency of RSK1 in AML in vivo and is consistent with the in vitro AML cell line DepMap CRISPR-KO data22, thereby validating RSK1 as a viable target for further clinical investigation.
The pro-inflammatory landscape of CD14+ monocytes in mediating MPN disease
The current paradigm of MPN disease progression highlights the role of underlying oncogenic drivers and sequential genetic alterations, with increasing evidence of critical contributions from a transformative pro-inflammatory milieu. We characterized inflammatory cytokine and chemokine profiles across the spectrum of MPN and sAML patients by combining plasma analyses from our previous studies18,33,34, overall encompassing PV (n = 37), MF (n = 107), and sAML (n = 6) patients and 30 age-matched healthy donors. Of the seven overlapping cytokines/chemokines common to all studies, six (interferon-gamma, IL-6, IL-8, IP-10/CXCL10, MIP-1α/CCL3, and TNF) increased with disease severity, while levels of VEGF were conserved (Fig. 3a). We also observed significant correlations in the pro-inflammatory cytokines between IL-6 with IL-8. IP-10/CXCL10, and MIP-1α/CCL3 in PV, and when profiling for additional cytokines, correlations with interferon-gamma, MIP-1β/CCL4, and TNF in MF samples (Supplementary Fig. 4a).
a Plasma cytokine expression across 180 samples representative of PV (n = 37), MF (n = 107), sAML (n = 6) patients, and healthy donors (n = 30). Statistics were assessed by a two-tailed Mann-Whitney U test. Data are presented as mean values +/− SD. b Schematic of the WashU Cohort CD14 + RNA-seq encompassing 68 primary samples representative of PV (n = 8), ET (n = 15), MF (n = 31), and NBM (n = 14). c Principal component analysis of primary samples in the WashU Cohort CD14 + RNA-seq by gene expression. d Volcano plot showing DEGs by DESeq2 analysis comparing MF to NBM CD14 + monocytes. Y-axis denoting -Log10 adj. p-value. X-axis denoting Log2 expression fold change. e Boxplot showing mRNA expression of genes of interest from the WashU Cohort CD14 + RNA-seq. Statistics assessed by two-tailed student’s t test between MF and NBM. Sample numbers are denoted in panel (b). Boxplot showing median, upper, and lower quartiles with whiskers representing highest and lowest values excluding outliers, and potential outliers. f Gene set enrichment analysis of top altered pathways comparing MF to NBM CD14 + monocytes. NES = normalized enrichment score. NOM p-value: Nominal p-value denoting the statistical significance of the enrichment score.
To interrogate if intracellular cytokine production was confined to specific populations within the peripheral blood and bone marrow compartments, we performed CyTOF on paired PB and BM samples from PV patient 760741 and a healthy donor (Supplementary Fig. 4b). We observed the predominant production of cytokines including IL-8, MIP-1β/CCL4, TNF, and TGFβ from a prevalent CD14 + monocyte population with greater production in PV than normal cells (Supplementary Fig. 4c–e). TGFβ was also highly produced by myeloid-derived suppressor cells (MDSCs; Supplementary Fig. 4e). These pro-inflammatory findings in monocytes from PV patients are consistent with our previous analyses in MF and sAML patients17,18.
Given these critical roles that monocytes play in disease progression, there remains a need for concerted profiling of these pro-inflammatory effectors across MPN subtypes. Thus, to better understand the underlying transcriptional landscape, we performed a second iteration of RNA-seq on paired CD34- CD14 + monocytes from the WashU Cohort encompassing PV (n = 8), ET (n = 15), and MF (n = 34) patients, and 14 healthy donors (Fig. 3b). Similar to the analysis of CD34 + cells, PCA revealed a gradient of disease progression in monocytes (Fig. 3c). We then performed DEG analysis between MF and normal CD14 + monocytes, which revealed elevated platelet markers PPBP, ITGB3, and PF4 markers, and downregulated lymphoid and erythroid markers DNTT, VPREB, various immunoglobulin transcripts, and GYPA (CD235a; Fig. 3d and Supplementary Data 4). In addition to most of these patients harboring mutations in JAK2 or CALR, there was also increased expression of these genes and upregulation of downstream effectors STAT1 and STAT5A. MPL mutations were only detected in one patient, but gene expression was universally elevated, especially in MF monocytes. These observations reiterate the importance of both the presence of gain-of-function driver mutations and increased expression. Notably, we also observed profound elevation of pro-inflammatory cytokines/chemokines such as TNF, IL6, CCL4, and CCL5, and NFκB pathway effectors including NFKB1, NFKB2, NFKBIE, RELA, RELB, and IRAK2 (Fig. 3e), consistent with elevated inflammatory plasma cytokines.
When comparing monocytes from the three different MPN disease phenotypes to those from healthy donors, 101 candidates shared across PV, ET, and MF were identified, whose roles primarily relate to coagulation, complement, and JAK-STAT and TNF/NFκB signaling (Supplementary Fig. 5a). Additional GSEA confirmed enrichment of interferon and inflammatory responses, and signaling through JAK-STAT and NFκB, with strongest enrichment seen in MF monocytes (Fig. 3f and Supplementary Fig. 5b). PV monocytes also exhibited greater heme metabolism and cell cycle potential relative to NBM, ET, and MF. We then examined CD14 + monocytes from our curated signaling CyTOF atlas which showed similar upregulated activity of JAK-STAT, NFκB, and S6 pathways across MPN and sAML samples compared to normal donors (Supplementary Fig. 5c, d), consistent with our RNA-seq analysis. Together with our CD34 + data, these sequencing and proteomic efforts establish a comprehensive transcriptional and signaling atlas across the spectrum of MPNs and provide insight into the loss of homeostatic function with simultaneous enhancement of oncogenic signaling and inflammatory capacity along disease progression.
To corroborate these bulk sequencing findings, we then performed high dimensional analyses on serial samples obtained from an MPN patient at early and late PV stages and at transformation to MF (Fig. 4a). CyTOF analyses showed expansion of Lin- CD34 + cells in the peripheral blood (0.3% to 1.1% to 3%; Fig. 4b). CD14 + monocytes demonstrated consistent elevation of inflammatory cytokines including TNF, IL-6 IL-8, MIP-1α/CCL3, and MIP-1β/CCL4 along disease progression and compared to normal monocytes (Fig. 4c, d). Cytokine production at the MF stage was also less inhibited by JAK2 inhibitor ruxolitinib treatment ex vivo relative to antecedent PV stages (Fig. 4e). Further CyTOF analysis of signaling pathways revealed elevated STAT and NFκB, heightened transcription and translation potential with increased CREB and p4E-BP1, and enhanced proliferative potential with increased Ki67 and decreased cleaved caspase 3 in both Lin- CD34+ cells and CD14 + monocytes at MF versus Early PV (Fig. 4f). Complementing these findings, we also performed single-cell RNA-seq (scRNA-seq) and observed similar upregulation of pro-inflammatory and signaling effectors in CD14 + monocytes (Fig. 4g, h and Supplementary Data 5). Given this mediating role of monocytes, we then interrogated intercellular communication networks with CellChat35. Relative to early PV, we observed augmented interaction networks particularly in CD14 + monocytes at the MF stage (Supplementary Fig. 6a, b). Using this approach, we observed significant inbound and outgoing signaling through TNF signaling in MF, which was both cell-autonomous and acting on other populations including HSPCs, as well as a stronger CCL3 network, overall suggesting an enhanced inflammatory response through disease transformation (Supplementary Fig. 6c–e). Additional trajectory modeling revealed similar bifurcation of CD14 + monocytes but not HSPCs by disease severity (Fig. 4i). Further pseudotime analysis highlighted increased expression of key mediators such as TNF, CCL3, and notably, RPS6KA1 through PV to MF transition (Fig. 4j and Supplementary Data 6). These data together delineate the distinct monocyte landscape characteristics in MPN and highlight their profound roles in establishing and enhancing a pro-inflammatory environment and mediating disease progression.
a Schematic of clinical information and sample collection of patient 551599 through disease transformation. b Cell population proportion of 551599 PBMC samples by CyTOF. c tSNE plots showing surface CD14 + protein and key cytokine expression of 551599 samples. d Percent positive cytokine expression in 551599 CD14 + monocytes across disease stages and healthy peripheral blood donor (NPB) control by CyTOF. e Fold change of cytokine expression in 551599 CD14 + monocytes following 4h ex vivo treatment with 5 μM ruxolitinib by CyTOF. f Expression of signaling proteins in 551599 CD34 + HSCs and CD14 + monocytes at MF pPV compared to Early PV by CyTOF. Values represent the median arcsinh ratio. g UMAPs of 551599 from scRNA-seq by disease state and cell identity. h Expression dot plot of key genes from 551599 across disease states by scRNA-seq. i Trajectory branching of HSPC and CD14 + monocyte populations. j Pseudotime trajectory analysis of TNF, CCL3, and RPS6KA1 expression across disease progression.
RSK inhibition dampens the monocyte-mediated pro-inflammatory milieu
The enrichment of RPS6KA1 through disease progression and linkage to mediators such as TNF suggest a contributory inflammatory role in pathogenesis. In our MPN scRNA-seq data, we observed high RPS6KA1 expression in antigen-presenting cells, including CD14 + and CD16 + monocytes, and dendritic cells, which was enriched through disease progression (Supplementary Fig. 7a, b). Similarly, by GSEA, we also found elevated JAK-STAT, TNF/NFκB, and inflammatory cytokine modules in RPS6KA1-high AML patients compared to RPS6KA1-low AML patients (Supplementary Fig. 7c, d). We further found that RPS6KA1 significantly correlated with RELA and NFKB1 in AML cell lines and patient samples at both mRNA and protein levels (Supplementary Fig. 7e–g). These observations, combined with the established hyperactive cytokine/chemokine profile and NFκB response in MPN/sAML patients provide a rationale that RSK1 may mediate NFκB pathway signaling and that targeting RSK1 may dampen these pro-inflammatory signatures.
There were significant correlations between RPS6KA1 and CD14 expression and with PB monocytes across the TCGA AML, BeatAML, and our WashU monocyte Cohorts (Fig. 5a, b), overall suggesting an important RSK1-mediated role in monocytes. To further examine this relationship, we sorted CD14 + monocytes from three MF patients, treated cells acutely with PMD-026 ex vivo, and then performed RNA-seq. GSEA of top 500 DEGs revealed TNF/NFκB, inflammatory response, and mTORC1 signaling pathways to be the most significantly altered after treatment (Fig. 5c). At the individual transcript level, there was profound suppression of cytokines and chemokines including TNF, IL1B, IL6, CCL3, CCL4, and CCL5 by PMD-026 (Fig. 5d and Supplementary Data 7). To validate our RNA-seq findings, we performed qRT-PCR analysis on CD14 + monocytes from seven additional MF patients treated with PMD-026 ex vivo which revealed consistently suppressed transcription of key pro-inflammatory mediators (Fig. 5e). PMD-026 also inhibited transcription of NFKB1 and NFKB2 (Fig. 5d, e), suggesting a larger regulatory role of RSK1 on transcription. These findings were recapitulated in the THP-1 monocytic AML cell line after RSK1 inhibition by PMD-026 and shRNA knockdown (Supplementary Fig. 7h–j), reiterating that effects were primarily due to RSK1 inhibition. Evidence of transcriptional suppression of NFκB by PMD-026 was also observed with luminescence assays in THP-1 cells expressing luciferase under the control of an NFκB promoter (Fig. 5f). A previous study identified RSK1 having the capacity to bind to p65/RELA and translocate to the nucleus upon IL-1β stimulation21. We validated the interaction of RSK1 with p65/RELA through co-immunoprecipitation (Supplementary Fig. 7k). Finally, in subcellular fractionation assays, PMD-026 treatment repressed TNF-induced p65/RELA trafficking to the nucleus (Fig. 5g). These data indicate that RSK inhibition profoundly impacts the transcriptional landscape of monocytes, in part through regulating NFκB activity.
a Spearman correlation of CD14 and RPS6KA1 expression from TCGA LAML (n = 165) and WashU Cohort CD14 + monocytes (n = 68). Correlation (blue line) and two-tailed P-values of the WashU Cohort CD14 + monocytes represent values across all MPN patients (n = 54). b Spearman correlation of percent PB monocyte and RPS6KA1 expression from the BeatAML cohort (n = 451). Correlation (blue line) and two-tailed P-value presented. c Top altered Hallmark pathways by enrichment from top 500 DEGs from RNA-seq analysis of CD14 + cells isolated from 3 MF patients treated with 5 μM PMD-026 compared to control for 6 h. Gene count represents the number of genes enriched across each pertinent gene set. Gene ratio represents the number of genes enriched/total number of genes in each gene set. d Heatmap showing changes to RNA-seq expression of key effectors from (d) after PMD-026 treatment relative to control. e qRT-PCR of key effectors from CD14+ monocytes seven additional MPN patients after 6 h 5 μM PMD-026 treatment relative to control. n = 7 patients for all effectors except CXCL10 (n = 6). Statistics were assessed by a two-tailed paired Student’s t test compared to control. Data are presented as mean values ± SD. f Luciferase assay of THP-1 NFκB-Luc2 cells treated with 5 μM PMD-026 or control for 24 h. n = 6 for each group. Statistics were assessed by a two-tailed Student’s t test. Data are presented as mean values ± SD. g Immunoblot of RELA localization upon TNF and/or PMD-026 treatment. THP-1 cells were pre-treated with 10 μM PMD-026 for 15 min and then stimulated with 20 ng/mL TNF for 15 min. h Transcription start site plot and profile heatmap of annotated peaks within 2000 base pairs from ATAC-seq analysis of THP-1 cells treated with 5 μM PMD-026 or DMSO control for 24 h. i Pie charts of gene section breakdown of annotated peaks from (h). j Enrichment bubble plots of top altered pathways from differentially annotated regions with PMD-026 treatment. Cut-off was denoted as p-value ≤ 0.05 and false discovery rate (FDR) step up ≤ 0.05. k Enrichment bubble plots of top-downregulated pathways from differentially annotated TSS regions with PMD-026 treatment. Cut-off was denoted as TSS annotation with p-value ≤ 0.05 and false discovery rate (FDR) step up ≤ 0.05. l Known vertebrate motifs from the JASPAR database are over-represented in the list of sequences in the significant regions list from j). Each motif name, matrix ID, and p-value are denoted. Analysis performed using Partek Flow pipeline. m CUT&Tag H3K27ac histone tracks at promoter regions of genes of interest. THP-1 cells were treated with 5 μM PMD-026 or DMSO control for 24 h. Genomic tracks were visualized using the WashU EpiGenome Browser.
We then performed an Assay for Transposase-Accessible Chromatin with sequencing (ATAC-seq) in THP-1 monocytes treated with PMD-026 to investigate alterations to chromatin accessibility. Annotated genomic region enrichment reads remained stable with treatment, with intronic, intergenic, and transcription start site (TSS) regions encompassing ~ 75% of reads (Fig. 5h, i). GSEA of differential peaks revealed alterations in mitotic spindle, TGFβ, and PI3K/AKT/mTOR signaling nodes across all genomic regions (Fig. 5j) and predominantly in MYC targets and G2M checkpoint nodes across downregulated TSS regions (Fig. 5k). Top motifs from differential peak sequences included binding sites for CREB1, MYNC (N-Myc), RELA, and E2F4, in addition to FOS/FOSL1/FOSL2/JUN/JUNB/JUND, hematopoiesis regulators CEPBA and SPI1 (PU.1), and highly conserved zinc finger effector CTCF (Fig. 5l). In tandem, we performed Cleavage Under Targets and Tagmentation (CUT&Tag) for active transcription histone marker H3K27ac. Upon PMD-026 treatment, there was a reduction of acetylation marks at the TSS for NFKB1, TNF, and YBX1, and cell cycle regulators CCNA1/2 and CDK1/2 (Fig. 5m). Taken together, these assays profiling altered global chromatin and epigenetic states highlight RSK1 inhibition to result in a consistent cell cycle arrest phenotype and concurrent impact on TNF/NFκB and RSK1-effector signatures.
The genomic and transcriptional regulation of inflammation by RSK1 was also consistently reflected at the protein level. PBMCs from MF patient #5 were stimulated with thrombopoietin (TPO) in the presence of PMD-026 ex vivo and the impact on pro-inflammatory cytokine production was assessed by CyTOF. PMD-026 nearly abrogated all induction of TNF, IL-6, IL-8, CCL3, and CCL4 in CD14 + monocytes, with inhibitory effects substantially below basal conditions (Fig. 6a). In contrast, ruxolitinib yielded either mild or minimal changes. Similar superior suppression of inflammatory cytokines by PMD-026 was observed with four additional MPN patients (Fig. 6b). In accordance with these findings, on-target inhibition of S6 and NFκB pathways was achieved with PMD-026 treatment of MPN CD14 + monocytes in a dose-dependent manner, which could be observed after acute treatment (Fig. 6c). Further profiling of the NFκB signaling cascade revealed that PMD-026 could also inhibit TNF-induced NFκB activation in THP-1 cells (Fig. 6d), as also denoted in reduced p65/RELA nuclear trafficking (Fig. 5h). Decreased phosphorylation of IKKα/β S176/180 and two key p65/RELA phosphorylation sites in addition to prevention of IκBα degradation suggest direct and multimodal NFκB pathway regulation by PMD-026 upstream of the IKK complex and at the IκBα/p65 complex. Similar NFκB signaling inhibition by PMD-026 was observed in JAK2 mutant cell lines (Supplementary Fig. 8a). To further extend our observations of RSK1 acting as a fundamental inflammatory signal transducer, we performed similar co-treatment experiments in THP-1 cells stimulated with toll-like receptor (TLR) agonizts lipopolysaccharide (LPS; TLR4) and Pam3CSK4 (TLR1/2) in the presence of PMD-026. Consistent with the suppressive effects on TNF-mediated NFκB activation, PMD-026 also inhibited TLR-mediated NFκB activation (Supplementary Fig. 8b). Together, these results further highlight RSK1 necessity in oncogenic NFκB function.
a CyTOF analysis of CD14 + monocytes from MF #5 under treatment conditions. Cells were treated with 5 μM ruxolitinib, 5 μM PMD-026, 10 ng/mL TPO, or combination for 4 h. b Changes to cytokine expression from (a) and in four additional MF patients. Comparisons were performed between TPO alone versus TPO + inhibitor. c Immunoblot of CD14 + monocytes from three unique MF patients treated with indicated PMD-026 doses and time points. d Immunoblot of THP-1 cells stimulated with 20 ng/mL TNF and treated with 10 μM PMD-026 alone or in combination for 30 min. Immunoblot is representative of two independent experiments. e Left: Venn diagram showing overlapping DEGs between WashU Cohort MF CD14 + monocytes compared to those from NBM and CMML CD14 + monocytes compared to old healthy controls from (GSE135902). Middle: Key shared candidates from 279 total shared DEGs between MF and CMML monocytes. Right: Top altered Hallmark pathways by enrichment analysis from 279 shared DEGs. Gene count represents the number of genes enriched across each pertinent gene set. Gene ratio represents the number of genes enriched/total number of genes in each gene set. f Immunoblot of CD14 + monocytes from two unique CMML patients treated with indicated PMD-026 doses for 4 h. g qRT-PCR of key effectors from CD14 + monocytes from three CMML patients after 6 h 5 μM PMD- 026 treatment relative to control.
Given that monocytes can differentiate into M1 pro-inflammatory macrophages in response to microenvironmental cues, we also assessed if RSK1 inhibition could blunt this response. THP-1 monocytes were induced into M1 macrophages with phorbol 12-myristate 13-acetate (PMA) treatment followed by interferon-gamma and LPS stimulation in the presence or absence of PMD-026. Indeed, PMD-026 treatment prevented M1 polarization as observed with decreased surface expression of mature macrophage marker CD71 (Supplementary Fig. 8c).
To extend our findings beyond conventional MPN and sAML, we assessed another disease model in CMML, a myelodysplastic syndrome/MPN overlap disease characterized by the overproduction of monocytes. CMML monocytes have been similarly described to be hyper-inflammatory in having elevated CXCL8 expression and IL-6 signatures that contribute to pathogenicity, clonal expansion, and morbidity in patients36,37,38. In comparing the transcriptional landscapes of CD14 + monocytes from MPN and CMML relative to their respective healthy controls, we found numerous overlapping genes mediating pro-inflammation and NFκB signaling including TNF, IL6, CCL4, CCL7, CXCL2, CXCL3, NFKB2, NFKBIB, and NFKBIZ which was also supported by GSEA of shared, positively upregulated DEGs (Fig. 6e). We then sorted CD14 + cells from CMML patients which after treatment with PMD-026 demonstrated similar suppression of p65/RELA and S6 phosphorylation (Fig. 6f). qRT-PCR analysis revealed potent transcriptional suppression of inflammatory cytokines mediated by PMD-026 (Fig. 6g). These findings collectively demonstrate the potency of RSK1 targeting in suppressing pro-inflammatory cytokine/chemokine production and NFκB signaling both at homeostasis and in response to inflammatory cues. RSK inhibition may, therefore, reduce disease burden and transformation potential in MPN and CMML through modulating the microenvironment and suppression of monocytes.
PMD-026 is efficacious in syngeneic MPN mouse models
We then tested the efficacy of PMD-026 across syngeneic models of MPN. In the Jak2 knock-in model39, we observed significant and dose-dependent suppression of white blood cells (WBCs) at 50 mg/kg and 100 mg/kg doses of PMD-026 (Fig. 7a). There was a trend of suppression of spleen weights, but not of red blood cells (RBCs) or platelets (Supplementary Fig. 9a). Further in vivo assessment included utilizing PMD-026 alone or in combination with ruxolitinib in the more aggressive MPL W515L mouse model40 that recapitulates MF features including leukocytosis, splenomegaly, and bone marrow fibrosis. Transplanted mice were randomized to receive 100 mg/kg PMD-026, 90 mg/kg ruxolitinib, or a combination of oral gavage twice a day. RSK1 inhibition prevented leukocytosis upsurgence including potent suppression of monocytosis and neutrophilia (Fig. 7b and Supplementary Fig. 9b). Hematocrit was not affected in the first two weeks of treatment relative to vehicle, while thrombocytosis was significantly blunted (Fig. 7b) Pathological end-organ spleen and liver weights were also reversed by PMD-026, and there was extended survival of animals beyond two weeks of treatment by which time all vehicle mice were exhausted (Fig. 7b, c). These disease-ameliorating effects mediated by PMD-026 were comparable to those of ruxolitinib, and their combination further reduced (WBC) counts. Bone marrow histopathological analysis revealed hypercellularity, dysmegakaryopoiesis, and fibrosis in vehicle-treated mice which was only mildly suppressed by ruxolitinib; in contrast, PMD-026 alone or in combination reverted disease burden (Fig. 7d).
a White blood cell count following drug treatment in the JAK2V617F mouse model. Mice were treated with control (n = 7), 50 mg/kg PMD-026 (n = 5), or 100 mg/kg PMD-026 (n = 5). Statistics were assessed by two-way ANOVA between treatment groups versus control. Data are presented as mean values +/− SD. b Altered hematopoietic and disease parameters following drug treatment in the MPLW515L mouse model. Mice were treated with control (n = 8), 90 mg/kg ruxolitinib (n = 8), 100 mg/kg PMD-026 (n = 8) or combination (n = 8). White blood cells, monocytes, hematocrit, and platelets were assessed by Hemavet. Statistics were assessed by two-way ANOVA between treatment groups as indicated. Data are presented as mean values +/− SD. Spleen and liver weights were normalized to mouse weights at the endpoint. Statistics were assessed by one-way ANOVA with Dunnett’s multiple comparisons test. c Kaplan-Meier survival analysis. Statistics were assessed by log-rank test between treatment groups as indicated. d Histology of femur bone marrow at endpoint. Images show representative H&E and reticulin staining from 3 mice per group. Scale bar = 100 μM. e Luminex multiplex assays detecting plasma cytokines of treated MPLW515L mice. Vehicle (n = 7), ruxolitinib (n = 5), PMD-026 (n = 7), combo (n = 7). Statistics assessed by two-tailed Student’s t test. Boxplot showing median, upper, and lower quartiles with whiskers representing highest and lowest values excluding outliers, and potential outliers.
To recapitulate that RSK1 inhibition could ameliorate the hyperinflammatory response in vivo, we performed Luminex multiplex cytokine assays on plasma collected at the endpoint from our MPL W515L experiment. Potent suppression of TNF, IL-6, and IL-1b was observed with monotreatment with ruxolitinib or PMD-026, but significant decreases of CCL3, RANTES/CCL5, IP-10, and IL-2 were observed by PMD-026 but not ruxolitinib (Fig. 7e and Supplementary Fig. 9c). Ruxolitinib and PMD-026 combination led to similar inhibition profiles as PMD-026 treatment alone and these results are consistent with overall suppression of total monocytes in the MPL W515L mice (Fig. 7e). These findings indicate that PMD-026 may have a similar and superior cytokine and chemokine suppression profile to current standard-of-care JAK inhibitor therapy, which is consistent with our ex vivo MPN patient CyTOF data.
We also evaluated the safety profile of PMD-026 in vivo by treating wild-type C57BL/6 mice with 100 mg/kg PMD-026 for four weeks. While PMD-026 treatment led to a reduction in WBC, in particular lymphocytes, all counts remained within healthy, reference ranges (Supplementary Fig. 9d). PMD-026 did not alter hematocrit or platelet counts in WT mice. Thus, myelosuppressive effects were mild compared to potent disease suppression in the counterpart MPL W515L model (Fig. 7b–e).
PMD-026 reduces disease burden in PDX models of myeloid malignancies
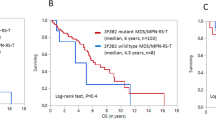
We lastly aimed to replicate anti-leukemic efficacy from the Jak2 and MPL W515L models by pharmacologically targeting RSK1 with PMD-026 in PDX models spanning the spectrum of myeloid malignancies. We sorted and transplanted CD34 + cells from one MF, three sAML, and one CMML patient, which together reflect a diverse landscape of driver and disease-modifying mutations (Fig. 8a). Mice transplanted with CD34 + cells from post-ET MF patient 201943 carrying JAK2 and ASXL1 mutations demonstrated robust engraftment with ~ 40% of hCD45 + cells in the peripheral blood within two weeks following transplantation (Fig. 8b). Treatment with 100 mg/kg PMD-026, 60 mg/kg ruxolitinib dose, or combination was then initiated. The rapid expansion of hCD45 + leukemic cells was observed in the vehicle group, and mice died within the next 5 days. In contrast, treatment with ruxolitinib, PMD-026, and combination were all able to significantly decelerate the slope of disease progression and extend survival, with the strongest effects seen with combination treatment. There was overwhelming hCD45 + engraftment in the control group bone marrow at the endpoint, which was decreased by two-fold with PMD-026 + ruxolitinib treatment (86.4% vs 44.6%, respectively). Reversal of pathological spleen and liver weights was also greatest with combination treatment. Deliberate usage of a lower ruxolitinib dose here (60 mg/kg versus 90 mg/kg in MPL W515L mice) led to relatively milder leukemic suppression but enabled demonstration of combinatorial benefit with PMD-026 in this MF PDX.
a Schematic of the PDX model setup and mutation burden of five patient samples utilized to establish PDX models. b Efficacy in the 201943 MF model. Plots show the percentage of hCD45 + cells in the PB and BM of transplanted mice treated with vehicle, 60 mg/kg ruxolitinib, 100 mg/kg PMD-026, or combination (n = 5 per group), spleen, liver weights of mice at endpoint normalized by mouse weight, and survival data. %hCD45 + in PB statistics comparing treatment group to control assessed by one-way ANOVA with Dunnett’s multiple comparisons; PMD-026 to combo by two-way Student’s t test. %hCD45 + in BM, and normalized spleen and liver weights statistics were assessed by one-way ANOVA with Dunnett’s multiple comparisons. Data are presented as mean values +/− SD. Kaplan-Meier survival analysis. Statistics were assessed by log-rank test between treatment groups as indicated. c Efficacy in the 290375 sAML model. Data are presented as mean values +/− SD. %hCD45 + in PB (n = 5 in vehicle; n = 6 in PMD-026) statistics assessed by two-way ANOVA. %hCD45 + in BM (n = 5 in vehicle; n = 6 in PMD-026) statistics assessed by two-tailed Student’s t test. d Efficacy in the 310914 sAML model. Data are presented as mean values +/− SD. %hCD45 + in PB (n = 5 per group) statistics assessed by two-way ANOVA. %hCD45 + in BM (n = 4 in vehicle; n = 5 in PMD-026) statistics assessed by two-tailed Student’s t test. e Efficacy in the 728508 sAML model. Data are presented as mean values +/− SD. %hCD45 + in PB (n = 5 per group) statistics assessed by two-way ANOVA. %hCD45 + in BM (n = 5 per group) statistics assessed by two-tailed Student’s t test. f Efficacy in the 676151 CMML model. Data are presented as mean values +/− SD. %hCD45 + in PB (n = 5 per group) statistics assessed by two-way ANOVA. %hCD45 + in BM assessed by two-tailed Student’s t test.
Strong inhibition of leukemic engraftment by PMD-026 was also observed across three sAML PDX exhibiting greater genetic diversity and harboring multiple high-risk mutations (Fig. 8c–e). Compared to vehicle, PMD-026 treatment led to a 67–96 percent reduction in PB engraftment and a 65–94 percent reduction in BM engraftment in these models, highlighting near-elimination of disease by RSK1 targeting. Significant reduction of PB engraftment was similarly observed with PMD-026 treatment in the CMML PDX (Fig. 8f). These data indicate the therapeutic efficacy of RSK inhibition across chronic MPNs and post-MPN sAML.
Discussion
Current approved JAK2 inhibitors for MPN lack specificity for mutant JAK2, consequently causing myelosuppression and pancytopenia in a large subset of patients. In addition, the insufficiency of JAK2 inhibitors failing to eliminate the underlying malignant clone or halt disease transformation makes it evident that intact oncogenic driver signaling is merely one avenue required for pathogenesis. A major contribution by a pro-inflammatory niche is a hallmark of MPN and other myeloid malignancies alike in fueling disease progression, outlining a cyclic relationship between driver dependencies and ripe microenvironment. In line with this relationship, exploration of therapeutic strategies is needed and essential for MPN.
MPNs serve as a model disease to study the interplay of oncogenic signaling and inflammation. We sequenced paired CD34 + HSPCs and CD14 + monocytes and established here a comprehensive transcriptome atlas totaling 158 primary samples that span the spectrum of MPN with PV, ET, MF, and post-MPN sAML samples with comparative cells of reference from healthy donors. In this WashU Cohort, there was clear evidence of enrichment in PI3K/AKT/mTOR, RAS/MAPK/ERK, JAK-STAT, and NFκB signaling pathways effectors at the transcriptional level, which was further reflected across signaling proteomic CyTOF assays encompassing 106 paired CD34 + and CD14 + samples. Furthermore, we observed augmented inflammatory cytokine profiles such as TNF in plasma and CD14 + monocytes increasing from early MPN phenotypes to MF/sAML validated at cohort level and in single-cell analysis of disease transformation from PV to MF. These efforts provide valuable resources to survey the landscape of hematopoiesis, disease progression, and dysregulated inflammatory networks with means to uncover candidates and stem future studies.
The pleiotropic functions in oncogenic signal transduction through effectors such as S6 bridged with the capacity to mobilize NFκB-mediated inflammation centers RSK1 as a pivotal node in myeloid pathogenesis. As such, we evaluated targeting RSK1 with PMD-026 across multiple leukemic models. In syngeneic MPLW515L mice, PMD-026 demonstrated potency in suppressing MF hallmarks, including reduction of bone marrow fibrosis superior to standard-of-care ruxolitinib. Disease amelioration was similarly observed across four PDX leukemia models that harbored a plethora of driver and disease-modifying mutations, indicative of shared dependency of RSK1 and downstream pathways. An effector of interest, YB-1, plays key roles in mediating therapeutic resistance in JAK2-mutated neoplasms and in survival maintenance of myeloid leukemias29,41. We observed a consistent reduction in YB-1 activity and a reduction in transcriptional accessibility by PMD-026, in addition to other canonical oncogenic targets including S6 and mTOR.
Suppression of RSK1 diminished NFκB-mediated signal transduction and cytokine production at transcriptional and post-transcriptional levels. Most striking, we observe PMD-026 treatment potently inhibiting cytokines including TNF, IL-6, IL-8, CCL3, and CCL4. In CyTOF analysis, cytokine levels were suppressed below basal levels, as compared to mild effects by ruxolitinib. Clinically, effectors such as IL-6 remain high in ruxolitinib-treated MF patients, and elevated IL-8 levels correlate with poor overall and leukemia-free survival and greater transformation potential to sAML34,42,43. Overall, we demonstrate that RSK1 inhibition to be two-pronged by suppressing driver signaling and dampening the pro-inflammatory milieu in MPN/sAML and other myeloid disorders, including CMML, that share congruent hyper-inflammatory monocytes. We further extend our findings to de novo leukemia driven by internal-tandem duplications in FLT3, where RSK1 inhibition led to a marked reduction in disease burden44. These findings suggest a conserved RSK1 dependency across myeloid malignancies.
Given that RSK1 is essential to leukemic propagation, this potentially enables a favorable therapeutic window. Through genetic perturbation assays, we also demonstrate that RSK1 deficiency does not affect the colony formation of normal hematopoietic cells. These results are consistent with previous studies by us and others showing a lack of hematopoietic alterations in colony formation of human bone marrow cells with BI-D1870 treatment22,24. In contrast, PMD-026 was efficacious across equivalent leukemia models, especially PDXs. PDX models were established through intra-tibial injection45 where patient cells are presumably seeded in a primary bone marrow niche, compared to other models where primary cells are introduced via tail-vein or retro-orbitally and may have to home to the bone marrow. Nonetheless, observed anti-leukemic effects could be in part due to engraftment disadvantage. In addition, while we adhered to previously established PDX models for PMD-026 dosage in vivo25, we cannot exclude the possibility that enhanced efficacy may be achievable via higher potency RSK inhibition.
Our KINOMEscan analysis of PMD-026 revealed preferential selectivity against RSK1-4 with minimal off-target effects against other kinases. Anti-proliferative signatures were reaffirmed with genomic inhibition of RPS6KA1, a major dependency in myeloid disease22. The current phase 1/1b/2 clinical trial of PMD-026 evaluating drug safety and tolerability in breast cancer has demonstrated favorable pharmacokinetic profiles with exposure being achieved in the desired preclinical range following continuous oral dosing46. PMD-026 monotherapy was generally well-tolerated with mostly low-grade TRAEs46.
Together, these findings demonstrate robust therapeutic efficacy across leukemia models with favorable safety profiles of targeting RSK in humans, thus supporting further evaluation across myeloid malignancies in the clinical setting.
Methods
Ethics statement
All research complies with all relevant ethical regulations. Patient and healthy donor control peripheral blood (PB) or bone marrow (BM) samples were obtained according to a protocol approved by the Washington University Human Studies Committee (WU no. 01-1014). All patients previously provided consent to have samples banked and were not newly recruited for this study. All in vivo procedures were conducted in accordance with the Institutional Animal Care and Use Committee (IACUC) of Washington University (no. 20-0463). Mice were handled according to standard care procedures including: bedding, cage change interval, light cycle, temperature, humidity, food type, water, and caging.
The maximal disease burden denoted by the ethics committee/institutional review board (e.g., moribund and sluggish appearance, hunched and with ruffled coats, harboring visible lesions) was not exceeded. Animals were humanely euthanized upon immobility or appearance with signs of distress.
Inclusion and ethics statement
We have carefully considered research contributions and authorship criteria in our multi-region collaborations to promote greater equity in research.
Cell culture
HEL (ATCC) cells were cultured in RPMI 1640 ATCC modification (ThermoFisher, Waltham, MA). MV4-11 (ATCC) cells were cultured in IMDM. THP-1 (ATCC) and THP-1 NFκB-Luc2 (ATCC) cells were cultured RPMI 1640 supplemented with 0.05 mM beta-mercaptoethanol. Ba/F3 (DSMZ) cells were cultured in RPMI 1640 supplemented with 1 ng/mL murine IL-3, while Ba/F3 transduced with MPL and JAK2 were cultured in the absence of murine IL-3. HEK293T (ATCC) was cultured in DMEM. All cell lines were maintained at 37 °C and 5% CO2, grown in media supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin, and regularly tested for mycoplasma.
Patient samples
Mononuclear cells were obtained by Ficoll gradient extraction and cryopreserved according to standard procedures. Additional BMMCs were purchased from STEMCELL Technologies (Vancouver, Canada) and AllCells (Alameda, CA). A list of patient samples utilized in this study is provided in Supplementary Data 1.
Compounds, immunoblot and flow cytometry antibodies
PMD-026 was provided by Phoenix Molecular Designs. Ruxolitinib was purchased from Selleck Chemicals (Houston, TX). BI-D1870 was purchased from MedChemExpress (Monmouth Junction, NJ). Human TPO, human IFN-gamma, murine IL-3, murine stem cell factor (SCF), murine TPO, and murine FLT3-ligand were purchased from Peprotech (Rocky Hill, NJ). Protein Transport Inhibitor Cocktail (500X), LPS, and PAM3CSK4 were purchased from ThermoFisher (Waltham, MA). Phorbol 12-myristate 13-acetate (PMA) was purchased from Sigma-Aldrich (St. Louis, MO).
Immunoblotting antibodies were used as follows: RSK1 (D6D5, #8408), p90RSK T359/S363 (#9344), p90RSK S573 (#9346), pS6 S235/236 (D57.2.2E, #4858), p-mTOR S2448 (#2971), mTOR (L27D4, #4517), IκBα (44D4, #4812), NFκB p65 (L8F6, #6956), pNFκB p65 S468 (#3039), pNFκB p65 S536 (93H1, #3033), pIKKα/β S176/180 (16A6, #2697), pYB-1 S102 (C34A2, #2900), Lamin A/C (#2032), α-Tubulin (DM1A; #3873), and pSTAT3 Y705 (#9131) from Cell Signaling Technology (Danvers, MA). HSP90 (sc-13119) and IKKβ (H-4, sc-8014) from Santa Cruz Biotechnology (Dallas, TX). Primary antibodies were utilized at a 1:1000 concentration except for HSP90 and Tubulin at 1:5000 dilution. Secondary antibodies were utilized at a 1:10000 dilution. Flow cytometry antibodies were used as follows: PE Mouse Anti-Human CD3 (UCHT1), PE Mouse Anti-Human CD19 (HIB19), and PE Mouse Anti-Human CD7 (M-T701), Anti-Human CD71-APC-Cy7 (CY1G4), Anti-Human CD14-PacBlue (M5E2) from BioLegend (San Diego, CA).
Plasmid constructs and transduction
Lentiviral and retroviral transduction with a low multiplicity of infection (MOI) was performed following the protocol described at http://www.broadinstitute.org/rnai/public/resources/protocols. Transduced cells were selected in puromycin for 2-3 days and plated for downstream assays immediately after selection.
Individual shRNA vectors utilized in cell line and PDX experiments used were from the Mission TRC library (Sigma) provided by the McGill Platform for Cellular Perturbation (MPCP) of Rosalind and Morris Goodman Cancer Research Center at McGill University as follows: control vector pLKO.5, shRPS6KA1 #1 (TRCN0000001386), shRPS6KA1 #2 (TRCN0000001388). In cord blood assays, shRNAs targeting RPS6KA1, RPS6KA2, and RPS6KA3 were generated using the p309-mCherry plasmid. Sequences are provided in Supplementary Data 8.
KINOMEscan and counter screen
398 kinases (Eurofins Discovery, San Diego, CA), including mutant kinases were screened at a fixed concentration of 30 nM PMD-026. Human kinome image created by TREEspot. Identified kinases were subsequently evaluated in an 11-concentration curve through counter-screening to establish an IC50.
Cell viability assays
Cell lines were seeded into 96-well plates at a density of 2 × 103 to 2 × 104 cells/well upon gene perturbation or treated with indicated doses of inhibitors and cultured for 72–96 h. At the endpoint, resazurin viability reagent was added and absorbance was read in a BioTek microplate reader.
CD34+ cell colony assay
Patient PBMC and NBM cells were thawed, filtered, and sorted using CD34 + beads. Cells were seeded at 1000 cells/mL in Methocult H4034 (StemCell) with the indicated inhibitors or following shRNA knockdown. Each condition was plated in duplicate or triplicate as indicated. Colonies were enumerated after 10–14 days post-seeding. Cord blood CD34 + hematopoietic stem and progenitor cells were isolated from cord blood (New York Blood Center) using MACS cell separation (Miltenyi Biotec). Cells were cultured in X-VIVO15 medium (Lonza) containing 10% fetal bovine serum, 1% penicillin-streptomycin-glutamine (Thermo Fisher Scientific), TPO, SCF, Flt3L, IL-3, IL-6, GM-CSF, and EPO (Miltenyi Biotec). Cells were sorted for mCherry at 5 days post-transduction. 1000 mCherry + cells were plated in methylcellulose media and enumerated after 2 weeks. Colonies were counted by an investigator blinded to the conditions.
Immunoblotting
Cells were collected and lysed with Laemmli Sample Buffer (BioRad, Hercules, CA) with beta-mercaptoethanol and heated at 95 °C. Samples were processed with NuPAGE Novex Gel Electrophoresis Systems (ThermoFisher) followed by a standard immunoblotting procedure.
Immunoprecipitation (IP) assay
Co-IPs were performed using the Pierce Classic Magnetic IP/Co-IP Kit (Thermofisher). In brief, cells were pelleted, washed with PBS, and lysed with IP lysis/wash buffer (pH 7.4, 0.025 M Tris, 0.15 M NaCl, 0.001 M EDTA, 1% NP40, and 5% glycerol). Cells were then centrifuged, an input fraction was collected, and the supernatant was incubated with 1 µg of RSK1 primary antibody or IgG control (Cell Signaling #2729) for 2 h at room temperature to form the immune complex. Immune complexes were then incubated with protein A/G magnetic beads, eluted, and collected following heating under reducing conditions.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
RNA from cell lines and primary samples were extracted using the RNeasy Mini Kit (Qiagen, Hilden, Germany). cDNA was synthesized using the Maxima First-Strand cDNA Synthesis Kit (ThermoFisher). Relative mRNA levels were measured through qRT-PCR using SYBR Green Master Mix (Roche) and normalized to the expression of β-actin (ACTB). Primer sequences are provided in Supplementary Data 9.
Flow cytometry, annexin, and cell cycle assays
Cells were stained in live cell buffer and run on the BD FACSCanto II Cell Analyzer utilizing unstained controls, and downstream analysis was performed with FlowJo (Ashland, OR).
Annexin V apoptosis assay
Cells were collected and stained per the PE Annexin V Apoptosis Detection Kit I (BD Biosciences). In brief, post-treatment, cells were washed with PBS and resuspended in binding buffer, incubated with PE-Annexin V, and stained with 7-AAD viability staining solution. Cells were then analyzed by flow cytometry, in which AnnexinV + /7-AAD + cells were identified as necrotic, AnnexinV + /7-AAD- as apoptotic, and AnnexinV-/7-AAD- as healthy.
Cell cycle assay
Cells were seeded and post-treatment, were washed with PBS and fixed in 70% ethanol for 24 h. Cells were washed with PBS and then treated with RNase, followed by staining with propidium iodide. Cells were then analyzed by flow cytometry.
Macrophage differentiation assay
THP-1 monocytes were differentiated into macrophages following established protocols47. In brief, cells were cultured in the presence of 150 nM phorbol 12-myristate 13-acetate (PMA) for 24 h followed by 24 h media recovery in RPMI. Cells were then polarized to M1 macrophages through culturing in the presence of 20 ng/ml of IFN-γ and 10 pg/ml of LPS.
Luciferase assay
Luciferase assays were performed using the Pierce Firefly Luc One-Step Glow Assay Kit (Thermofisher). In brief, THP-1 NFκB-Luc2 cells were seeded into a 96-well plate and treated with PMD-026 for 24 h. Following, cells were incubated with the luciferase working solution and read on a BioTek microplate reader.
Sequencing and downstream analyses
RNA-sequencing (RNA-seq)
For the MPN/sAML atlas, bone marrow and peripheral samples were sorted using CD34 + UltraPure beads (Miltenyi Biotec, Germany) on an autoMACS Separator. Following, the CD34- fraction was sorted using CD14 + beads (Miltenyi Biotec, Germany). For other assays, patient samples were sorted with either CD34 + or CD14 + beads. RNA was extracted using the RNeasy Mini Kit (Qiagen) with DNase treatment. For cell lines, RNA was isolated in duplicate. cDNA synthesis was performed using Clontech SMARTer for primary samples and PolyA selection for cell lines. Samples were then indexed, pooled, and sequenced by Illumina NovaSeq 6000. Basecalls and demultiplexing were performed with Illumina’s bcl2fastq software and a custom Python demultiplexing program with a maximum of one mismatch in the indexing read. RNA-seq reads were then aligned to the Ensembl release 76 primary assembly with STAR version 2.5.1a48. TMM normalization size factors in EdgeR49 were calculated to adjust for samples for differences in library size. Ribosomal genes and genes not expressed in the smallest group size minus one sample greater than one count-per-million were excluded from further analysis. The TMM size factors and the matrix of counts were then imported into the R/Bioconductor package Limma50. Additional analysis was performed using DEseq251, using GSEA52 software with the Hallmark gene set, or pathway analysis at https://www.gsea-msigdb.org/.
scRNA-sequencing (scRNA-seq)
PBMCs from 551599 Early PV and 551599 MFpPV were thawed and sorting for non-lymphoid cells denoted as 7AAD- CD3-CD7- CD19-. Following the 10x genomics protocol, oligonucleotide labeled cells were subjected to droplet bead capture, followed by cell lysis, reverse transcription, and amplification following protocol. Single cell-derived cDNA libraries were sequenced on NovaSeq 6000 S4 cell sequencer (Illumina, San Diego, CA). Cellranger (10x Genomics, version 3.0.1) was used for transcript alignment, counting, and inter-library normalization of single-cell samples. Seurat53 (version 4.0.0) was used for cluster analysis following the data processing described previously22. Cell-cell communication analysis was performed using CellChat35. Trajectory analysis was performed using Monocle 254.
Assay for transposase-accessible chromatin with sequencing (ATAC-seq)
ATAC-seq was performed using the ATAC-seq Kit (Active Motif) and following protocols. In brief, 100,000 THP-1 cells were treated with PMD-026 or DMSO in triplicate for 24 h, collected, washed, and tagmented. Following, tagmented DNA was amplified with PCR indexed with included i7 and i5 primers and sequenced with standard paired-end reads (2 × 150) on a NovaSeq6000 with a target of 50 M reads. Following, samples were demultiplexed, underwent adapter trimming, filtering, and alignment to the hg38 reference genome following the ENCODE ATAC-seq pipeline (https://github.com/ENCODE-DCC/atac-seq-pipeline, v2.2.0). Consistency among replicates was assessed based on Irreproducible Discovery Rates55. Processed samples were then imported into Partek Flow (Partek) following the ATAC-seq pipeline. In brief, peaks were generated with MACS, and regions were annotated to hg38. Following quantification, normalization, and differential expression analysis, GSEA and motif calling using the JASPAR 2018 CORE vertebrate collection were performed.
Cleavage under targets and tagmentation (CUT&Tag)
CUT&Tag was performed using the Complete CUT&Tag Assay Kit (Active Motif) and following protocols. In brief, 500,000 THP-1 cells were treated with PMD-026 or DMSO control for 24 h in duplicate or triplicate, collected, washed, and bound to Concanavalin A beads. Cells were then incubated with a histone H3K27ac antibody (Active Motif 39135) or IgG control (Cell Signaling, DAIE, #66362) overnight. Following, cells were incubated with secondary antibodies and washed before binding to pA-Tn5 transposomes. Cells were then tagmented, after which extracted DNA underwent PCR amplification and indexed with included i7 and i5 primers. Samples were then sequenced with standard paired-end reads (2 × 150) on a NovaSeq6000 with a target of 10 M reads. Post-sequencing, samples were demultiplexed, and reads were aligned to the human genome hg38 following adapter trimming and filtering using the ENCODE ChIP-seq pipeline (https://github.com/ENCODE-DCC/chip-seq-pipeline2, v2.1.6), with a custom alignment script to use Bowtie2 parameters as recommended56. The ENCODE ChIP-seq pipeline was then used to generate peaks with MACS2 and evaluate peak consistency among replicates based on Irreproducible Discovery Rates55. Signal tracks were visualized within the WashU Epigenome browser57.
Mass cytometry (CyTOF)
Signaling and cytokine CyTOF experiments were conducted on a CyTOF2 mass cytometer (Fluidigm) with validated antibody panels following protocol as previously described16,17,18,22. For cytokine CyTOF, in brief, cells were then labeled with cisplatin, washed, and incubated with indicated stimulant or inhibitor for the indicated duration. Cells were then fixed with paraformaldehyde, permeabilized, and barcoded. Following, cells were stained with surface marker antibodies, permeabilized with methanol, stained with intracellular antibodies, and resuspended in DNA IR-intercalator. For cytokine CyTOF experiments, primary cells were incubated with 5 µM ruxolitinib, PMD-026, and/or TPO for 4 h, and a protein transport inhibitor cocktail (eBioscience) was added at the 2-hour treatment mark. For signaling CyTOF experiments, primary cells were labeled with cisplatin, treated with indicated inhibitors for 1 h, fixed, barcoded, and permeabilized with methanol. Following, cells were stained with intracellular antibodies resuspended in an Ir-intercalator. Cell identities were demultiplexed through debarcoding and data was analyzed in Cytobank (https://wustl.cytobank.org/). For CD34 + and CD14 + atlas generation, analysis was combined with our previously published studies17,22,58.
Plasma cytokine analysis
Human cytokine panel
Plasma cytokine concentrations from our previously published studies18,33,34 generated from custom U-PLEX 10plex or V-PLEX 30plex human cytokine kits (Meso Scale Discovery, Rockville, MD, USA) were combined with additional assays to compare PV, MF, and sAML plasma samples compared to age-matched donor plasma.
Luminex multiplex panel for MPL W515L mouse model
Plasma was extracted from MPLW515L mice treated with vehicle, ruxolitinib, PMD-026, or combination and run using the MILLIPLEX MAP Mouse Cytokine/Chemokine Magnetic Bead Panel (MCYTOMAG-70K) for analysis of ten cytokines. Data was generated using Belysa software. 5 parameter curve was used with standard and background controls.
Animal models
Jak2 V617F mice
The Jak2W515L syngeneic model was established following a previous experimental setup22,39. Briefly, Kit + cells were isolated from CD45.2 + mice Jak2V617F donor mice (6–12 weeks old) and transplanted via tail vein injection into female CD45.1+ lethally irradiated recipient mice. Recipient mice were irradiated with a total of 1100 cGy, given as two separate doses of 550 cGy with 3–5 h between doses. Four weeks following transplant, mice were treated with vehicle or PMD-026 at 50 mg/kg or 100 mg/kg twice daily, six times a week. Mice were treated for 4 weeks with bleeds being performed weekly, and cells were analyzed by Hemavet. Mice were sacrificed at the endpoint, and body, spleen, and liver weights were recorded.
MPL W515L mice
The MPLW515L retroviral model that reflects myelofibrosis was established following a previous experimental setup40. 5 × 105 bead-enriched Kit + cells were retrovirally transduced with MPLW515L were transplanted via tail vein into female C57BL/6 J CD45.1 lethally irradiated recipient mice (7-8 weeks old). Two weeks after transplant, peripheral blood was collected and hematologic parameters were measured by Hemavet and flow cytometry. Mice were treated with vehicle, ruxolitinib (90 mg/kg), PMD-026 (100 mg/kg), or combination by oral gavage intraperitonially twice a day. Bleeds were performed weekly, and cells were analyzed by Hemavet. Survival was monitored daily, and moribund mice were humanely sacrificed. Spleens were weighed at the endpoint and normalized to the mouse's body weight at the endpoint.
Wild-type mice
Vehicle or PMD-026 (100 mg/kg) was administered by oral gavage to 7-week-old female C57BL/6 J mice follow treatment protocol for MPLW515L mice described above. Bleeds were performed weekly, and cells were analyzed by Hemavet. Survival was monitored daily, and moribund mice were humanely sacrificed. Spleens were weighed at endpoint and normalized to mouse body weight at endpoint.
Patient-derived xenograft (PDX) models
PDX models were established as described previously16,45. In brief, PBMCs from cancer patients were isolated by Ficoll gradient extraction. All patients provided informed written consent. CD34 + cells were enriched by CD34 bead purification and transplanted into sub-lethally irradiated (200 cGy) male and female 6–9 week-old NOD-scid-Il2rg-null-3/GM/SF (NSGS; The Jackson Laboratory #013062) by intra-tibial injection under anesthesia. For drug assays, mice were treated 2-3 weeks post-transplant with vehicle, 60 mg/kg ruxolitinib, 100 mg/kg PMD-026, or combination by oral gavage twice a day for 4 weeks or until endpoint in which mice were sacrificed. For knockdown experiments, CD34 + cells were transduced with control or shRNAs targeting RPS6KA1 and then transplanted into NSGS mice. Transplanted mice were then followed for 12 weeks. Peripheral blood was collected weekly, and engraftment was evaluated by flow cytometry. Bone marrow, spleens, and livers were collected at the endpoint, and organ weights were normalized to mouse body weight at the endpoint.
Histology
Femurs were extracted from MPLW515L mice at the endpoint and fixed in 10% neutral buffered formalin overnight at 4 °C. Post fixation, femurs were decalcified in 14% EDTA for 12 days and then rehydrated with serial exposure to 20, 30, 50, and 70% ethanol, all for 1 h. Femurs were then rinsed in PBS and processed for paraffin embedding and sectioned at 5 μM. Hematoxylin & Eosin (H&E) and reticulin staining were performed by the Washington University Musculoskeletal Histology and Morphometry Core.
Statistics and reproducibility
Statistical analyses were performed using GraphPad Prism 9 (San Diego, CA) and R software. All measurements were taken from distinct samples. All relevant assays were performed independently at least 3 times. For in vivo experiments, randomization and blinding were used. No data were excluded from the analyses. In all in vivo experiments, the sample size was a minimum of five animals per condition, where feasible, to ensure statistical reliability. All immunoblots are representatives of independent experiments. Figures were generated with GraphPad Prism 9, R software, and with BioRender.com.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The interactive WashU CD34 + and CD14 + cell MPN Cohort RNA-seq dataset can be accessed at https://stohlab.shinyapps.io/RNAseq/. The bulk RNA-seq, scRNA-seq, and CUT&Tag data generated in this study have been deposited in the Gene Expression Omnibus (GEO) with accession number GSE228760. RNA-seq datasets of HEL cells treated with RSK inhibitor BI-D1870 used for this study were deposited in GEO under accession code GSE19051722. RNA-seq count dataset of CD14 + monocytes from CMML patients, age-matched and young controls used for this study deposited in GEO under accession code GSE13590236. Phospho-proteomic data of 34 AML cell lines was accessed from CCLE 201959. RNA-seq data of AML cell lines were accessed from CCLE28 (public 21Q1; https://depmap.org/portal/download/). Gene effect scores (CERES)32 from CRISPR (Avana) public 21Q1 were accessed from the DepMap portal by the Broad Institute. OHSU BeatAML60 and BeatAML261 clinical and expression data were obtained from http://vizome.org. Pan-cancer TCGA gene expression and TCGA LAML mutational and clinical data were accessed from cBioPortal62. LFQ protein abundance was obtained from source publication63. Mass cytometry data is available upon request on https://wustl.cytobank.org/. Source data are provided in this paper.
Code availability
No new code was developed in this study. R scripts utilized in this study are available from the corresponding author upon request.
References
Tefferi, A. & Pardanani, A. Myeloproliferative neoplasms: A contemporary review. JAMA Oncol. 1, 97–105 (2015).
Cancer Genome Atlas Research, N. et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074 (2013).
Murati, A. et al. Myeloid malignancies: mutations, models and management. BMC Cancer 12, 304 (2012).
Harrison, C. et al. JAK inhibition with ruxolitinib versus best available therapy for myelofibrosis. N. Engl. J. Med. 366, 787–798 (2012).
Verstovsek, S. et al. A double-blind, placebo-controlled trial of ruxolitinib for myelofibrosis. N. Engl. J. Med. 366, 799–807 (2012).
Vannucchi, A. M. et al. Mutations and prognosis in primary myelofibrosis. Leukemia 27, 1861–1869 (2013).
Dunbar, A. J., Rampal, R. K. & Levine, R. Leukemia secondary to myeloproliferative neoplasms. Blood 136, 61–70 (2020).
Miles, L. A. et al. Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature 587, 477–482 (2020).
Rodriguez-Meira, A. et al. Unravelling intratumoral heterogeneity through high-sensitivity single-cell mutational analysis and parallel RNA sequencing. Mol. Cell 73, 1292–1305 (2019).
Hasselbalch, H. C. & Bjorn, M. E. MPNs as inflammatory diseases: The evidence, consequences, and perspectives. Mediators Inflamm. 2015, 102476 (2015).
Fisher, D. A. C., Fowles, J. S., Zhou, A. & Oh, S. T. Inflammatory pathophysiology as a contributor to myeloproliferative neoplasms. Front. Immunol. 12, 683401 (2021).
Trowbridge, J. J., Starczynowski, D. T. Innate immune pathways and inflammation in hematopoietic aging, clonal hematopoiesis, and MDS. J. Exp. Med. 218, https://doi.org/10.1084/jem.20201544 (2021).
Muto, T. et al. Adaptive response to inflammation contributes to sustained myelopoiesis and confers a competitive advantage in myelodysplastic syndrome HSCs. Nat. Immunol. 21, 535–545 (2020).
Kleppe, M. et al. Dual targeting of oncogenic activation and inflammatory signaling increases therapeutic efficacy in myeloproliferative neoplasms. Cancer Cell 33, 29–43 (2018).
Laranjeira, A. B. A. et al. In vivo ablation of NFkappaB cascade effectors alleviates disease burden in myeloproliferative neoplasms. Blood 143, 2414–2424. (2024).
Kong, T. et al. Pevonedistat targets malignant cells in myeloproliferative neoplasms in vitro and in vivo via NFkappaB pathway inhibition. Blood Adv. 6, 611–623 (2022).
Fisher, D. A. C. et al. Mass cytometry analysis reveals hyperactive NF Kappa B signaling in myelofibrosis and secondary acute myeloid leukemia. Leukemia 31, 1962–1974 (2017).
Fisher, D. A. C. et al. Cytokine production in myelofibrosis exhibits differential responsiveness to JAK-STAT, MAP kinase, and NFkappaB signaling. Leukemia 33, 1978–1995 (2019).
Roux, P. P., Richards, S. A. & Blenis, J. Phosphorylation of p90 ribosomal S6 kinase (RSK) regulates extracellular signal-regulated kinase docking and RSK activity. Mol. Cell Biol. 23, 4796–4804 (2003).
Hu, Y. et al. 90-kDa ribosomal S6 kinase is a direct target for the nuclear fibroblast growth factor receptor 1 (FGFR1). J. Biol. Chem. 279, 29325–29335 (2004).
Xu, S., Bayat, H., Hou, X. & Jiang, B. Ribosomal S6 kinase-1 modulates interleukin-1beta-induced persistent activation of NF-kappaB through phosphorylation of IkappaBbeta. Am. J. Physiol. Cell Physiol. 291, C1336–C1345 (2006).
Kong, T. et al. DUSP6 mediates resistance to JAK2 inhibition and drives leukemic progression. Nat. Cancer 4, 108–127 (2023).
Watanabe, D. et al. FLT3-ITD activates RSK1 to enhance proliferation and survival of AML cells by activating mTORC1 and eIF4B cooperatively with PIM or PI3K and by inhibiting bad and BIM. Cancers 11, https://doi.org/10.3390/cancers11121827 (2019).
Chae, H. D. et al. RSK inhibitor BI-D1870 inhibits acute myeloid leukemia cell proliferation by targeting mitotic exit. Oncotarget 11, 2387–2403 (2020).
Dunn, S. E. et al. Abstract P3-10-10: PMD-026, a first-in-class oral p90 ribosomal S6 kinase (RSK) inhibitor for triple negative breast cancer (TNBC). Cancer Res. 80, https://doi.org/10.1158/1538-7445.SABCS19-P3-10-10 (2020).
Jayanthan, A. et al. Abstract 1038: PMD-026, a first in class oral RSK inhibitor, demonstrates synergy when combined with standard of care in breast cancer tumor models. Cancer Res. 81, 1038–1038 (2021).
Erdem, A. et al. The glycolytic gatekeeper PDK1 defines different metabolic states between genetically distinct subtypes of human acute myeloid leukemia. Nat. Commun. 13, 1105 (2022).
Barretina, J. et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012).
Jayavelu, A. K. et al. Splicing factor YBX1 mediates persistence of JAK2-mutated neoplasms. Nature 588, 157–163 (2020).
Perner, F. et al. YBX1 mediates translation of oncogenic transcripts to control cell competition in AML. Leukemia 36, 426–437 (2022).
Stratford, A. L. et al. Y-box binding protein-1 serine 102 is a downstream target of p90 ribosomal S6 kinase in basal-like breast cancer cells. Breast Cancer Res. 10, R99 (2008).
Meyers, R. M. et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 49, 1779–1784 (2017).
Fowles, J. S., How, J., Allen, M. J. & Oh, S. T. Young versus old age at diagnosis confers distinct genomic profiles in patients with polycythemia vera. Leukemia 33, 1522–1526 (2019).
Zhou, A. et al. Hepcidin is elevated in primary and secondary myelofibrosis and remains elevated in patients treated with ruxolitinib. Br. J. Haematol. 197, e49–e52 (2022).
Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12, 1088 (2021).
Franzini, A. et al. The transcriptome of CMML monocytes is highly inflammatory and reflects leukemia-specific and age-related alterations. Blood Adv. 3, 2949–2961 (2019).
Ferrall-Fairbanks, M. C. et al. Progenitor hierarchy of chronic myelomonocytic Leukemia identifies inflammatory monocytic-biased trajectory linked to worse outcomes. Blood Cancer Discov. 3, 536–553 (2022).
Niyongere, S. et al. Comprehensive inflammatory cytokine profiling pdentifies IL-8/CXCL8 As elevated, associated with proliferative features, and independently prognostic in chronic myelomonocytic Leukemia (CMML). Blood 128, 109 (2016).
Mullally, A. et al. Physiological Jak2V617F expression causes a lethal myeloproliferative neoplasm with differential effects on hematopoietic stem and progenitor cells. Cancer Cell 17, 584–596 (2010).
Pikman, Y. et al. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med. 3, e270 (2006).
Feng, M. et al. YBX1 is required for maintaining myeloid leukemia cell survival by regulating BCL2 stability in an m6A-dependent manner. Blood 138, 71–85 (2021).
Fisher, D. A. C. et al. Cytokine production in myelofibrosis exhibits differential responsiveness to JAK-STAT, MAP kinase, and NFκB signaling. Leukemia 33, 1978–1995 (2019).
Tefferi, A. et al. Circulating interleukin (IL)-8, IL-2R, IL-12, and IL-15 levels are independently prognostic in primary myelofibrosis: a comprehensive cytokine profiling study. J. Clin. Oncol. 29, 1356–1363 (2011).
Kong, T. et al. RSK1 dependency in FLT3-ITD acute myeloid leukemia. Blood Cancer J. 14, 207 (2024).
Celik, H. et al. A humanized animal model predicts clonal evolution and therapeutic vulnerabilities in myeloproliferative neoplasms. Cancer Discov. 11, 3126–3141 (2021).
Wang, J. S. et al. High levels of RSK2 in breast cancer patients is associated with longer PFS in patients treated with PMD-026, a first in class RSK inhibitor. SABCS https://aacrjournals.org/cancerres/article/83/5_Supplement/P4-01-16/718165/Abstract-P4-01-16-High-levels-of-RSK2-in-breast (2022).
Genin, M., Clement, F., Fattaccioli, A., Raes, M. & Michiels, C. M1 and M2 macrophages derived from THP-1 cells differentially modulate the response of cancer cells to etoposide. BMC Cancer 15, 577 (2015).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Qiu, X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017).
Li, Q., Brown, J. B., Huang, H. & Bickel, P. J. Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 5, 1752–1779, 1728 (2011).
Zheng, Y., Ahmad, K. & Henikoff, S. CUT&Tag Data Processing and Analysis Tutorial v1 (2020).
Li, D., Hsu, S., Purushotham, D., Sears, R. L. & Wang, T. WashU epigenome browser update 2019. Nucleic Acids Res. 47, W158–W165 (2019).
Kong, T. et al. Comprehensive profiling of clinical JAK2 inhibitors in myeloproliferative neoplasms. Blood 140, 3951–3952 (2022).
Ghandi, M. et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019).
Tyner, J. W. et al. Functional genomic landscape of acute myeloid Leukaemia. Nature 562, 526–531 (2018).
Bottomly, D. et al. Integrative analysis of drug response and clinical outcome in acute myeloid Leukemia. Cancer Cell 40, 850–864.e859 (2022).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Kramer, M. H. et al. Proteomic and phosphoproteomic landscapes of acute myeloid Leukemia. Blood 140, 1533–1548 (2022).
Acknowledgements
This work was supported by NIH grants R01HL134952 (S.T.O.) and T32HL007088 (J.S.F.), a Canadian Institutes of Health Research (CIHR) Doctoral Foreign Study Award (T.K.), and the MPN Research Foundation (S.T.O.). Additional support was provided by NIH grants R01HL147978 (G.A.C), R01HL163274 (S.L.R.), T32HL134635 (S.N.), the Leukemia and Lymphoma Society Translational Research Program (S.T.O.), When Everyone Survives Foundation (S.T.O.), Edward P. Evans Foundation (G.A.C.), and Gabrielle’s Angel Foundation (G.A.C.), Stanford Maternal Child Health Research Institute (K.M.S.), Department of Defense HT94252311008 (K.M.S.), and Hyundai Hope on Wheels (K.M.S.). G.A.C. is a scholar of the Leukemia and Lymphoma Society. Technical support was provided by the Alvin J. Siteman Cancer Center Tissue Procurement Core Facility, Biostatistics Shared Resource, Flow Cytometry Core, Barnes-Jewish Hospital, Institute of Clinical and Translational Sciences, Barnard Cancer Institute, and Immunomonitoring Laboratory, which are supported by NCATS Clinical and Translational Sciences Award, #UL1 TR002345 and NCI Cancer Center Support Grant P30CA91842 and by the Andrew M and Jane M Bursky Center for Human Immunology and Immunotherapy Programs. We thank Diane Bender, Roderick Lin, and Kristin Link for assistance with mass cytometry and Luminex cytokine experiments. We are grateful to Ross Levine (MSKCC) for providing the MPL W515L retroviral construct. We thank the Genetic Perturbation Service of Goodman Cancer Research Center at McGill University for access to and preparation of functional genetic tools.
Author information
Authors and Affiliations
Contributions
T.K., A.B.A.L., C.T.L., L.Y., S.L., J.S.F., D.A.C.F., S.N., W.Y., F.H., M.Y., K.M., and A.S.J. performed experiments and/or data analysis. J.L., A.B.K., M.J.C., M.C.F., A.J., G.L., S.L.R., L.D., S.E.D., and G.A.C. provided technical and/or clinical support. T.K., K.M.S., and S.T.O. designed and supervised the experiments. T.K. and S.T.O. wrote the manuscript. All authors read and approved of the manuscript.
Corresponding author
Ethics declarations
Competing interests
S.T.O. has served as a consultant for Kartos Therapeutics, CTI BioPharma, Celgene/Bristol Myers Squibb, Disc Medicine, Protagonist, Blueprint Medicines, Cogent, PharmaEssentia, Constellation, Geron, Abbvie, Sierra Oncology, and Incyte. A.J., G.L., and S.E.D. are employees of Phoenix Molecular Designs. S.E.D. holds patents on utilizing RSK inhibitors for the treatment of cancer. G.A.C. has received funding for consulting and/or sponsored research from Incyte, Ajax Therapeutics, and Renegade Therapeutics. All other authors disclose no competing interests.
Peer review
Peer review information
Nature Communications thanks Ghayas Issa, and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kong, T., Laranjeira, A.B.A., Letson, C.T. et al. RSK1 is an exploitable dependency in myeloproliferative neoplasms and secondary acute myeloid leukemia. Nat Commun 16, 492 (2025). https://doi.org/10.1038/s41467-024-55643-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-55643-7
This article is cited by
-
A novel anoikis resistance-associated gene model for prognostic prediction and immune microenvironment characterization in lung squamous cell carcinoma
Discover Oncology (2026)
-
Biomarkers of response to neoadjuvant palbociclib plus anastrozole in endocrine-resistant estrogen receptor-positive/HER2-negative breast cancer: a phase 2 trial
Nature Communications (2026)
-
A proteogenomic gene signature defines prognostic subgroups highlighting PI3K/AKT/mTOR signaling pathway as a therapeutic vulnerability in myeloid malignancies
Cell Communication and Signaling (2025)
-
Elucidating the risk factors and oncogene drivers of acute myeloid leukemia
Human Cell (2025)
-
Expansion characteristics of CD34+ cells derived from umbilical cord blood HS/PCs in single-parameter culture under serum-free static suspension condition
Human Cell (2025)